New Circulating Variants of SARS-CoV-2 in Asturias During the Period (2022–2024)
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. WGS
2.3. Classification/Characterisation
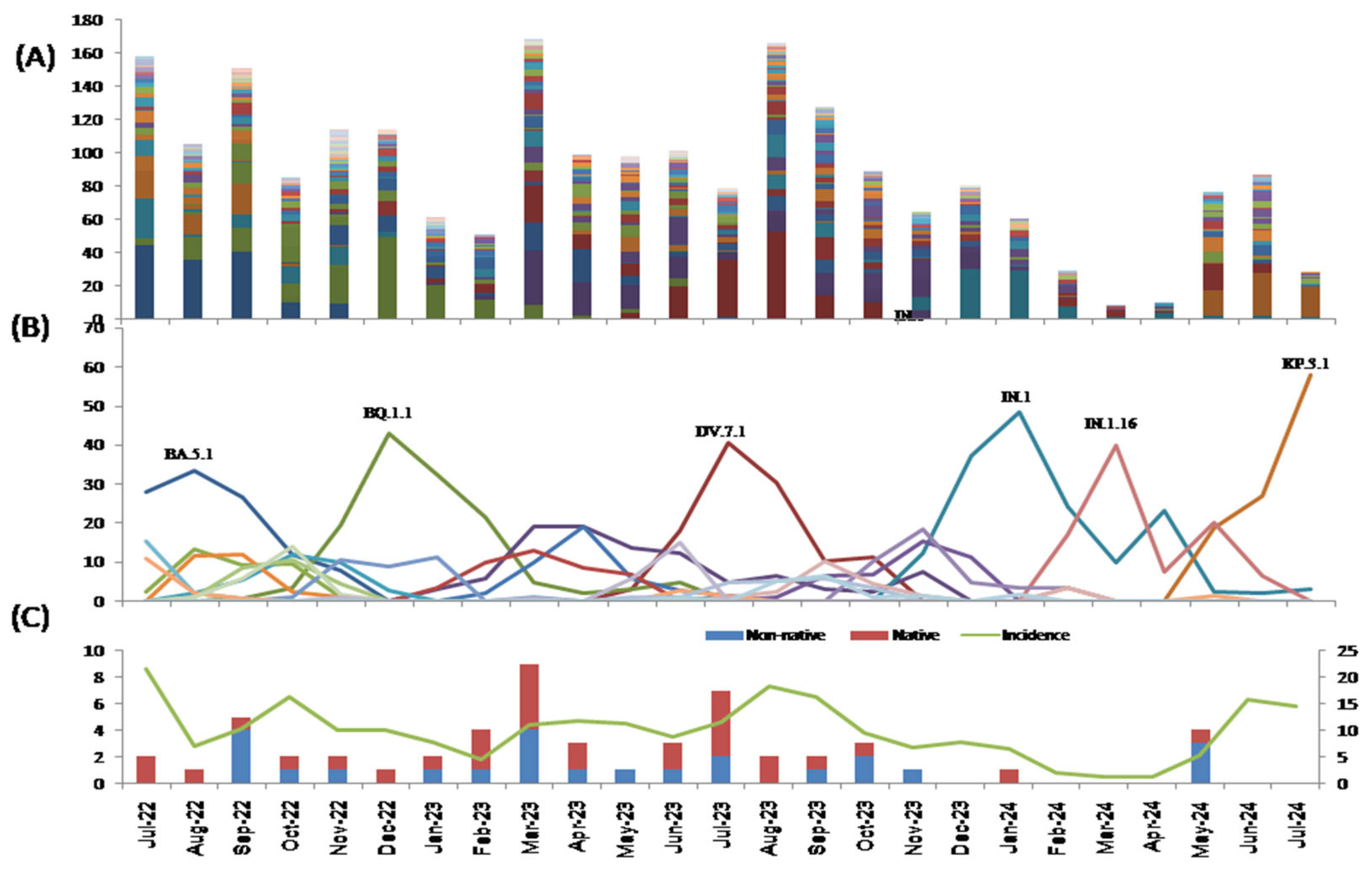
3. Results
3.1. Mutations
3.2. Lineages
3.3. Variants of Interest
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhou, C.-M.; Qin, X.-R.; Yan, L.-N.; Jiang, Y.; Yu, X.-J. Global trends in COVID-19. Infect. Med. 2022, 1, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Tian, D.; Sun, Y.; Xu, H.; Ye, Q. The emergence and epidemic characteristics of the highly mutated SARS-CoV-2 Omicron variant. J. Med. Virol. 2022, 94, 2376–2383. [Google Scholar] [CrossRef] [PubMed]
- Lino, A.; Cardoso, M.A.; Martins-Lopes, P.; Gonçalves, H.M.R. Omicron—The new SARS-CoV-2 challenge? Rev. Med. Virol. 2022, 32, e2358. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Alba, J.M.; Rojo-Alba, S.; Perez-Martinez, Z.; Boga, J.A.; Alvarez-Arguelles, M.E.; Gomez, J.; Herrero, P.; Costales, I.; Alba, L.M.; Martin-Rodriguez, G.; et al. Monitoring and tracking the spread of SARS-CoV-2 in Asturias, Spain. Access Microbiol. 2023, 5, 000573.v4. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Alba, J.M.G.; Pérez-Martínez, Z.; Boga, J.A.; Rojo-Alba, S.; de Oña, J.G.; Alvarez-Argüelles, M.E.; Rodríguez, G.M.; Gonzalez, I.C.; González, I.H.; Coto, E.; et al. Emergence of New SARS-CoV2 Omicron Variants after the Change of Surveillance and Control Strategy. Microorganisms 2022, 10, 1954. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ministerio de Sanidad. Salud Publica. Available online: https://www.sanidad.gob.es/areas/alertasEmergenciasSanitarias/alertasActuales/nCov/variantesSARS-COV-2/home.htm (accessed on 1 July 2024).
