Acute Respiratory Infections (ARIs): Current Etiological Perspectives and Advances in Viral Metagenomics—A Review
Abstract
1. Introduction
2. Acute Respiratory Infection
3. Viral Respiratory Infections
4. Commonly Diagnosed Viral Causes of ARI
4.1. Adenoviruses
4.2. Human Bocaviruses
4.3. Coronaviruses
4.4. Influenza Viruses
4.5. Parainfluenza Viruses
4.6. Human Metapneumovirus (HMPV)
4.7. Respiratory Syncytial Virus
4.8. Enteroviruses
4.9. Human Rhinovirus (HRV)
5. Multiplex Virus Panels for Diagnosis of ARI
6. Viral Metagenomics
7. Complementary Genomic Approaches to Metagenomic Next Generation Sequencing
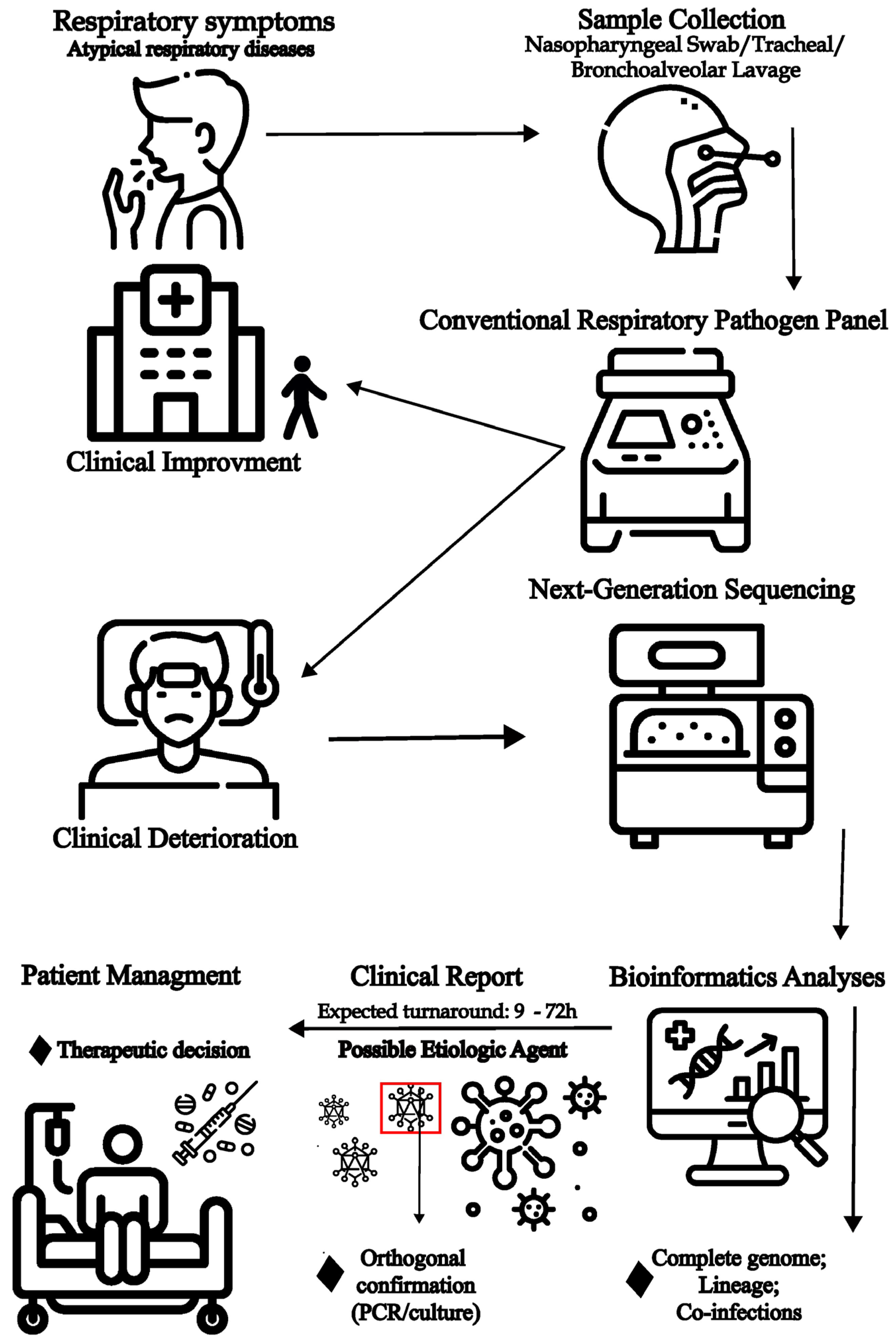
8. Metagenomic Identification of Etiological Causes of Acute Respiratory Disease
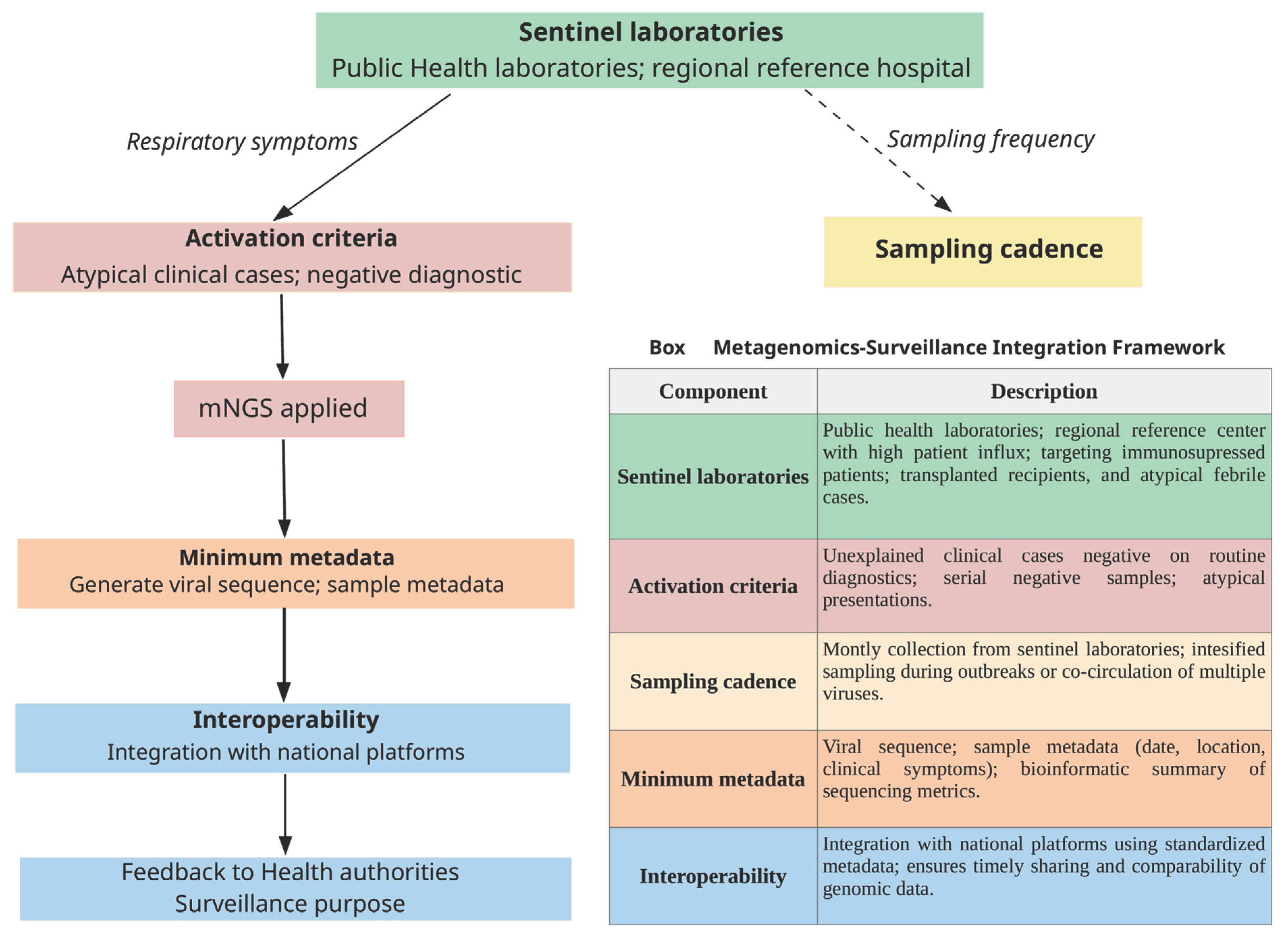
9. Role of Metagenomics for Viral Genomic Surveillance and Investigation of Transmission Clusters
10. Challenges of the Use of Metagenomics for Respiratory Pathogen Diagnosis
11. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ARI | Acute respiratory infection |
| PCR | Polymerase chain reaction |
| mNGS | Metagenomic next-generation sequencing |
| ARDS | acute respiratory distress syndrome |
| SARS-CoV-2 | Severe acute respiratory syndrome coronavirus 2 |
| COVID-19 | Coronavirus disease 2019 |
| RSV | Respiratory syncytial virus |
| HAdV | Human adenovirus |
| HBoV | Human bocavirus |
| HPIV | Human parainfluenza virus |
| HMPV | Human metapneumovirus |
| EV-D68 | Enterovirus D68 |
| HRV | Human rhinoviruses |
| NAATs | Nucleic acid amplification tests |
| CMV | Cytomegalovirus |
| EBV | Epstein–Barr virus |
| HCoV | Human coronavirus |
| HHV | Human herpes virus |
| HSV-1 | Herpes simplex 1 |
References
- Bender, R.G.; Sirota, S.B.; Swetschinski, L.R.; Dominguez, R.-M.V.; Novotney, A.; Wool, E.E.; Ikuta, K.S.; Vongpradith, A.; Rogowski, E.L.B.; Doxey, M.; et al. Global, Regional, and National Incidence and Mortality Burden of Non-COVID-19 Lower Respiratory Infections and Aetiologies, 1990–2021: A Systematic Analysis from the Global Burden of Disease Study 2021. Lancet Infect. Dis. 2024, 24, 974–1002. [Google Scholar] [CrossRef] [PubMed]
- Goins, W.P.; Talbot, H.K.; Talbot, T.R. Health Care–Acquired Viral Respiratory Diseases. Infect. Dis. Clin. N. Am. 2011, 25, 227–244. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A New Coronavirus Associated with Human Respiratory Disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.R.; Sample, H.A.; Zorn, K.C.; Arevalo, S.; Yu, G.; Neuhaus, J.; Federman, S.; Stryke, D.; Briggs, B.; Langelier, C.; et al. Clinical Metagenomic Sequencing for Diagnosis of Meningitis and Encephalitis. N. Engl. J. Med. 2019, 380, 2327–2340. [Google Scholar] [CrossRef]
- De Campos, G.M.; Santos, H.d.O.; Lima, A.R.J.; Leite, A.B.; Ribeiro, G.; Todão Bernardino, J.d.S.; do Nascimento, J.P.M.; Souza, J.V.C.; de Lima, L.P.O.; Lima, M.B.Z.; et al. Unveiling Viral Pathogens in Acute Respiratory Disease: Insights from Viral Metagenomics in Patients from the State of Alagoas, Brazil. PLoS Negl. Trop. Dis. 2024, 18, e0012536. [Google Scholar] [CrossRef]
- de Campos, G.M.; de La-Roque, D.G.L.; Lima, A.R.J.; Zucherato, V.S.; de Carvalho, E.; de Lima, L.P.O.; de Queiroz Cattony Neto, P.; dos Santos, M.M.; Ciccozzi, M.; Giovanetti, M.; et al. Exploring Viral Metagenomics in Pediatric Patients with Acute Respiratory Infections: Unveiling Pathogens beyond SARS-CoV-2. Microorganisms 2023, 11, 2744. [Google Scholar] [CrossRef]
- Buddle, S.; Forrest, L.; Akinsuyi, N.; Martin Bernal, L.M.; Brooks, T.; Venturini, C.; Miller, C.; Brown, J.R.; Storey, N.; Atkinson, L.; et al. Evaluating Metagenomics and Targeted Approaches for Diagnosis and Surveillance of Viruses. Genome Med. 2024, 16, 111. [Google Scholar] [CrossRef]
- Cui, C.; Timbrook, T.T.; Polacek, C.; Heins, Z.; Rosenthal, N.A. Disease Burden and High-Risk Populations for Complications in Patients with Acute Respiratory Infections: A Scoping Review. Front. Med. 2024, 11, 1325236. [Google Scholar] [CrossRef]
- Rogan, M. Respiratory Infections, Acute. In International Encyclopedia of Public Health; Elsevier: Amsterdam, The Netherlands, 2017; pp. 332–336. [Google Scholar]
- Troeger, C.; Blacker, B.; Khalil, I.A.; Rao, P.C.; Cao, J.; Zimsen, S.R.M.; Albertson, S.B.; Deshpande, A.; Farag, T.; Abebe, Z.; et al. Estimates of the Global, Regional, and National Morbidity, Mortality, and Aetiologies of Lower Respiratory Infections in 195 Countries, 1990–2016: A Systematic Analysis for the Global Burden of Disease Study 2016. Lancet Infect. Dis. 2018, 18, 1191–1210. [Google Scholar] [CrossRef]
- Flint, J.; Racaniello, V.R.; Rall, G.F.; Hatziioannou, T.; Skalka, A.M. Principles of Virology; Wiley: Hoboken, NJ, USA; ASM Press: Washington, DC, USA, 2020; ISBN 9781683673583. [Google Scholar]
- Jacobs, S.E.; Lamson, D.M.; St. George, K.; Walsh, T.J. Human Rhinoviruses. Clin. Microbiol. Rev. 2013, 26, 135–162. [Google Scholar] [CrossRef]
- Bizot, E.; Bousquet, A.; Charpié, M.; Coquelin, F.; Lefevre, S.; Le Lorier, J.; Patin, M.; Sée, P.; Sarfati, E.; Walle, S.; et al. Rhinovirus: A Narrative Review on Its Genetic Characteristics, Pediatric Clinical Presentations, and Pathogenesis. Front. Pediatr. 2021, 9, 643219. [Google Scholar] [CrossRef] [PubMed]
- Ljubin-Sternak, S.; Meštrović, T. Rhinovirus—A True Respiratory Threat or a Common Inconvenience of Childhood? Viruses 2023, 15, 825. [Google Scholar] [CrossRef] [PubMed]
