Comparative Analysis of Codon Usage Patterns and Host Adaptation in Merbecoviruses
Abstract
1. Introduction
2. Materials and Methods
2.1. Source of Target Sequences
2.2. Recombination Analysis
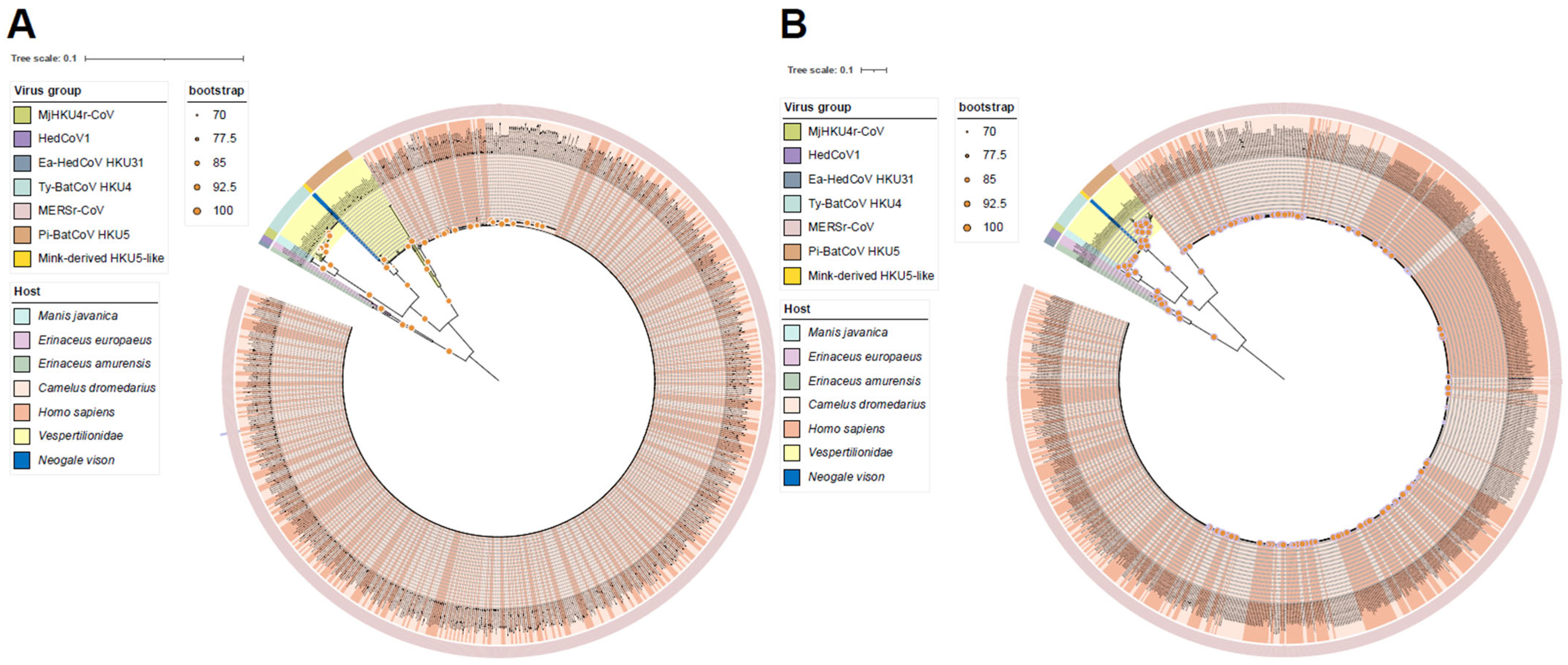
2.3. Phylogenetic Analysis