- Peck, K.M.; Laurin, A.S. Complexities of viral mutation rates. J. Virol. 2018, 92, 1031–1037. [Google Scholar] [CrossRef]
- Ministerio de Sanidad. Salud Publica. Available online: https://www.sanidad.gob.es/profesionales/saludPublica/ccayes/alertasActual/nCov/documentos/Nueva_estrategia_vigilancia_y_control.pdf (accessed on 17 June 2022).
- Colson, P.; Bader, W.; Fantini, J.; Dudouet, P.; Levasseur, A.; Pontarotti, P.; Devaux, C.; Raoult, D. From viral democratic genomes to viral wild bunch of quasispecies. J. Med. Virol. 2023, 95, e29209. [Google Scholar] [CrossRef]
- Messali, S.; Rondina, A.; Giovanetti, M.; Bonfanti, C.; Ciccozzi, M.; Caruso, A.; Caccuri, F. Traceability of SARS-CoV-2 transmission through quasispecies analysis. J. Med. Virol. 2023, 95, e28848. [Google Scholar] [CrossRef]
- Liu, Y. Is SARS-CoV-2 facing constraints in its adaptive evolution? Biomol. Biomed. 2025, 25, 2407–2415. [Google Scholar] [CrossRef] [PubMed]
- Bloom, J.D.; Neher, R.A. Fitness effects of mutations to SARS-CoV-2 proteins. Virus Evol. 2023, 9, vead055, Erratum in Virus Evol. 2024, 10, veae026. https://doi.org/10.1093/ve/veae026. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Maiti, A.K. Progressive Evolutionary Dynamics of Gene-Specific? Led to the Emergence of Novel SARS-CoV-2 Strains Having Super-Infectivity and Virulence with Vaccine Neutralization. Int. J. Mol. Sci. 2024, 25, 6306. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Emam, M.; Oweda, M.; Antunes, A.; El-Hadidi, M. Positive selection as a key player for SARS-CoV-2 pathogenicity: Insights into ORF1ab, S and E genes. Virus Res. 2021, 302, 198472. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cocherie, T.; Zafilaza, K.; Leducq, V.; Marot, S.; Calvez, V.; Marcelin, A.-G.; Todesco, E. Epidemiology and Characteristics of SARS-CoV-2 Variants of Concern: The Impacts of the Spike Mutations. Microorganisms 2022, 11, 30. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cao, Y.; Wang, J.; Jian, F.; Xiao, T.; Song, W.; Yisimayi, A.; Huang, W.; Li, Q.; Wang, P.; An, R.; et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 2022, 602, 657–663. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yang, S.; Yu, Y.; Jian, F.; Song, W.; Yisimayi, A.; Chen, X.; Xu, Y.; Wang, P.; Wang, J.; Yu, L.; et al. Antigenicity and infectivity characterisation of SARS-CoV-2 BA.2.86. Lancet Infect. Dis. 2023, 23, e457–e459. [Google Scholar] [CrossRef] [PubMed]
- Barrett, C.T.; Neal, H.E.; Edmonds, K.; Moncman, C.L.; Thompson, R.; Branttie, J.M.; Boggs, K.B.; Wu, C.Y.; Leung, D.W.; Dutch, R.E. Effect of clinical isolate or cleavage site mutations in the SARS-CoV-2 spike protein on protein stability, cleavage, and cell-cell fusion. J. Biol. Chem. 2021, 297, 100902. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zandi, M.; Shafaati, M.; Kalantar-Neyestanaki, D.; Pourghadamyari, H.; Fani, M.; Soltani, S.; Kaleji, H.; Abbasi, S. The role of SARS-CoV-2 accessory proteins in immune evasion. Biomed. Pharmacother. 2022, 156, 113889. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Li, X.; Hou, P.; Ma, W.; Wang, X.; Wang, H.; Yu, Z.; Chang, H.; Wang, T.; Jin, S.; Wang, X.; et al. SARS-CoV-2 ORF10 suppresses the antiviral innate immune response by degrading MAVS through mitophagy. Cell. Mol. Immunol. 2022, 19, 67–78, Erratum in Cell. Mol. Immunol. 2023, 20, 686. https://doi.org/10.1038/s41423-023-01023-y. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wong, L.R.; Perlman, S. Immune dysregulation and immunopathology induced by SARS-CoV-2 and related coronaviruses—Are we our own worst enemy? Nat. Rev. Immunol. 2022, 22, 47–56. [Google Scholar] [CrossRef]
- Cornillez-Ty, C.T.; Liao, L.; Yates, J.R., 3rd; Kuhn, P.; Buchmeier, M.J. Severe acute respiratory syndrome coronavirus nonstructural protein 2 interacts with a host protein complex involved in mitochondrial biogenesis and intracellular signaling. J Virol. 2009, 83, 10314–10318. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lei, J.; Kusov, Y.; Hilgenfeld, R. Nsp3 of coronaviruses: Structures and functions of a large multi-domain protein. Antivir. Res. 2018, 149, 58–74. [Google Scholar] [CrossRef] [PubMed]
- Moustaqil, M.; Ollivier, E.; Chiu, H.P.; Van Tol, S.; Rudolffi-Soto, P.; Stevens, C.; Bhumkar, A.; Hunter, D.J.B.; Freiberg, A.N.; Jacques, D.; et al. SARS-CoV-2 proteases PLpro and 3CLpro cleave IRF3 and critical modulators of inflammatory pathways (NLRP12 and TAB1): Implications for disease presentation across pecies. Emerg. Microbes. Infect. 2021, 10, 178–195. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gohari, K.; Kazemnejad, A.; Sheidaei, A.; Hajari, S. Clustering of countries according to the COVID-19incidence and mortality rates. BMC Public Health 2022, 22, 632. [Google Scholar] [CrossRef] [PubMed]
- Darques, R.; Trottier, J.; Gaudin, R.; Ait-Mouheb, N. Clustering and mapping the first COVID-19 outbreak in France. BMC Public Health 2022, 22, 1279. [Google Scholar] [CrossRef]