- Su, S.; Wong, G.; Shi, W.; Liu, J.; Lai, A.C.K.; Zhou, J.; Liu, W.; Bi, Y.; Gao, G.F. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends Microbiol. 2016, 24, 490–502. [Google Scholar] [CrossRef] [PubMed]
- Uyeki, T.M.; Hui, D.S.; Zambon, M.; Wentworth, D.E.; Monto, A.S. Influenza. Lancet 2022, 400, 693–706. [Google Scholar] [CrossRef]
- Domachowske, J.B.; Rosenberg, H.F. Respiratory Syncytial Virus Infection: Immune Response, Immunopathogenesis, and Treatment. Clin. Microbiol. Rev. 1999, 12, 298–309. [Google Scholar] [CrossRef]
- Jiang, M.-Y.; Duan, Y.-P.; Tong, X.-L.; Huang, Q.-R.; Jia, M.-M.; Yang, W.-Z.; Feng, L.-Z. Clinical Manifestations of Respiratory Syncytial Virus Infection and the Risk of Wheezing and Recurrent Wheezing Illness: A Systematic Review and Meta-Analysis. World J. Pediatr. 2023, 19, 1030–1040. [Google Scholar] [CrossRef]
- Maykowski, P.; Smithgall, M.; Zachariah, P.; Oberhardt, M.; Vargas, C.; Reed, C.; Demmer, R.T.; Stockwell, M.S.; Saiman, L. Seasonality and Clinical Impact of Human Parainfluenza Viruses. Influenza Other Respir. Viruses 2018, 12, 706–716. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical Features of Patients Infected with 2019 Novel Coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Han, S.; Xu, B.; Feng, Q.; Feng, Z.; Zhu, Y.; Ai, J.; Deng, L.; Li, C.; Cao, L.; Sun, Y.; et al. Multicenter Analysis of Epidemiological and Clinical Features of Pediatric Acute Lower Respiratory Tract Infections Associated with Common Human Coronaviruses in China, 2014–2019. Virol. J. 2023, 20, 229. [Google Scholar] [CrossRef]
- Ison, M.G.; Hayden, R.T. Adenovirus. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef]
- Kajon, A.E. Adenovirus Infections: New Insights for the Clinical Laboratory. J. Clin. Microbiol. 2024, 62, e00836-22. [Google Scholar] [CrossRef]
- MacNeil, K.M.; Dodge, M.J.; Evans, A.M.; Tessier, T.M.; Weinberg, J.B.; Mymryk, J.S. Adenoviruses in Medicine: Innocuous Pathogen, Predator, or Partner. Trends Mol. Med. 2023, 29, 4–19. [Google Scholar] [CrossRef]
- Stalkup, J.R.; Chilukuri, S. Enterovirus Infections: A Review of Clinical Presentation, Diagnosis, and Treatment. Dermatol. Clin. 2002, 20, 217–223. [Google Scholar] [CrossRef]
- Mohammadi, K.; Faramarzi, S.; Yaribash, S.; Valizadeh, Z.; Rajabi, E.; Ghavam, M.; Samiee, R.; Karim, B.; Salehi, M.; Seifi, A.; et al. Human Metapneumovirus (HMPV) in 2025: Emerging Trends and Insights from Community and Hospital-Based Respiratory Panel Analyses—A Comprehensive Review. Virol. J. 2025, 22, 150. [Google Scholar] [CrossRef]
- Lindner, J.; Karalar, L.; Schimanski, S.; Pfister, H.; Struff, W.; Modrow, S. Clinical and Epidemiological Aspects of Human Bocavirus Infection. J. Clin. Virol. 2008, 43, 391–395. [Google Scholar] [CrossRef]
- Gamiño-Arroyo, A.E.; Arellano-Galindo, J.; Del Carmen Guerra-de-Blas, P.; Ortega-Villa, A.M.; Mateja, A.; Llamosas-Gallardo, B.; Ortíz-Hernández, A.A.; Valdéz-Vázquez, R.; Ramírez-Venegas, A.; Galindo-Fraga, A.; et al. Clinical and Molecular Characterization of Children and Adults with Respiratory Bocavirus Infection in Mexico: A Cross-Sectional Nested Study within the ILI002 Prospective Observational Study. Lancet Reg. Health—Am. 2024, 29, 100647. [Google Scholar] [CrossRef] [PubMed]
- Cowling, B.J.; Chan, K.H.; Fang, V.J.; Lau, L.L.H.; So, H.C.; Fung, R.O.P.; Ma, E.S.K.; Kwong, A.S.K.; Chan, C.-W.; Tsui, W.W.S.; et al. Comparative Epidemiology of Pandemic and Seasonal Influenza A in Households. N. Engl. J. Med. 2010, 362, 2175–2184. [Google Scholar] [CrossRef] [PubMed]
- Gradisteanu Pircalabioru, G.; Iliescu, F.S.; Mihaescu, G.; Cucu, A.I.; Ionescu, O.N.; Popescu, M.; Simion, M.; Burlibasa, L.; Tica, M.; Chifiriuc, M.C.; et al. Advances in the Rapid Diagnostic of Viral Respiratory Tract Infections. Front. Cell Infect. Microbiol. 2022, 12, 807253. [Google Scholar] [CrossRef] [PubMed]
- Bos, L.D.J.; Ware, L.B. Acute Respiratory Distress Syndrome: Causes, Pathophysiology, and Phenotypes. Lancet 2022, 400, 1145–1156. [Google Scholar] [CrossRef]
- Jain, S.; Self, W.H.; Wunderink, R.G.; Fakhran, S.; Balk, R.; Bramley, A.M.; Reed, C.; Grijalva, C.G.; Anderson, E.J.; Courtney, D.M.; et al. Community-Acquired Pneumonia Requiring Hospitalization among U.S. Adults. N. Engl. J. Med. 2015, 373, 415–427. [Google Scholar] [CrossRef]
- Andersen, O.; Ernberg, I.; Hedström, A.K. Treatment Options for Epstein-Barr Virus-Related Disorders of the Central Nervous System. Infect. Drug Resist. 2023, 16, 4599–4620. [Google Scholar] [CrossRef]
- Current ICTV Taxonomy Release|ICTV. Available online: https://ictv.global/taxonomy (accessed on 19 October 2025).
- Khales, P.; Razizadeh, M.H.; Ghorbani, S.; Moattari, A.; Sarvari, J.; Saadati, H.; Sayyahfar, S.; Salavatiha, Z.; Hasanabad, M.H.; Poortahmasebi, V.; et al. Human Adenoviruses in Children with Gastroenteritis: A Systematic Review and Meta-Analysis. BMC Infect. Dis. 2024, 24, 478. [Google Scholar] [CrossRef] [PubMed]
- Kesson, A.M. Respiratory Virus Infections. Paediatr. Respir. Rev. 2007, 8, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhang, L.; Nie, X.; Wang, L.; Kang, L.; Zhang, Y.; Chen, Z.; Li, Y.; Wu, Y. Epidemiology and Mortality Risk of Severe Viral Pneumonia During the Pre-Pandemic, COVID-19 Pandemic and Post-Pandemic Era: A Retrospective Study of Hospitalized Children in ShenZhen, China Between 2017 and 2023. J. Epidemiol. Glob. Health 2025, 15, 53. [Google Scholar] [CrossRef] [PubMed]