- Arora, P.; Mrig, S.; Goldust, Y.; Kroumpouzos, G.; Karadağ, A.S.; Rudnicka, L.; Galadari, H.; Szepietowski, J.C.; Di Lernia, V.; Goren, A.; et al. New Coronavirus (SARS-CoV-2) Crossing Borders Beyond Cities, Nations, and Continents: Impact of International Travel. Balk. Med. J. 2021, 38, 205–211. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Xiang, L.; Ma, S.; Yu, L.; Wang, W.; Yin, Z. Modeling the global dynamic contagion of COVID-19. Front. Public Health 2022, 9, 809987. [Google Scholar] [CrossRef]
- Findlater, A.; Bogoch, I.I. Human mobility and the global spread of infectious diseases: A focuson air travel. Trends Parasitol. 2018, 34, 772–783. [Google Scholar] [CrossRef]
- World Health Organization. Available online: www.who.int/activities/tracking-SARS-CoV-2-variants (accessed on 15 March 2023).
- Parsons, R.J.; Acharya, P. Evolution of the SARS-CoV-2 Omicron spike. Cell Rep. 2023, 42, 113444. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Syed, A.M.; Ciling, A.; Khalid, M.M.; Sreekumar, B.; Chen, P.Y.; Kumar, G.R.; Silva, I.; Milbes, B.; Kojima, N.; Hess, V.; et al. Omicron mutations enhance infectivity and reduce antibody neutralization of SARS-CoV-2 virus-like particles. medRxiv 2022. medRxiv:2021.12.20.21268048 Update in Proc. Natl. Acad. Sci. USA 2022, 119, e2200592119. https://doi.org/10.1073/pnas.2200592119. [Google Scholar] [CrossRef] [PubMed]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827.e19. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hartley, P.D.; Tillett, R.L.; AuCoin, D.P.; Sevinsky, J.R.; Xu, Y.; Gorzalski, A.; Pandori, M.; Buttery, E.; Hansen, H.; Picker, M.A.; et al. Genomic surveillance of Nevada patients revealed prevalence of unique SARS-CoV-2 variants bearing mutations in the RdRp gene. J. Genet. Genomics 2021, 48, 40–51. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, P.; Casner, R.G.; Nair, M.S.; Wang, M.; Yu, J.; Cerutti, G.; Liu, L.; Kwong, P.D.; Huang, Y.; Shapiro, L.; et al. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. Cell Host Microbe 2021, 29, 747–751.e4. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Minami, S.; Kotaki, T.; Sakai, Y.; Okamura, S.; Torii, S.; Ono, C.; Motooka, D.; Hamajima, R.; Nouda, R.; Nurdin, J.A.; et al. Vero cell-adapted SARS-CoV-2 strain shows increased viral growth through furin-mediated efficient spike cleavage. Microbiol.Spectr. 2024, 12, e0285923. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Qu, P.; Evans, J.P.; Kurhade, C.; Zeng, C.; Zheng, Y.M.; Xu, K.; Shi, P.Y.; Xie, X.; Liu, S.L. Determinants and Mechanisms of the Low Fusogenicity and High Dependence on Endosomal Entry of Omicron Subvariants. mBio 2023, 14, e0317622. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Subramoney, K.; Mtileni, N.; Giandhari, J.; Naidoo, Y.; Ramphal, Y.; Pillay, S.; Ramphal, U.; Maharaj, A.; Tshiabuila, D.; Tegally, H.; et al. Molecular Epidemiology of SARS-CoV-2 during Five COVID-19 Waves and the Significance of Low-Frequency Lineages. Viruses 2023, 15, 1194, Erratum in Viruses 2023, 15, 1502. https://doi.org/10.3390/v15071502. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wu, H.; Xing, N.; Meng, K.; Fu, B.; Xue, W.; Dong, P.; Tang, W.; Xiao, Y.; Liu, G.; Luo, H.; et al. Nucleocapsid mutations R203K/G204R increase the infectivity, fitness, and virulence of SARS-CoV-2. Cell Host Microbe 2021, 29, 1788–1801.e6. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Alsuwairi, F.A.; Alsaleh, A.N.; Alsanea, M.S.; Al-Qahtani, A.A.; Obeid, D.; Almaghrabi, R.S.; Alahideb, B.M.; AlAbdulkareem, M.A.; Mutabagani, M.S.; Althawadi, S.I.; et al. Association of SARS-CoV-2 Nucleocapsid Protein Mutations with Patient Demographic and Clinical Characteristics during the Delta and Omicron Waves. Microorganisms 2023, 11, 1288. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nguyen, A.; Zhao, H.; Myagmarsuren, D.; Srinivasan, S.; Wu, D.; Chen, J.; Piszczek, G.; Schuck, P. Modulation of biophysical properties of nucleocapsid protein in the mutant spectrum of SARS-CoV-2. Elife 2024, 13, RP94836. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yang, W.T.; Huang, W.H.; Liao, T.L.; Hsiao, T.H.; Chuang, H.N.; Liu, P.Y. SARS-CoV-2 E484K Mutation Narrative Review: Epidemiology, Immune Escape, Clinical Implications, and Future Considerations. Infect. Drug Resist. 2022, 15, 373–385. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lin, X.; Sha, Z.; Trimpert, J.; Kunec, D.; Jiang, C.; Xiong, Y.; Xu, B.; Zhu, Z.; Xue, W.; Wu, H. The NSP4 T492I mutation increases SARS-CoV-2 infectivity by altering non-structural protein cleavage. Cell Host Microbe 2023, 31, 1170–1184.e7. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Nie, Y.; Penny, M. Transmission dynamics of the COVID-19 outbreak and effectiveness of government interventions: A data-driven analysis. J. Med. Virol. 2020, 92, 645–659. [Google Scholar] [CrossRef]
- Brand, S.P.C.; Ojal, J.; Aziza, R.; Were, V.; Okiro, E.A.; Kombe, I.K.; Mburu, C.; Ogero, M.; Agweyu, A.; Warimwe, G.M.; et al. COVID-19 transmission dynamics underlying epidemic waves in Kenya. Science 2021, 374, 989–994. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
 ) and lost (
) and lost ( ) in the population in each gene during COVID-19 (March 2020 to June 2022) and the post-COVID-19 period (July 2022 to July 2024).