- Calderaro, A.; Buttrini, M.; Farina, B.; Montecchini, S.; De Conto, F.; Chezzi, C. Respiratory Tract Infections and Laboratory Diagnostic Methods: A Review with A Focus on Syndromic Panel-Based Assays. Microorganisms 2022, 10, 1856. [Google Scholar] [CrossRef]
- Qiu, J.; Söderlund-Venermo, M.; Young, N.S. Human Parvoviruses. Clin. Microbiol. Rev. 2017, 30, 43–113. [Google Scholar] [CrossRef]
- Liao, J.; Yang, Z.; He, Y.; Wei, J.; Ren, L.; Liu, E.; Zang, N. Respiratory Tract Infection of Fatal Severe Human Bocavirus 1 in a 13-Month-Old Child: A Case Report and Literature Review. Front. Pediatr. 2022, 10, 949817. [Google Scholar] [CrossRef]
- Polo, D.; Lema, A.; Gándara, E.; Romalde, J.L. Prevalence of Human Bocavirus Infections in Europe. A Systematic Review and Meta-analysis. Transbound. Emerg. Dis. 2022, 69, 2451–2461. [Google Scholar] [CrossRef]
- Trapani, S.; Caporizzi, A.; Ricci, S.; Indolfi, G. Human Bocavirus in Childhood: A True Respiratory Pathogen or a “Passenger” Virus? A Comprehensive Review. Microorganisms 2023, 11, 1243. [Google Scholar] [CrossRef]
- Tang, G.; Liu, Z.; Chen, D. Human Coronaviruses: Origin, Host and Receptor. J. Clin. Virol. 2022, 155, 105246. [Google Scholar] [CrossRef]
- Tosta, S.; Moreno, K.; Schuab, G.; Fonseca, V.; Segovia, F.M.C.; Kashima, S.; Elias, M.C.; Sampaio, S.C.; Ciccozzi, M.; Alcantara, L.C.J.; et al. Global SARS-CoV-2 Genomic Surveillance: What We Have Learned (so Far). Infect. Genet. Evol. 2023, 108, 105405. [Google Scholar] [CrossRef] [PubMed]
- Neumann, G.; Kawaoka, Y. Transmission of Influenza A Viruses. Virology 2015, 479–480, 234–246. [Google Scholar] [CrossRef] [PubMed]
- Han, A.X.; de Jong, S.P.J.; Russell, C.A. Co-Evolution of Immunity and Seasonal Influenza Viruses. Nat. Rev. Microbiol. 2023, 21, 805–817. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y. Pathogenicity and Virulence of Influenza. Virulence 2023, 14, 2223057. [Google Scholar] [CrossRef]
- Nayak, J.; Hoy, G.; Gordon, A. Influenza in Children. Cold Spring Harb. Perspect. Med. 2021, 11, a038430. [Google Scholar] [CrossRef]
- Branche, A.; Falsey, A. Parainfluenza Virus Infection. Semin. Respir. Crit. Care Med. 2016, 37, 538–554. [Google Scholar] [CrossRef]
- Schuster, J.E.; Williams, J.V. Human Metapneumovirus. Microbiol. Spectr. 2014, 2. [Google Scholar] [CrossRef]
- Tulloch, R.L.; Kok, J.; Carter, I.; Dwyer, D.E.; Eden, J.-S. An Amplicon-Based Approach for the Whole-Genome Sequencing of Human Metapneumovirus. Viruses 2021, 13, 499. [Google Scholar] [CrossRef]
- Yi, L.; Zou, L.; Peng, J.; Yu, J.; Song, Y.; Liang, L.; Guo, Q.; Kang, M.; Ke, C.; Song, T.; et al. Epidemiology, Evolution and Transmission of Human Metapneumovirus in Guangzhou China, 2013–2017. Sci. Rep. 2019, 9, 14022. [Google Scholar] [CrossRef]
- Feng, Y.; He, T.; Zhang, B.; Yuan, H.; Zhou, Y. Epidemiology and Diagnosis Technologies of Human Metapneumovirus in China: A Mini Review. Virol. J. 2024, 21, 59. [Google Scholar] [CrossRef]
- Twumasi, S.; Ansah, R.O.; Ayirebi, A.A.; Antonio, D.N.M.; Asafoakaa, Y.A.; Tawiah, E.; Opoku, A. Human Metapneumovirus: A Review of Its Epidemiology, Clinical Features, Public Health Implications and Treatment Options. Rev. Med. Virol. 2025, 35, e70043. [Google Scholar] [CrossRef] [PubMed]
- Nam, H.H.; Ison, M.G. Respiratory Syncytial Virus Infection in Adults. BMJ 2019, 366, l5021. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, X.; Blau, D.M.; Caballero, M.T.; Feikin, D.R.; Gill, C.J.; Madhi, S.A.; Omer, S.B.; Simões, E.A.F.; Campbell, H.; et al. Global, Regional, and National Disease Burden Estimates of Acute Lower Respiratory Infections Due to Respiratory Syncytial Virus in Children Younger than 5 Years in 2019: A Systematic Analysis. Lancet 2022, 399, 2047–2064. [Google Scholar] [CrossRef] [PubMed]
- Alfano, F.; Bigoni, T.; Caggiano, F.P.; Papi, A. Respiratory Syncytial Virus Infection in Older Adults: An Update. Drugs Aging 2024, 41, 487–505. [Google Scholar] [CrossRef]
- Langedijk, A.C.; Bont, L.J. Respiratory Syncytial Virus Infection and Novel Interventions. Nat. Rev. Microbiol. 2023, 21, 734–749. [Google Scholar] [CrossRef]
- Fall, A.; Norton, J.M.; Abdullah, O.; Pekosz, A.; Klein, E.; Mostafa, H.H. Enhanced Genomic Surveillance of Enteroviruses Reveals a Surge in Enterovirus D68 Cases, the Johns Hopkins Health System, Maryland, 2024. J. Clin. Microbiol. 2025, 63, e00469-25. [Google Scholar] [CrossRef]
- Esposito, S.; Bosis, S.; Niesters, H.; Principi, N. Enterovirus D68 Infection. Viruses 2015, 7, 6043–6050. [Google Scholar] [CrossRef]
- Esneau, C.; Duff, A.C.; Bartlett, N.W. Understanding Rhinovirus Circulation and Impact on Illness. Viruses 2022, 14, 141. [Google Scholar] [CrossRef]
- Morelli, T.; Freeman, A.; Staples, K.J.; Wilkinson, T.M.A. Hidden in Plain Sight: The Impact of Human Rhinovirus Infection in Adults. Respir. Res. 2025, 26, 120. [Google Scholar] [CrossRef]
- Berginc, N.; Sočan, M.; Prosenc Trilar, K.; Petrovec, M. Seasonality and Genotype Diversity of Human Rhinoviruses during an Eight-Year Period in Slovenia. Microorganisms 2024, 12, 341. [Google Scholar] [CrossRef]
- Grech, A.K.; Foo, C.T.; Paul, E.; Aung, A.K.; Yu, C. Epidemiological Trends of Respiratory Tract Pathogens Detected via MPCR in Australian Adult Patients before COVID-19. BMC Infect. Dis. 2024, 24, 38. [Google Scholar] [CrossRef] [PubMed]
- Lima, A.R.J.; Santos, H.d.O.; Pereira, J.S.; Leite, A.B.; Nascimento, J.P.M.d.; Souza, J.V.C.; Lima, M.B.Z.; de Araújo, M.A.; Giovanetti, M.; Kallas, E.G.; et al. Enterovirus D68 Subgenotype B3 Circulation in Children with Acute Respiratory Illness in the State of Alagoas, Brazil. Viruses 2025, 17, 242. [Google Scholar] [CrossRef] [PubMed]
- Moscona, A. Entry of Parainfluenza Virus into Cells as a Target for Interrupting Childhood Respiratory Disease. J. Clin. Investig. 2005, 115, 1688–1698. [Google Scholar] [CrossRef]
- Hanson, K.E.; Azar, M.M.; Banerjee, R.; Chou, A.; Colgrove, R.C.; Ginocchio, C.C.; Hayden, M.K.; Holodiny, M.; Jain, S.; Koo, S.; et al. Molecular Testing for Acute Respiratory Tract Infections: Clinical and Diagnostic Recommendations from the IDSA’s Diagnostics Committee. Clin. Infect. Dis. 2020, 71, 2744–2751. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liang, S.; Zhang, D.; He, M.; Zhang, H. The Clinical Application of Metagenomic Next-Generation Sequencing in Sepsis of Immunocompromised Patients. Front. Cell Infect. Microbiol. 2023, 13, 1170687. [Google Scholar] [CrossRef]
- Nair, H.; Nokes, D.J.; Gessner, B.D.; Dherani, M.; Madhi, S.A.; Singleton, R.J.; O’Brien, K.L.; Roca, A.; Wright, P.F.; Bruce, N.; et al. Global Burden of Acute Lower Respiratory Infections Due to Respiratory Syncytial Virus in Young Children: A Systematic Review and Meta-Analysis. Lancet 2010, 375, 1545–1555. [Google Scholar] [CrossRef]
- Roberts, A.L.; Sammons, J.S.; Mourani, P.M.; Thomas, N.J.; Yehya, N. Specific Viral Etiologies Are Associated with Outcomes in Pediatric Acute Respiratory Distress Syndrome. Pediatr. Crit. Care Med. 2019, 20, e441–e446. [Google Scholar] [CrossRef]
- Huang, H.-S.; Tsai, C.-L.; Chang, J.; Hsu, T.-C.; Lin, S.; Lee, C.-C. Multiplex PCR System for the Rapid Diagnosis of Respiratory Virus Infection: Systematic Review and Meta-Analysis. Clin. Microbiol. Infect. 2018, 24, 1055–1063. [Google Scholar] [CrossRef]
- Touma, M. COVID-19: Molecular Diagnostics Overview. J. Mol. Med. 2020, 98, 947–954. [Google Scholar] [CrossRef]
- Choi, Q.; Kim, H. Respiratory Viruses Co-Detection on a Multiplex RT-PCR Panel: A Comparative Clinical Study. Clin. Lab. 2024, 70. [Google Scholar] [CrossRef]
- Rheem, I.; Park, J.; Kim, T.-H.; Kim, J.W. Evaluation of a Multiplex Real-Time PCR Assay for the Detection of Respiratory Viruses in Clinical Specimens. Ann. Lab. Med. 2012, 32, 399–406. [Google Scholar] [CrossRef]
- Sanghavi, S.K.; Bullotta, A.; Husain, S.; Rinaldo, C.R. Clinical Evaluation of Multiplex Real-time PCR Panels for Rapid Detection of Respiratory Viral Infections. J. Med. Virol. 2012, 84, 162–169. [Google Scholar] [CrossRef] [PubMed]
- Rojahn, S.; Hambuch, T.; Adrian, J.; Gafni, E.; Gileta, A.; Hatchell, H.; Johnson, B.; Kallman, B.; Karfilis, K.; Kautzer, C.; et al. Scalable Detection of Technically Challenging Variants through Modified Next-generation Sequencing. Mol. Genet. Genom. Med. 2022, 10, e2072. [Google Scholar] [CrossRef] [PubMed]
- Madi, N.; Al-Nakib, W.; Mustafa, A.S.; Habibi, N. Metagenomic Analysis of Viral Diversity in Respiratory Samples from Patients with Respiratory Tract Infections in Kuwait. J. Med. Virol. 2018, 90, 412–420. [Google Scholar] [CrossRef]
- Thomas, T.; Gilbert, J.; Meyer, F. Metagenomics—A Guide from Sampling to Data Analysis. Microb. Inform. Exp. 2012, 2, 3. [Google Scholar] [CrossRef]
- Duarte, V.d.S.; Porcellato, D. Host DNA Depletion Methods and Genome-Centric Metagenomics of Bovine Hindmilk Microbiome. mSphere 2024, 9, e00470-23. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, J.; Hou, H.; Song, L.; Cheng, X.; Liu, Y. Advancing Plant Microbiome Research Through Host DNA Depletion Techniques. Plant Biotechnol. J. 2025. [Google Scholar] [CrossRef]
- Thurber, R.V.; Haynes, M.; Breitbart, M.; Wegley, L.; Rohwer, F. Laboratory Procedures to Generate Viral Metagenomes. Nat. Protoc. 2009, 4, 470–483. [Google Scholar] [CrossRef]
- Zhang, D.; Lou, X.; Yan, H.; Pan, J.; Mao, H.; Tang, H.; Shu, Y.; Zhao, Y.; Liu, L.; Li, J.; et al. Metagenomic Analysis of Viral Nucleic Acid Extraction Methods in Respiratory Clinical Samples. BMC Genom. 2018, 19, 773. [Google Scholar] [CrossRef]
- Ko, K.K.K.; Chng, K.R.; Nagarajan, N. Metagenomics-Enabled Microbial Surveillance. Nat. Microbiol. 2022, 7, 486–496. [Google Scholar] [CrossRef]
- Dugan, V.G.; Saira, K.; Ghedin, E. Large-Scale Sequencing and the Natural History of Model Human RNA Viruses. Future Virol. 2012, 7, 563–573. [Google Scholar] [CrossRef] [PubMed]
- Van Poelvoorde, L.A.E.; Karlsson, E.A.; Dupont-Rouzeyrol, M.; Roosens, N.H.C.J. Can Wastewater Surveillance Enhance Genomic Tracking of Climate-Driven Pathogens? Microorganisms 2025, 13, 294. [Google Scholar] [CrossRef] [PubMed]
- Castro, C.J.; Marine, R.L.; Ramos, E.; Ng, T.F.F. The Effect of Variant Interference on de Novo Assembly for Viral Deep Sequencing. BMC Genom. 2020, 21, 421. [Google Scholar] [CrossRef]
- Yang, Z.; Guarracino, A.; Biggs, P.J.; Black, M.A.; Ismail, N.; Wold, J.R.; Merriman, T.R.; Prins, P.; Garrison, E.; de Ligt, J. Pangenome Graphs in Infectious Disease: A Comprehensive Genetic Variation Analysis of Neisseria Meningitidis Leveraging Oxford Nanopore Long Reads. Front. Genet. 2023, 14, 1225248. [Google Scholar] [CrossRef] [PubMed]