) in the population in each gene during COVID-19 (March 2020 to June 2022) and the post-COVID-19 period (July 2022 to July 2024).
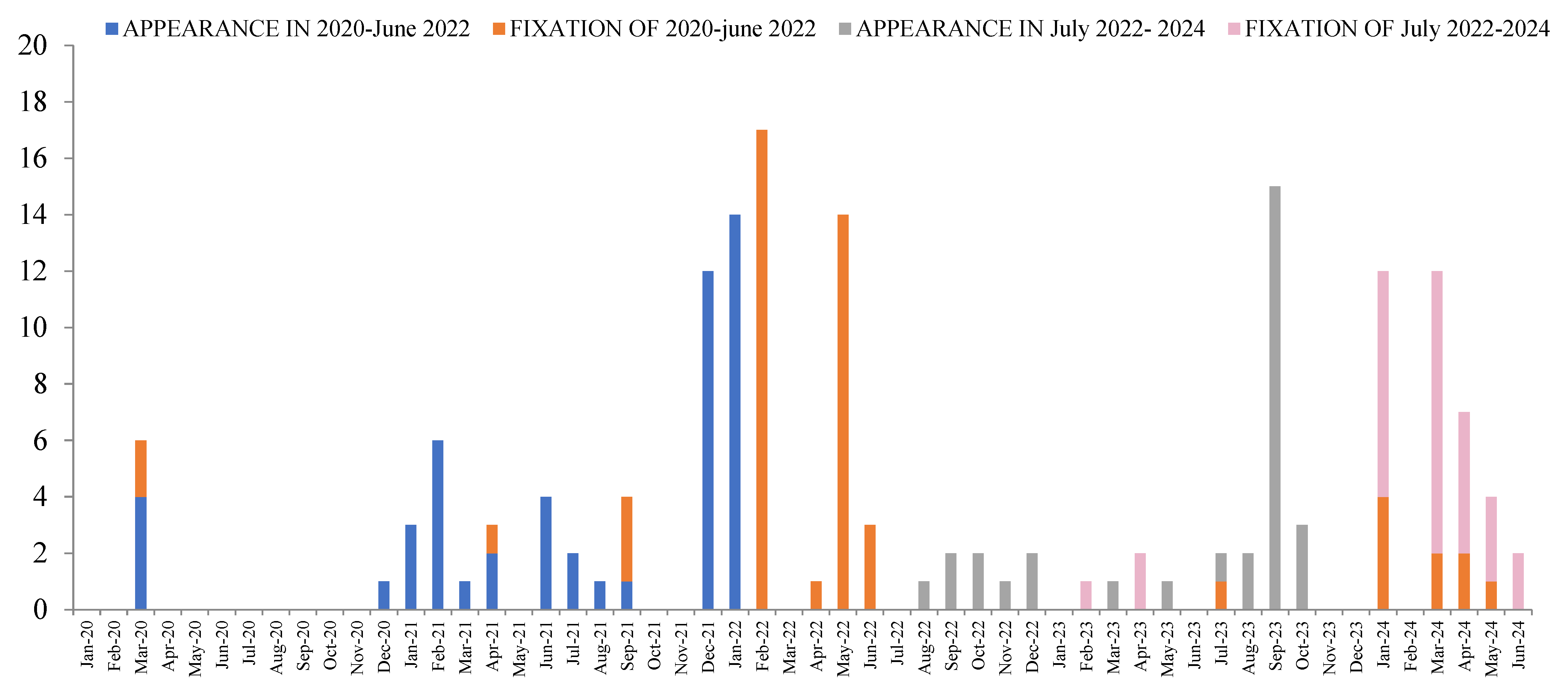
 ) and lost (
) and lost ( ) in the population in each gene during COVID-19 (March 2020 to June 2022) and the post-COVID-19 period (July 2022 to July 2024).
) in the population in each gene during COVID-19 (March 2020 to June 2022) and the post-COVID-19 period (July 2022 to July 2024).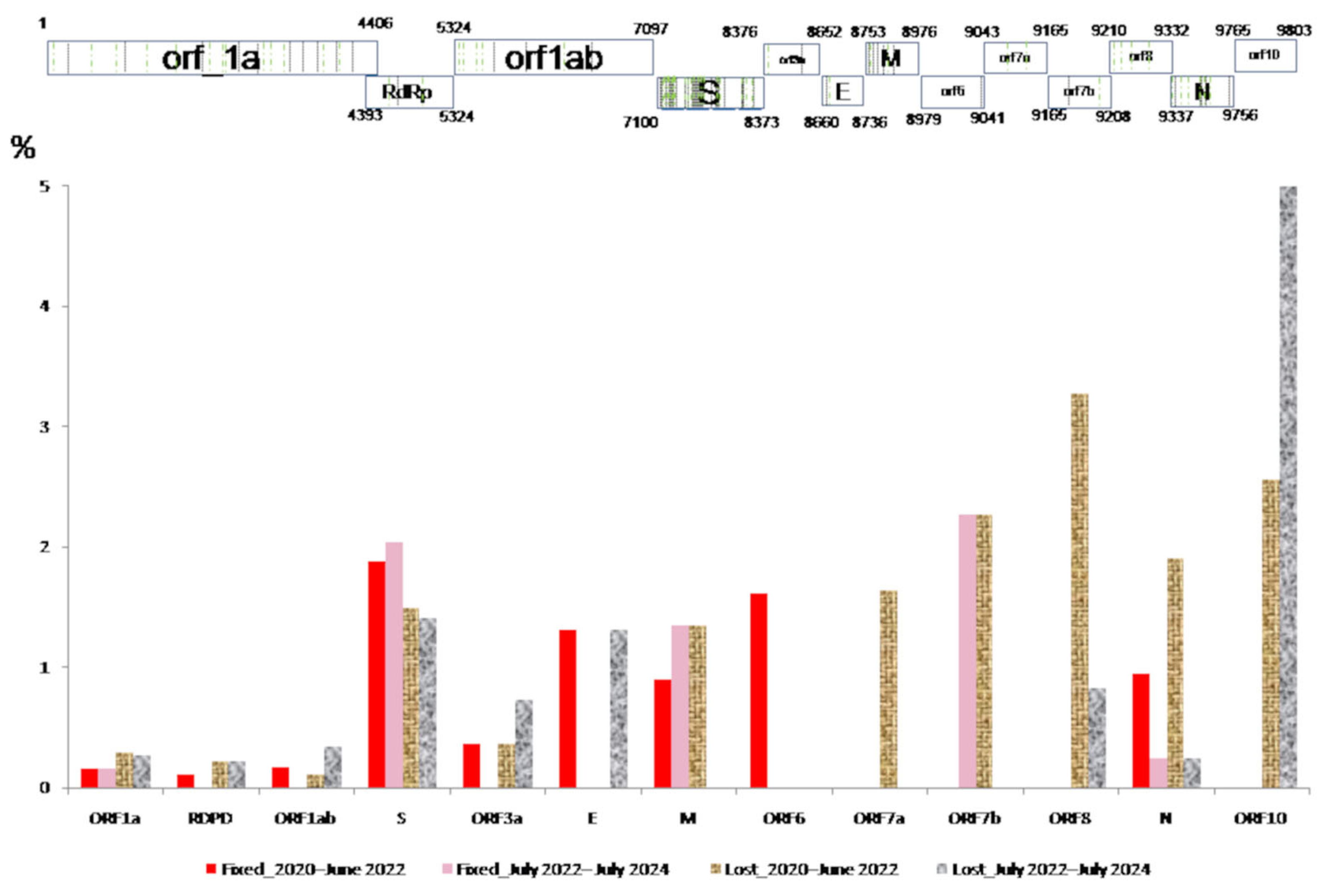

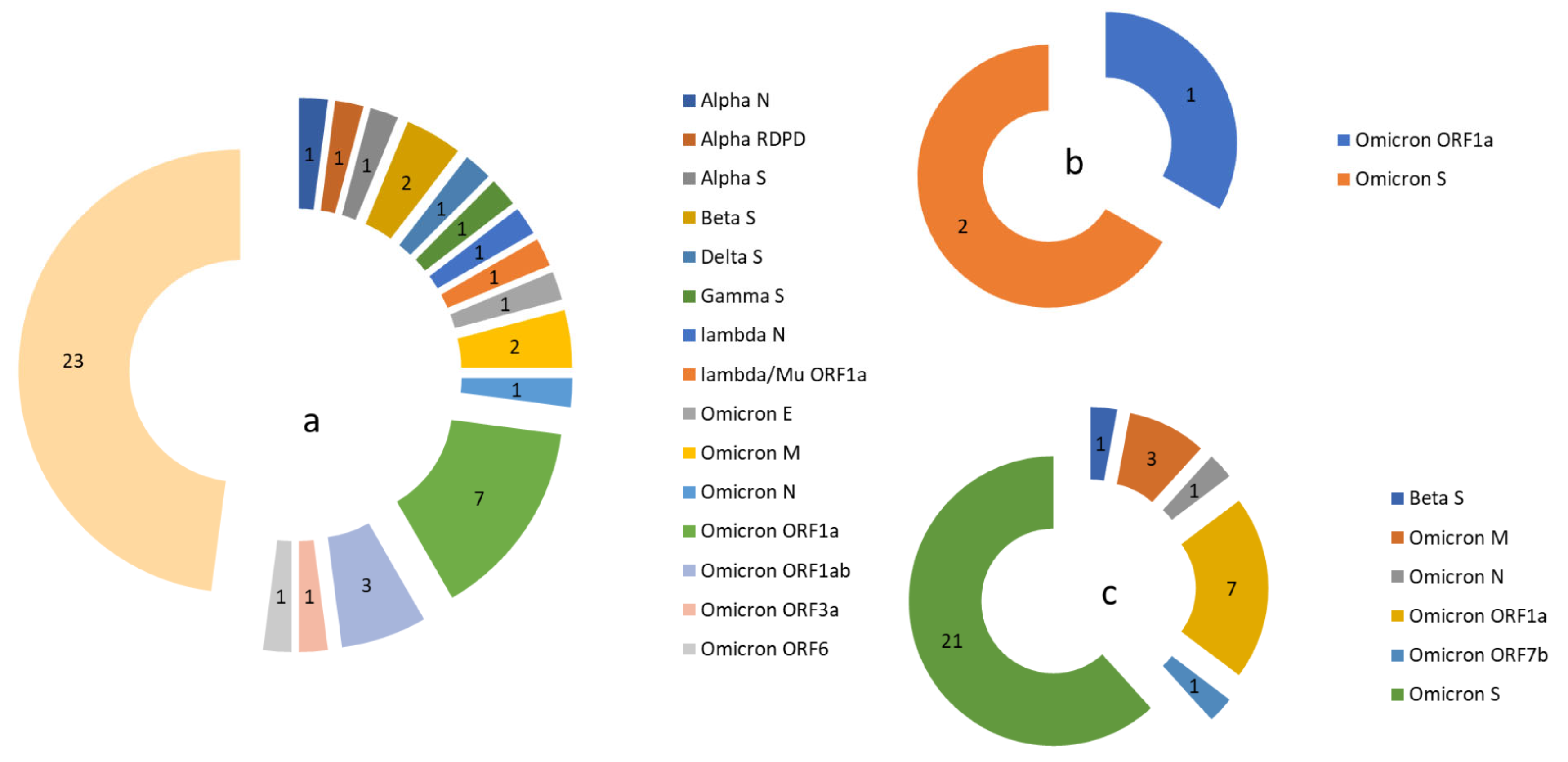
| Pre-Omicron Period | Omicron Period | |||
|---|---|---|---|---|
| Age (Years) | Sampling | Positivity (%) | Sampling | Positivity (%) |
| <1 | 3481 | 2.7 | 4136 | 12 |
| 1–5 | 19,605 | 2.9 | 10,494 | 2.7 |
| 6–20 | 61,017 | 6.5 | 12,499 | 4.5 |
| 21–65 | 253,653 | 5.5 | 36,489 | 19.9 |
| >65 | 100,920 | 4.8 | 31,475 | 16.1 |
| ORF1a | RdRp | ORF1ab | S | ORF3a | E | M | ORF6 | ORF7a | ORF7b | ORF8 | N | ORF10 | Total | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GISAID | 21 | 2 | 6 | 59 | 2 | 2 | 5 | 1 | 2 | 1 | 4 | 11 | 116 | |
| SPAIN | 29 | 4 | 7 | 82 | 2 | 2 | 8 | 1 | 2 | 2 | 4 | 14 | 1 | 158 |
| ASTURIAS | 31 | 3 | 8 | 82 | 2 | 2 | 7 | 1 | 2 | 2 | 4 | 12 | 1 | 157 |
| Mutations | ORF1a | RdRp | ORF1ab | S | ORF3a | E | M | ORF6 | ORF7a | ORF7b | ORF8 | N | ORF10 | Total | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2020–2022 | Occurred | 21 | 3 | 5 | 47 | 2 | 1 | 5 | 1 | 2 | 1 | 4 | 12 | 1 | 105 |
| Fixed | 8 | 1 | 3 | 29 | 1 | 1 | 2 | 1 | 5 | 51 | |||||
| Days or Mean± CI | 62/124(x4)/217/775,930 | 0 | 62 124 124 | 358 ± 132 (0–1209) | 496 | 372 | 62 62 | 868 | 124/310 403/713,961 | 344 ± 94 (0–1209) | |||||
| 2022–2024 | Ocsurred | 20 | 3 | 6 | 47 | 2 | 1 | 3 | 1 | 1 | 2 | 2 | 88 | ||
| Fixed | 6 | 21 | 3 | 1 | 31 | ||||||||||
| Days or Mean± CI | 93/124(x2) 217(x3) | 257 ± 53 (124–527) | 248 278 620 | 217 | 250 ± 46 (93–620) | ||||||||||
| ORF_1a | RdRp | ORF_1ab | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NSP1 | NSP2 | NSP3 | NSP4 | NSP5 | NSP6 | NSP8 | NSP9 | NSP10 | NSP12 | NSP13 | NSP14 | NSP15 | NSP16 | S | ORF3a | E | ORF7a | ORF8 | N | ORF10 |
| G94C | A357S | D112N | A128V | K90R | F235L | P10S | G38V | T115I | A4774V | V157L | A320T | D36G | K160R | A67V | D155Y | T9V | L5F | D119Y | P151S | I27T |
| P6S | E574D | D1764G | V13I | V35L | L37F | T21I | I4563M | I15T | P205S | P215L | A701V | D27H | Q94L | P38S | ||||||
| V121A | G265S | E119K | M5021V | N71S | A845S | L140F | T28I | |||||||||||||
| G285S | E387D | S4621N | P203L | D253G | M260K | |||||||||||||||
| L24F | I541V | Q22H | D80E | Q185H | ||||||||||||||||
| N254S | M988L | V328F | F456L | Q57H | ||||||||||||||||