- Matsvay, A.; Kiselev, D.; Ayginin, A.; Abramov, I.; Dedkov, V.; Shipulin, G.; Khafizov, K. Metabarcoding-Like Approach for High Throughput Detection and Identification of Viral Nucleic Acids. Proceedings 2020, 50, 136. [Google Scholar] [CrossRef]
- Gaudin, M.; Desnues, C. Hybrid Capture-Based Next Generation Sequencing and Its Application to Human Infectious Diseases. Front. Microbiol. 2018, 9, 02924. [Google Scholar] [CrossRef]
- Wylie, T.N.; Wylie, K.M.; Herter, B.N.; Storch, G.A. Enhanced Virome Sequencing Using Targeted Sequence Capture. Genome Res. 2015, 25, 1910–1920. [Google Scholar] [CrossRef]
- Briese, T.; Kapoor, A.; Mishra, N.; Jain, K.; Kumar, A.; Jabado, O.J.; Lipkin, W.I. Virome Capture Sequencing Enables Sensitive Viral Diagnosis and Comprehensive Virome Analysis. mBio 2015, 6, e01491-15. [Google Scholar] [CrossRef]
- Mah, A.H.; Qi, X.; Zhao, J.; Wiseman, K.; Edoli, L.; Metcalfe, K.; Donohoe, K.; Ojeda, M.; Billings, S.; Moreno, J.; et al. A Simplified Hybrid Capture Approach Retains High Specificity and Enables PCR-Free Workflow. BMC Genom. 2025, 26, 799. [Google Scholar] [CrossRef]
- Bianconi, I.; Pellecchia, G.V.; Incrocci, E.M.; Vittadello, F.; Nicoletti, M.; Pagani, E. Comparative Performance of Digital PCR and Real-Time RT-PCR in Respiratory Virus Diagnostics. Viruses 2025, 17, 1259. [Google Scholar] [CrossRef]
- Charalampous, T.; Alcolea-Medina, A.; Snell, L.B.; Alder, C.; Tan, M.; Williams, T.G.S.; Al-Yaakoubi, N.; Humayun, G.; Meadows, C.I.S.; Wyncoll, D.L.A.; et al. Routine Metagenomics Service for ICU Patients with Respiratory Infection. Am. J. Respir. Crit. Care Med. 2024, 209, 164–174. [Google Scholar] [CrossRef]
- Chapman, R.; Jones, L.; D’Angelo, A.; Suliman, A.; Anwar, M.; Bagby, S. Nanopore-Based Metagenomic Sequencing in Respiratory Tract Infection: A Developing Diagnostic Platform. Lung 2023, 201, 171–179. [Google Scholar] [CrossRef]
- Greninger, A.L.; Naccache, S.N.; Federman, S.; Yu, G.; Mbala, P.; Bres, V.; Stryke, D.; Bouquet, J.; Somasekar, S.; Linnen, J.M.; et al. Rapid Metagenomic Identification of Viral Pathogens in Clinical Samples by Real-Time Nanopore Sequencing Analysis. Genome Med. 2015, 7, 99. [Google Scholar] [CrossRef]
- Bull, R.A.; Adikari, T.N.; Ferguson, J.M.; Hammond, J.M.; Stevanovski, I.; Beukers, A.G.; Naing, Z.; Yeang, M.; Verich, A.; Gamaarachchi, H.; et al. Analytical Validity of Nanopore Sequencing for Rapid SARS-CoV-2 Genome Analysis. Nat. Commun. 2020, 11, 6272. [Google Scholar] [CrossRef]
- Kafetzopoulou, L.E.; Pullan, S.T.; Lemey, P.; Suchard, M.A.; Ehichioya, D.U.; Pahlmann, M.; Thielebein, A.; Hinzmann, J.; Oestereich, L.; Wozniak, D.M.; et al. Metagenomic Sequencing at the Epicenter of the Nigeria 2018 Lassa Fever Outbreak. Science 2019, 363, 74–77. [Google Scholar] [CrossRef] [PubMed]
- Otron, D.H.; Filloux, D.; Brousse, A.; Hoareau, M.; Fenelon, B.; Hoareau, C.; Fernandez, E.; Tiendrébéogo, F.; Lett, J.-M.; Pita, J.S.; et al. Improvement of Nanopore Sequencing Provides Access to High Quality Genomic Data for Multi-Component CRESS-DNA Plant Viruses. Virol. J. 2025, 22, 78. [Google Scholar] [CrossRef] [PubMed]
- Lewandowski, K.; Xu, Y.; Pullan, S.T.; Lumley, S.F.; Foster, D.; Sanderson, N.; Vaughan, A.; Morgan, M.; Bright, N.; Kavanagh, J.; et al. Metagenomic Nanopore Sequencing of Influenza Virus Direct from Clinical Respiratory Samples. J. Clin. Microbiol. 2019, 58, e00963-19. [Google Scholar] [CrossRef]
- Charalampous, T.; Kay, G.L.; Richardson, H.; Aydin, A.; Baldan, R.; Jeanes, C.; Rae, D.; Grundy, S.; Turner, D.J.; Wain, J.; et al. Nanopore Metagenomics Enables Rapid Clinical Diagnosis of Bacterial Lower Respiratory Infection. Nat. Biotechnol. 2019, 37, 783–792. [Google Scholar] [CrossRef]
- Briese, T.; Paweska, J.T.; McMullan, L.K.; Hutchison, S.K.; Street, C.; Palacios, G.; Khristova, M.L.; Weyer, J.; Swanepoel, R.; Egholm, M.; et al. Genetic Detection and Characterization of Lujo Virus, a New Hemorrhagic Fever–Associated Arenavirus from Southern Africa. PLoS Pathog. 2009, 5, e1000455. [Google Scholar] [CrossRef]
- Grard, G.; Fair, J.N.; Lee, D.; Slikas, E.; Steffen, I.; Muyembe, J.-J.; Sittler, T.; Veeraraghavan, N.; Ruby, J.G.; Wang, C.; et al. A Novel Rhabdovirus Associated with Acute Hemorrhagic Fever in Central Africa. PLoS Pathog. 2012, 8, e1002924. [Google Scholar] [CrossRef]
- Frémond, M.-L.; Pérot, P.; Muth, E.; Cros, G.; Dumarest, M.; Mahlaoui, N.; Seilhean, D.; Desguerre, I.; Hébert, C.; Corre-Catelin, N.; et al. Next-Generation Sequencing for Diagnosis and Tailored Therapy: A Case Report of Astrovirus-Associated Progressive Encephalitis. J. Pediatr. Infect. Dis. Soc. 2015, 4, e53–e57. [Google Scholar] [CrossRef]
- Klompas, M.; Imrey, P.B.; Yu, P.-C.; Rhee, C.; Deshpande, A.; Haessler, S.; Zilberberg, M.D.; Rothberg, M.B. Respiratory Viral Testing and Antibacterial Treatment in Patients Hospitalized with Community-Acquired Pneumonia. Infect. Control Hosp. Epidemiol. 2021, 42, 817–825. [Google Scholar] [CrossRef]
- Joelsons, D.; Alencar, C.S.; Pinho, J.R.R.; Ho, Y.-L. Investigation of Etiology of Community-Acquired Pneumonia in Hospitalized Patients in a Tertiary Hospital of São Paulo City, Brazil. Braz. J. Infect. Dis. 2023, 27, 103690. [Google Scholar] [CrossRef] [PubMed]