| S591I | N1322S | F486V | S171L | |||||||||||||||||
| T103I | N1680K | F59S | T270I | |||||||||||||||||
| P153L | G252V | V273L | ||||||||||||||||||
| Q167R | L249F | |||||||||||||||||||
| R1297G | N354K | |||||||||||||||||||
| S126L | P1263Q | |||||||||||||||||||
| S1428L | Q675H | |||||||||||||||||||
| S454G | T547I | |||||||||||||||||||
| T1203I | T572I | |||||||||||||||||||
| T424N | V1264L | |||||||||||||||||||
| T720I | V445P | |||||||||||||||||||
| T970M | ||||||||||||||||||||
| V1385I | ||||||||||||||||||||
| V1673I | ||||||||||||||||||||
| Y1535H | ||||||||||||||||||||
| NCV | N (%Total Identified) | First Detection Date | Last Detection Date | Time of Detection (Days) |
|---|---|---|---|---|
| Frst detected in the world | ||||
| BA.5.2.1 + NSP3_S454G | 6 (43) | 17/09/2022 | 22/10/2022 | 35 |
| BA.5.2.6 + NSP3_M988L | 7 (3) | 22/09/2022 | 06/11/2022 | 45 |
| BF.5 + S_D80E + S_A701V + NSP1_G94C | 8 (62) | 22/09/2022 | 08/11/2022 | 47 |
| BA.5.1 + NSP12_M5021V | 5 (7) | 24/09/2022 | 16/11/2022 | 53 |
| BF.7 + NSP14_P203L | 13 (29) | 19/10/2022 | 07/12/2022 | 49 |
| BQ.1.1.18 + S_T547I | 5 (28) | 16/11/2022 | 11/01/2023 | 56 |
| XBB.1.5 + NSP3_T1203I | 12 (6) | 24/01/2023 | 01/05/2023 | 97 |
| FL.5 + NSP14_Q22H + NSP3_P153L | 6 (2) | 15/02/2023 | 24/05/2023 | 98 |
| XBB.1.5.8 + NSP14_V328F | 10 (10) | 01/03/2023 | 29/03/2023 | 28 |
| BQ.1.18 + NSP14_I15T | 20 (17) | 04/03/2023 | 29/06/2023 | 117 |
| XBB.1.5 + NSP2_S591I + NSP3_N1322S | 12 (1) | 04/03/2023 | 20/08/2023 | 169 |
| XBB.1.5 + NSP5_V35L | 6 (1) | 09/03/2023 | 07/07/2023 | 120 |
| XBB.1.5.71 + NSP1_P6S + ORF3a_Q57H + NSP12_I4563M | 5 (4) | 28/04/2023 | 13/06/2023 | 46 |
| XBB.1.5.71 + NSP1_P6S + NSP12_I4563M | 6 (2) | 12/05/2023 | 03/06/2023 | 22 |
| EG.5.1 + ORF3a_D27H | 5 (1) | 20/06/2023 | 24/08/2023 | 65 |
| XBB.1.16.11 + NSP2_N254S | 6 (1) | 20/07/2023 | 16/10/2023 | 88 |
| DV.7.1 + NSP3_E119K | 6 (18) | 26/07/2023 | 08/09/2023 | 44 |
| EG.5.1.3 + NSP3_I541V | 5 (42) | 05/09/2023 | 14/09/2023 | 9 |
| JD.1.1 + ORF3a_M260K | 5 (2) | 13/10/2023 | 28/11/2023 | 46 |
| JG.3 + ORF3a_S171L | 7 (14) | 27/10/2023 | 04/12/2023 | 38 |
| JN.1.31 + S_T572I | 6 (23) | 29/11/2023 | 13/03/2024 | 105 |
| JN.1.16 + S_A67V + S_L249F + S_V445P | 6 (10) | 07/05/2024 | 26/05/2024 | 19 |
| JN.1.32 + S_F456L | 7 (1) | 12/05/2024 | 06/06/2024 | 25 |
| JN.1.16.1 + NSP3_S1428L + NSP5_K90R + NSP6_L37F + S_F59S | 5 (83) | 13/05/2024 | 19/05/2024 | 6 |
| First detected in Asturias | ||||
| BA.4.6 + NSP2_G265S + NSP3_D112N + ORF7a_Q94L | 7 (100) ** | 10/06/2022 | 17/10/2022 | 129 |
| CH.1.1.28 + NSP16_K160R | 14 (100) ** | 22/12/2022 | 22/03/2023 | 90 |
| XBB.1.5 + NSP3_T970M + NSP12_A4774V | 6 (100) ** | 27/01/2023 | 13/04/2023 | 76 |
| XBB.1.5.77 + S_G252V + NSP2_E574D + ORF7a_L5F + ORF7a_T28I | 6 (100) ** | 14/02/2023 | 07/04/2023 | 52 |
| XBB.1.5.1 + NSP15_D36G + NSP3_S126LL + NSP1_V121A + ORF8_P38S | 7 (100) ** | 03/04/2023 | 04/05/2023 | 31 |
| XBB.2.3 + E_T9V + NSP3_N1680K + NSP3_Y1535H + ORF10_I27T | 5 (100) ** | 16/04/2023 | 11/08/2023 | 117 |
| EG.1.4 + NSP12_S4621N + S_A845S | 6 (100) ** | 09/05/2023 | 03/08/2023 | 86 |
| DV.7.1 + NSP3_R1297G + ORF3a_Q185H | 6 (100) ** | 21/05/2023 | 31/08/2023 | 102 |
| JG.3 + NSP3_V1385I | 8 (100) ** | 04/10/2023 | 22/01/2024 | 110 |
| JN.1.16.2 + S_A67V + S_V445P + S_L249F | 8 (100) ** | 24/01/2024 | 25/05/2024 | 122 |
| BQ.1.1.15 + NSP3_T1203I | 6 (55) * | 26/07/2022 | 27/12/2022 | 154 |
| BQ.1.1.66 + NSP3_E387D + NSP9_G38V + NSP10_T115II | 10 (83) * | 23/11/2022 | 15/03/2023 | 112 |
| XBB.1.5.37 + NSP9_T21I | 7 (78) * | 15/12/2022 | 23/04/2023 | 129 |
| XBB.2.3.13 + NSP3_Q167R | 6 (60) * | 28/02/2023 | 31/07/2023 | 153 |
| XBB.2.3.13 + NSP2_A357S + NSP6_F235L + S_A701V | 13 (93) * | 12/03/2023 | 24/04/2023 | 43 |
| XBB.1.5.71 + NSP15_P205S + NSP2_L24F | 5 (83) * | 10/05/2023 | 27/09/2023 | 140 |
| BA.4.1 + NSP13_V157L | 7 (88) * | 09/06/2022 | 15/08/2022 | 67 |
| BE.1 + ORF3a_T270I + ORF3a_V273L | 6 (86) * | 10/07/2022 | 08/09/2022 | 60 |
| BF.7 + NSP4_A128V + S_Q675H + S_P1263Q | 10 (19) | 29/06/2022 | 01/12/2022 | 155 |
| CK.2.1.1 + S_V1264L + ORF8_D119Y | 5 (63) | 03/10/2022 | 21/12/2022 | 79 |
| EL.1 + NSP2_T103I + NSP3_T720I | 35 (90) | 16/11/2022 | 19/05/2023 | 184 |
| EF.1.2 + S_D253G + NSP4_V13I + ORF3a_D155Y | 10 (67) | 20/12/2022 | 16/03/2023 | 86 |
| XBB.2.3.13 + NSP2_A357S | 12 (16) | 05/02/2023 | 09/06/2023 | 124 |
| EL.1 + NSP14_A320T + NSP2_G265S + NSP3_T970M | 6 (75) | 14/02/2023 | 19/05/2023 | 94 |
| DV.7.1 + NSP14_N71S | 8 (57) | 28/04/2023 | 02/09/2023 | 127 |
| XBB.1.5 + NSP3_V1673I + ORF3a_L140F | 13 (81) | 08/06/2023 | 19/09/2023 | 103 |
| EG.6.1 + NSP3_S1428L | 11 (79) | 16/06/2023 | 02/10/2023 | 108 |