- Sandybayev, N.; Beloussov, V.; Strochkov, V.; Solomadin, M.; Granica, J.; Yegorov, S. Metagenomic Profiling of Nasopharyngeal Samples from Adults with Acute Respiratory Infection. R. Soc. Open Sci. 2024, 11, 240108. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Greninger, A.L.; Naccache, S.N.; Messacar, K.; Clayton, A.; Yu, G.; Somasekar, S.; Federman, S.; Stryke, D.; Anderson, C.; Yagi, S.; et al. A Novel Outbreak Enterovirus D68 Strain Associated with Acute Flaccid Myelitis Cases in the USA (2012–14): A Retrospective Cohort Study. Lancet Infect. Dis. 2015, 15, 671–682. [Google Scholar] [CrossRef]
- Li, H.; Xiao, Y.; Lü, J.; Zhang, W.; Lu, H. Viral Metagenomic Analysis Reveals a Human Rhinovirus from Hospitalized Neonates. Microbiol. Resour. Announc. 2021, 10, e00106-21. [Google Scholar] [CrossRef]
- Chen, X.-H.; Zhou, S.-J.; Liu, Y.-Y.; Cao, H.; Zheng, Y.-R.; Chen, Q. Application Value of Metagenomics Next-Generation Sequencing in the Diagnosis of Respiratory Virus Infection after Congenital Heart Surgery. Transl. Pediatr. 2024, 13, 260–270. [Google Scholar] [CrossRef]
- Gong, Y.-N.; Yang, S.-L.; Chen, G.-W.; Chen, Y.-W.; Huang, Y.-C.; Ning, H.-C.; Tsao, K.-C. A Metagenomics Study for the Identification of Respiratory Viruses in Mixed Clinical Specimens: An Application of the Iterative Mapping Approach. Arch. Virol. 2017, 162, 2003–2012. [Google Scholar] [CrossRef]
- Babiker, A.; Bradley, H.L.; Stittleburg, V.D.; Ingersoll, J.M.; Key, A.; Kraft, C.S.; Waggoner, J.J.; Piantadosi, A. Metagenomic Sequencing to Detect Respiratory Viruses in Persons under Investigation for COVID-19. J. Clin. Microbiol. 2020, 59, e02142-20. [Google Scholar] [CrossRef]
- Tan, J.K.; Servellita, V.; Stryke, D.; Kelly, E.; Streithorst, J.; Sumimoto, N.; Foresythe, A.; Huh, H.J.; Nguyen, J.; Oseguera, M.; et al. Laboratory Validation of a Clinical Metagenomic Next-Generation Sequencing Assay for Respiratory Virus Detection and Discovery. Nat. Commun. 2024, 15, 9016. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.; Zhang, D.Y. Artificial Intelligence and Computational Pathology. Lab. Investig. 2021, 101, 412–422. [Google Scholar] [CrossRef] [PubMed]
- Graf, E.H.; Simmon, K.E.; Tardif, K.D.; Hymas, W.; Flygare, S.; Eilbeck, K.; Yandell, M.; Schlaberg, R. Unbiased Detection of Respiratory Viruses by Use of RNA Sequencing-Based Metagenomics: A Systematic Comparison to a Commercial PCR Panel. J. Clin. Microbiol. 2016, 54, 1000–1007. [Google Scholar] [CrossRef] [PubMed]
- Downie, D.L.; Rao, P.; David-Ferdon, C.; Courtney, S.; Lee, J.S.; Quiner, C.; MacDonald, P.D.M.; Barnes, K.; Fisher, S.; Andreadis, J.L.; et al. Surveillance for Emerging and Reemerging Pathogens Using Pathogen Agnostic Metagenomic Sequencing in the United States: A Critical Role for Federal Government Agencies. Health Secur. 2024, 22, 85–92. [Google Scholar] [CrossRef]
- Makoni, M. Launch of Genomic Surveillance System for Respiratory Viruses. Lancet Microbe 2023, 4, e214. [Google Scholar] [CrossRef]
- Ali, J.; Johansen, W.; Ahmad, R. Short Turnaround Time of Seven to Nine Hours from Sample Collection until Informed Decision for Sepsis Treatment Using Nanopore Sequencing. Sci. Rep. 2024, 14, 6534. [Google Scholar] [CrossRef]
- Meredith, L.W.; Hamilton, W.L.; Warne, B.; Houldcroft, C.J.; Hosmillo, M.; Jahun, A.S.; Curran, M.D.; Parmar, S.; Caller, L.G.; Caddy, S.L.; et al. Rapid Implementation of SARS-CoV-2 Sequencing to Investigate Cases of Health-Care Associated COVID-19: A Prospective Genomic Surveillance Study. Lancet Infect. Dis. 2020, 20, 1263–1271. [Google Scholar] [CrossRef]
- Šúri, T.; Pfeiferová, L.; Bezdíček, M.; Svatoň, J.; Hampl, V.; Berka, K.; Jiřincová, H.; Lengerová, M.; Kolísko, M.; Nagy, A.; et al. Developing Molecular Surveillance of SARS-CoV-2 in the Czech Republic (2021–2022). Sci. Rep. 2025, 15, 19690. [Google Scholar] [CrossRef]
- Sahadeo, N.S.D.; Nicholls, S.; Moreira, F.R.R.; O’Toole, Á.; Ramkissoon, V.; Whittaker, C.; Hill, V.; McCrone, J.T.; Mohammed, N.; Ramjag, A.; et al. Implementation of Genomic Surveillance of SARS-CoV-2 in the Caribbean: Lessons Learned for Sustainability in Resource-Limited Settings. PLoS Glob. Public Health 2023, 3, e0001455. [Google Scholar] [CrossRef]
- Tan, M.; Xia, J.; Luo, H.; Meng, G.; Zhu, Z. Applying the Digital Data and the Bioinformatics Tools in SARS-CoV-2 Research. Comput. Struct. Biotechnol. J. 2023, 21, 4697–4705. [Google Scholar] [CrossRef]
- Wang, Y.; Zhu, N.; Li, Y.; Lu, R.; Wang, H.; Liu, G.; Zou, X.; Xie, Z.; Tan, W. Metagenomic Analysis of Viral Genetic Diversity in Respiratory Samples from Children with Severe Acute Respiratory Infection in China. Clin. Microbiol. Infect. 2016, 22, 458.e1–458.e9. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, A.; Nakielny, S.; Hsu, J.; Kyohere, M.; Byaruhanga, O.; de Bourcy, C.; Egger, R.; Dimitrov, B.; Juan, Y.-F.; Sheu, J.; et al. Metagenomic Next-Generation Sequencing of Samples from Pediatric Febrile Illness in Tororo, Uganda. PLoS ONE 2019, 14, e0218318. [Google Scholar] [CrossRef] [PubMed]
- Jerome, H.; Taylor, C.; Sreenu, V.B.; Klymenko, T.; Filipe, A.D.S.; Jackson, C.; Davis, C.; Ashraf, S.; Wilson-Davies, E.; Jesudason, N.; et al. Metagenomic Next-Generation Sequencing Aids the Diagnosis of Viral Infections in Febrile Returning Travellers. J. Infect. 2019, 79, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Torres Montaguth, O.E.; Buddle, S.; Morfopoulou, S.; Breuer, J. Clinical Metagenomics for Diagnosis and Surveillance of Viral Pathogens. Nat. Rev. Microbiol. 2025. [Google Scholar] [CrossRef]
- Casto, A.M.; Adler, A.L.; Makhsous, N.; Crawford, K.; Qin, X.; Kuypers, J.M.; Huang, M.-L.; Zerr, D.M.; Greninger, A.L. Prospective, Real-Time Metagenomic Sequencing During Norovirus Outbreak Reveals Discrete Transmission Clusters. Clin. Infect. Dis. 2019, 69, 941–948. [Google Scholar] [CrossRef]
- Lemieux, J.E.; Siddle, K.J.; Shaw, B.M.; Loreth, C.; Schaffner, S.F.; Gladden-Young, A.; Adams, G.; Fink, T.; Tomkins-Tinch, C.H.; Krasilnikova, L.A.; et al. Phylogenetic Analysis of SARS-CoV-2 in Boston Highlights the Impact of Superspreading Events. Science 2021, 371, eabe3261. [Google Scholar] [CrossRef]
- Borges, V.; Duque, M.P.; Martins, J.V.; Vasconcelos, P.; Ferreira, R.; Sobral, D.; Pelerito, A.; de Carvalho, I.L.; Núncio, M.S.; Borrego, M.J.; et al. Viral Genetic Clustering and Transmission Dynamics of the 2022 Mpox Outbreak in Portugal. Nat. Med. 2023, 29, 2509–2517. [Google Scholar] [CrossRef]
- Jurasz, H.; Pawłowski, T.; Perlejewski, K. Contamination Issue in Viral Metagenomics: Problems, Solutions, and Clinical Perspectives. Front. Microbiol. 2021, 12, 745076. [Google Scholar] [CrossRef]
- Sabatier, M.; Bal, A.; Destras, G.; Regue, H.; Quéromès, G.; Cheynet, V.; Lina, B.; Bardel, C.; Brengel-Pesce, K.; Navratil, V.; et al. Comparison of Nucleic Acid Extraction Methods for a Viral Metagenomics Analysis of Respiratory Viruses. Microorganisms 2020, 8, 1539. [Google Scholar] [CrossRef]
- Chang, W.-S.; Harvey, E.; Mahar, J.E.; Firth, C.; Shi, M.; Simon-Loriere, E.; Geoghegan, J.L.; Wille, M. Improving the Reporting of Metagenomic Virome-Scale Data. Commun. Biol. 2024, 7, 1687. [Google Scholar] [CrossRef]
- Duan, J.; Keeler, E.; McFarland, A.; Scott, P.; Collman, R.G.; Bushman, F.D. The Virome of the Kitome: Small Circular Virus-like Genomes in Laboratory Reagents. Microbiol. Resour. Announc. 2024, 13, e01261-23. [Google Scholar] [CrossRef]
- Uphoff, C.C.; Lange, S.; Denkmann, S.A.; Garritsen, H.S.P.; Drexler, H.G. Prevalence and Characterization of Murine Leukemia Virus Contamination in Human Cell Lines. PLoS ONE 2015, 10, e0125622. [Google Scholar] [CrossRef]
- Wally, N.; Schneider, M.; Thannesberger, J.; Kastner, M.T.; Bakonyi, T.; Indik, S.; Rattei, T.; Bedarf, J.; Hildebrand, F.; Law, J.; et al. Plasmid DNA Contaminant in Molecular Reagents. Sci. Rep. 2019, 9, 1652. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, J.A.; Richard, A.C.; Bach, K.; Lun, A.T.L.; Marioni, J.C. Detection and Removal of Barcode Swapping in Single-Cell RNA-Seq Data. Nat. Commun. 2018, 9, 2667. [Google Scholar] [CrossRef] [PubMed]
- Brandariz-Fontes, C.; Camacho-Sanchez, M.; Vilà, C.; Vega-Pla, J.L.; Rico, C.; Leonard, J.A. Effect of the Enzyme and PCR Conditions on the Quality of High-Throughput DNA Sequencing Results. Sci. Rep. 2015, 5, 8056. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Leung, P.M.; Lappan, R.; Eisenhofer, R.; Ricci, F.; Holland, S.I.; Dragone, N.; Blackall, L.L.; Dong, X.; Dorador, C.; et al. Guidelines for Preventing and Reporting Contamination in Low-Biomass Microbiome Studies. Nat. Microbiol. 2025, 10, 1570–1580. [Google Scholar] [CrossRef]
- Thoendel, M.; Jeraldo, P.; Greenwood-Quaintance, K.E.; Yao, J.; Chia, N.; Hanssen, A.D.; Abdel, M.P.; Patel, R. Impact of Contaminating DNA in Whole-Genome Amplification Kits Used for Metagenomic Shotgun Sequencing for Infection Diagnosis. J. Clin. Microbiol. 2017, 55, 1789–1801. [Google Scholar] [CrossRef]
- Davis, N.M.; Proctor, D.M.; Holmes, S.P.; Relman, D.A.; Callahan, B.J. Simple Statistical Identification and Removal of Contaminant Sequences in Marker-Gene and Metagenomics Data. Microbiome 2018, 6, 226. [Google Scholar] [CrossRef]
- Knights, D.; Kuczynski, J.; Charlson, E.S.; Zaneveld, J.; Mozer, M.C.; Collman, R.G.; Bushman, F.D.; Knight, R.; Kelley, S.T. Bayesian Community-Wide Culture-Independent Microbial Source Tracking. Nat. Methods 2011, 8, 761–763. [Google Scholar] [CrossRef]
- McKnight, D.T.; Huerlimann, R.; Bower, D.S.; Schwarzkopf, L.; Alford, R.A.; Zenger, K.R. MicroDecon: A Highly Accurate Read-subtraction Tool for the Post-sequencing Removal of Contamination in Metabarcoding Studies. Environ. DNA 2019, 1, 14–25. [Google Scholar] [CrossRef]
- Austin, G.I.; Park, H.; Meydan, Y.; Seeram, D.; Sezin, T.; Lou, Y.C.; Firek, B.A.; Morowitz, M.J.; Banfield, J.F.; Christiano, A.M.; et al. Contamination Source Modeling with SCRuB Improves Cancer Phenotype Prediction from Microbiome Data. Nat. Biotechnol. 2023, 41, 1820–1828. [Google Scholar] [CrossRef]
- Liu, Y.; Elworth, R.A.L.; Jochum, M.D.; Aagaard, K.M.; Treangen, T.J. De Novo Identification of Microbial Contaminants in Low Microbial Biomass Microbiomes with Squeegee. Nat. Commun. 2022, 13, 6799. [Google Scholar] [CrossRef]
- Di Gloria, L.; Casbarra, L.; Lotti, T.; Ramazzotti, M. Testing the Limits of Short-Reads Metagenomic Classifications Programs in Wastewater Treating Microbial Communities. Sci. Rep. 2025, 15, 23997. [Google Scholar] [CrossRef]
- Langenfeld, K.; Hegarty, B.; Vidaurri, S.; Crossette, E.; Duhaime, M.B.; Wigginton, K.R. Development of a Quantitative Metagenomic Approach to Establish Quantitative Limits and Its Application to Viruses. Nucleic Acids Res. 2025, 53, gkaf118. [Google Scholar] [CrossRef]
- Bhardwaj, K.; Niharika; Garg, A.; Jain, A.; Kumar, M.; Datt, M.; Singh, V.; Vrati, S. Identification and Profiling of Novel Metagenome Assembled Uncultivated Virus Genomes from Human Gut. Virol. J. 2025, 22, 254. [Google Scholar] [CrossRef]