| DV.7.1 + NSP8_P10S + NSP3_D1764G | 5 (16) | 23/06/2023 | 10/09/2023 | 79 |
| EG.5.1.5 + S_N354K + N_P151S | 12 (46) | 13/07/2023 | 26/08/2023 | 44 |
| HV.1 + NSP14_P203L | 6 (29) | 05/08/2023 | 13/10/2023 | 69 |
| JN.1 + S_F486V | 6 (75) | 29/10/2023 | 25/01/2024 | 88 |
| Variant | Alpha | Beta | Gamma | Delta | Zeta | Eta | Theta | Iota | Kappa | Lambda | Mu | Epsilon | Omicron | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutation | ||||||||||||||
| E_T9I | X | X | X | X | ||||||||||
| M_A63T | X | |||||||||||||
| N_G204R | X | X | X | X | X | X | ||||||||
| N_P13L | X | X | ||||||||||||
| N_R203K | X | X | X | X | X | X | ||||||||
| ORF1a_T3255I | X | X | X | |||||||||||
| RdRp_P323L | X | X | X | X | X | X | X | X | X | X | X | X | X | |
| S_D614G | X | X | X | X | X | X | X | X | X | X | X | X | X | |
| S_E484K | X | X | X | X | X | X | ||||||||
| S_H655Y | X | X | ||||||||||||
| S_K417N | X | |||||||||||||
| S_N501Y | X | X | X | X | ||||||||||
| S_N679K | X | |||||||||||||
| S_P681R | X | X | ||||||||||||
| S_Q954H | X | |||||||||||||
| S_S373P | X | |||||||||||||
| S_T478K | X | X | ||||||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
González-Alba, J.M.; Martínez, Z.P.; Rojo-Alba, S.; Varela, C.O.; Oña, J.G.d.; Pérez, M.R.; García, S.M.; Álvarez-Argüelles, M.E. New Circulating Variants of SARS-CoV-2 in Asturias During the Period (2022–2024). Viruses 2025, 17, 1531. https://doi.org/10.3390/v17121531
González-Alba JM, Martínez ZP, Rojo-Alba S, Varela CO, Oña JGd, Pérez MR, García SM, Álvarez-Argüelles ME. New Circulating Variants of SARS-CoV-2 in Asturias During the Period (2022–2024). Viruses. 2025; 17(12):1531. https://doi.org/10.3390/v17121531
Chicago/Turabian StyleGonzález-Alba, José María, Zulema Pérez Martínez, Susana Rojo-Alba, Cristina Ochoa Varela, Juan Gómez de Oña, Mercedes Rodríguez Pérez, Santiago Melón García, and Marta Elena Álvarez-Argüelles. 2025. "New Circulating Variants of SARS-CoV-2 in Asturias During the Period (2022–2024)" Viruses 17, no. 12: 1531. https://doi.org/10.3390/v17121531
APA StyleGonzález-Alba, J. M., Martínez, Z. P., Rojo-Alba, S., Varela, C. O., Oña, J. G. d., Pérez, M. R., García, S. M., & Álvarez-Argüelles, M. E. (2025). New Circulating Variants of SARS-CoV-2 in Asturias During the Period (2022–2024). Viruses, 17(12), 1531. https://doi.org/10.3390/v17121531