- Kosmopoulos, J.C.; Klier, K.M.; Langwig, M.V.; Tran, P.Q.; Anantharaman, K. Viromes vs. Mixed Community Metagenomes: Choice of Method Dictates Interpretation of Viral Community Ecology. Microbiome 2024, 12, 195. [Google Scholar] [CrossRef]
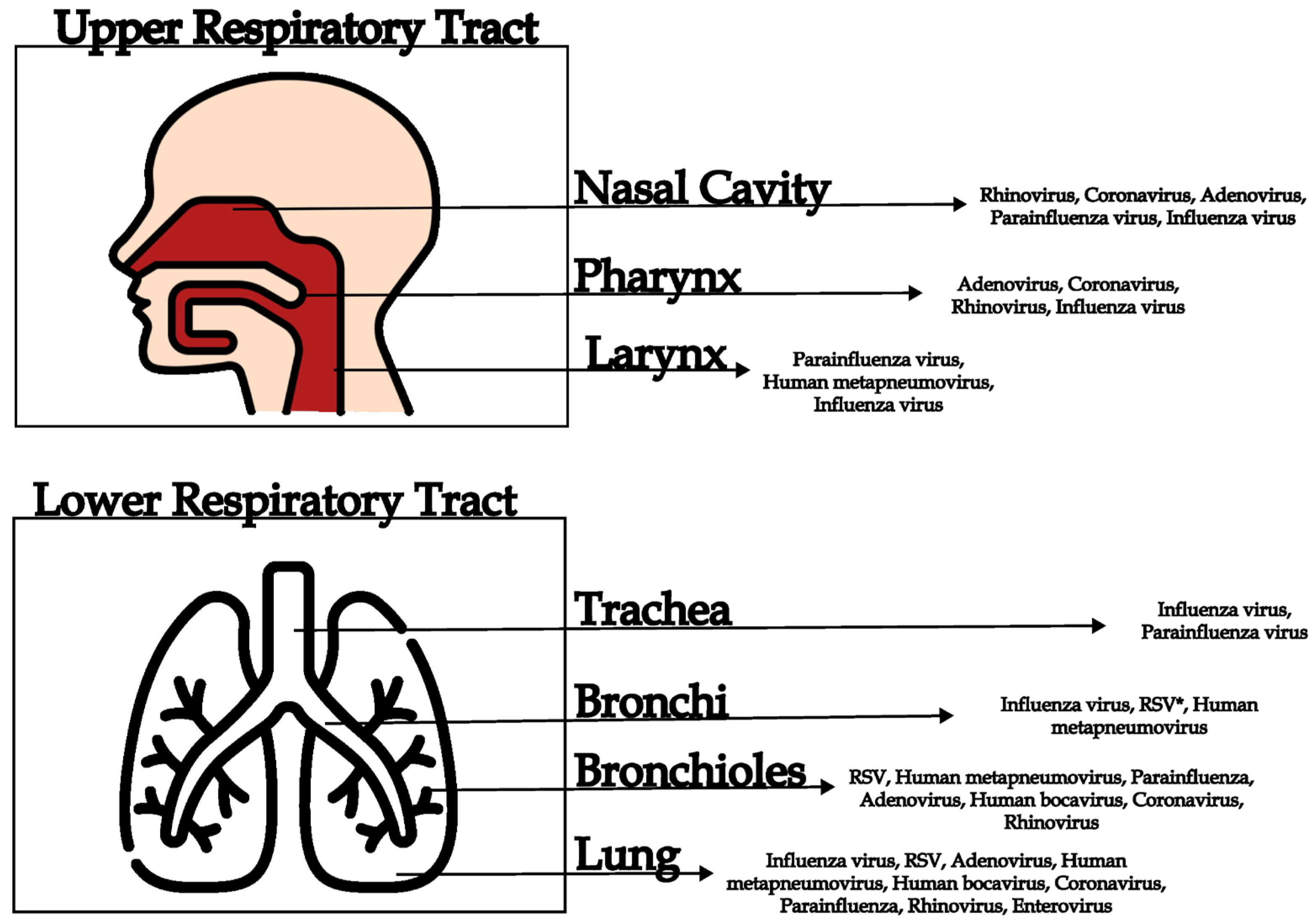
| Virus | Genome | Respiratory Syndromes |
|---|---|---|
| Rhinoviruses | RNA | The most common causes of the common cold, otitis media, rhinosinusitis, lower tract respiratory infections [12,13,14]. |
| Influenza viruses (A and B) | RNA (segmented) | Uncomplicated upper respiratory disease, upper and lower respiratory complications, cardiac and gastrointestinal complications [15,16]. |
| Respiratory syncytial virus (RSV) | RNA | An important cause of respiratory illness in infants and young children, might lead to severe clinical manifestations including respiratory failure [17,18] |
| Parainfluenza viruses | RNA | Causative agents of croup and other respiratory conditions, especially in children [19] |
| Coronaviruses | RNA | Fever, myalgia, fatigue, cough, headache. Include SARS-CoV-2 (COVID-19), as well as other coronaviruses causing milder respiratory illnesses [20,21]. |
| Adenoviruses | DNA | Acute respiratory infection, pneumonia, conjunctivitis, gastrointestinal infections [22,23,24]. |
| Enteroviruses | RNA | Respiratory illness, aseptic meningitis, encephalitis, meningitis, myocarditis, paralysis, multiorgan failure [25] |
| Human metapneumovirus (hMPV) | RNA | Cough, fever, nasal congestion, wheezing, mild to moderate respiratory disease. The most affected groups are children and the elderly [26] |
| Human bocavirus (HBoV) | DNA | Causative agents of acute respiratory tract infections related to fever, cough, and shortness of breath, especially in children [27,28]. |
| Species | Popular Name |
|---|---|
| Enterovirus alphacoxsackie | Coxsackievírus A |
| Enterovirus betacoxsackie | Coxsackievírus B |
| Enterovirus coxsackiepol | Poliovirus and Enterovirus C |
| Enterovirus deconjuncti | Enterovírus D |
| Enterovirus alpharhino | Rhinovirus A |
| Enterovirus betarhino | Rhinovirus B |
| Enterovirus cerhino | Rhinovirus C |
| Enterovirus eibovi | Enterovirus E |
| Enterovirus fitauri | Enterovirus F |
| Enterovirus geswini | Enterovirus G (Porcine enterovirus B) |
| Enterovirus hesimi | Enterovirus H (Simian enterovirus A) |
| Enterovirus idromi | Enterovirus I |
| Enterovirus jesimi | Enterovirus J |
| Enterovirus krodeni | Enterovirus K |
| Enterovirus lesimide | Enterovirus L |
| Site of Infection | Clinical Manifestation | Symptoms | Virus | References |
|---|---|---|---|---|
| Upper respiratory tract | Rhinitis | Sneezing, runny nose, nasal congestion | Rhinovirus, coronavirus, adenovirus | [2,11,30,65] |
| Sinusitis | Facial pain, nasal congestion, thick nasal discharge | Rhinovirus, parainfluenza viruses, adenovirus | [11,49,65] | |
| Pharyngitis | Sore throat, difficulty in swallowing, fever | Adenovirus, coronavirus, rhinovirus | [11,30,65] | |
| Laryngitis | Hoarseness, dry cough, pain when speaking | Parainfluenza virus, Human metapneumovirus | [11,36,65,66] | |
| Tonsillitis | Inflammation of the tonsils, sore throat, fever | Adenovirus, coronavirus, rhinovirus | [11,30,43] | |
| Lower respiratory tract | Tracheitis | Persistent dry cough, retrosternal pain | Influenza virus, parainfluenza virus | [11,29,30,36] |
| Bronchitis | Cough with or without mucus, wheezing, chest pain | Influenza virus, respiratory syncytial virus (RSV), human metapneumovirus | [11,29,50,58,67] | |
| Bronchiolitis | Cough, difficulty breathing, wheezing (common in infants), low-grade fever, rib retractions | RSV, human metapneumovirus, parainfluenza, adenovirus, human bocavirus, coronavirus, rhinovirus | [29,37,39,41,42,50,58] | |
| Pneumonia | High fever, cough with sputum, chest pain, shortness of breath | Influenza virus, RSV, adenovirus, human metapneumovirus, human bocavirus, coronavirus (SARS-CoV, MERS-CoV, SARS-CoV-2), parainfluenza, rhinovirus, enterovirus | [15,29,37,45,46,47,50,60,68,69,70] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
dos Santos, M.M.; Salles, T.S.; de Campos, G.M.; Brcko, I.C.; Lima, A.R.J.; Sampaio, S.C.; Elias, M.C.; Giovanetti, M.; Slavov, S.N. Acute Respiratory Infections (ARIs): Current Etiological Perspectives and Advances in Viral Metagenomics—A Review. Viruses 2025, 17, 1554. https://doi.org/10.3390/v17121554
dos Santos MM, Salles TS, de Campos GM, Brcko IC, Lima ARJ, Sampaio SC, Elias MC, Giovanetti M, Slavov SN. Acute Respiratory Infections (ARIs): Current Etiological Perspectives and Advances in Viral Metagenomics—A Review. Viruses. 2025; 17(12):1554. https://doi.org/10.3390/v17121554
Chicago/Turabian Styledos Santos, Murilo Marconi, Tiago Souza Salles, Gabriel Montenegro de Campos, Isabela Carvalho Brcko, Alex Ranieri Jerônimo Lima, Sandra Coccuzzo Sampaio, Maria Carolina Elias, Marta Giovanetti, and Svetoslav Nanev Slavov. 2025. "Acute Respiratory Infections (ARIs): Current Etiological Perspectives and Advances in Viral Metagenomics—A Review" Viruses 17, no. 12: 1554. https://doi.org/10.3390/v17121554
APA Styledos Santos, M. M., Salles, T. S., de Campos, G. M., Brcko, I. C., Lima, A. R. J., Sampaio, S. C., Elias, M. C., Giovanetti, M., & Slavov, S. N. (2025). Acute Respiratory Infections (ARIs): Current Etiological Perspectives and Advances in Viral Metagenomics—A Review. Viruses, 17(12), 1554. https://doi.org/10.3390/v17121554