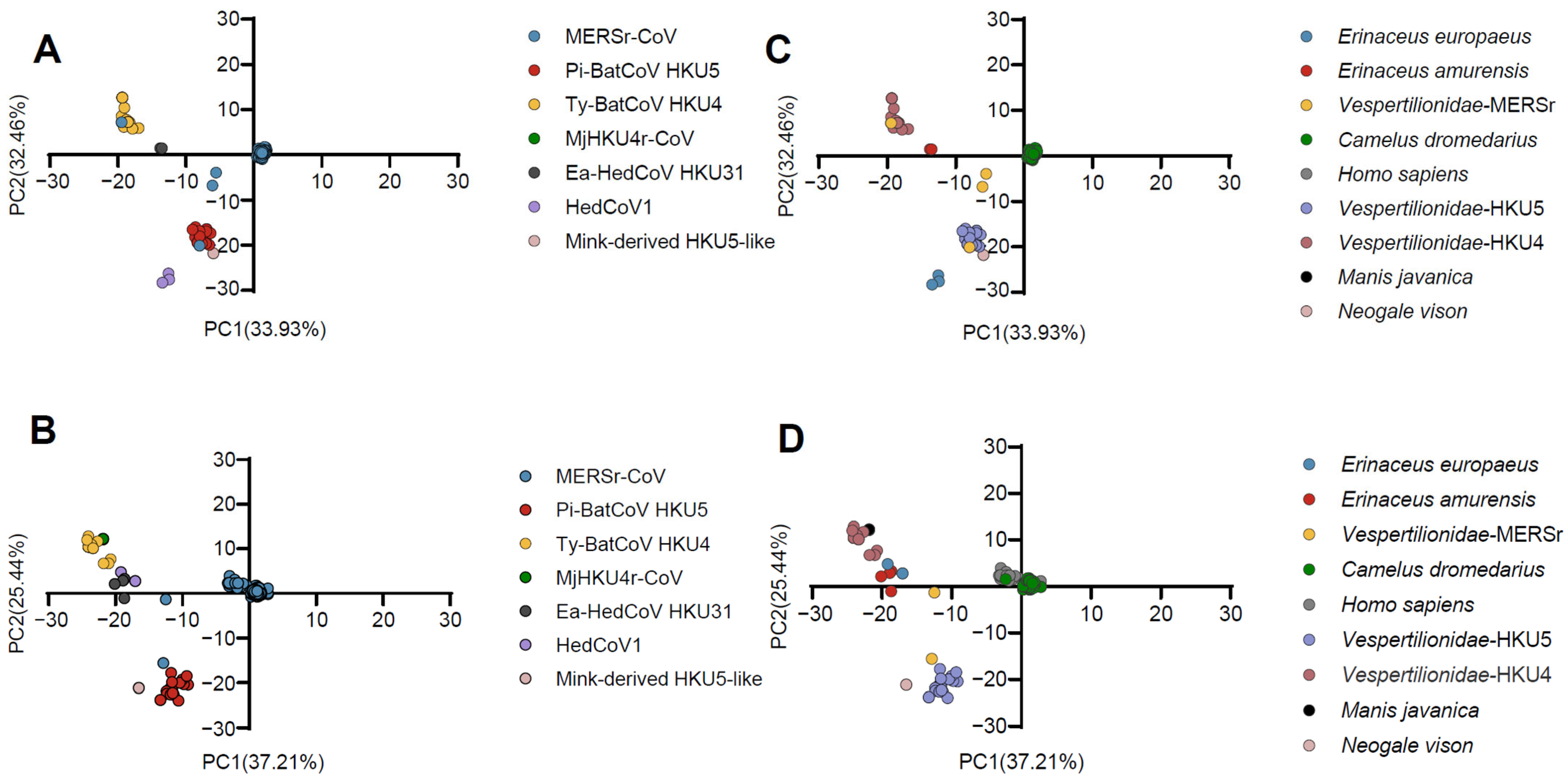
2.4. Principal Component Analysis (PCA)
2.5. Nucleotide Composition Analysis
2.6. Analysis of Relative Synonymous Codon Usage (RSCU)
2.7. Analysis of Dinucleotide Relative Abundance and Characterization
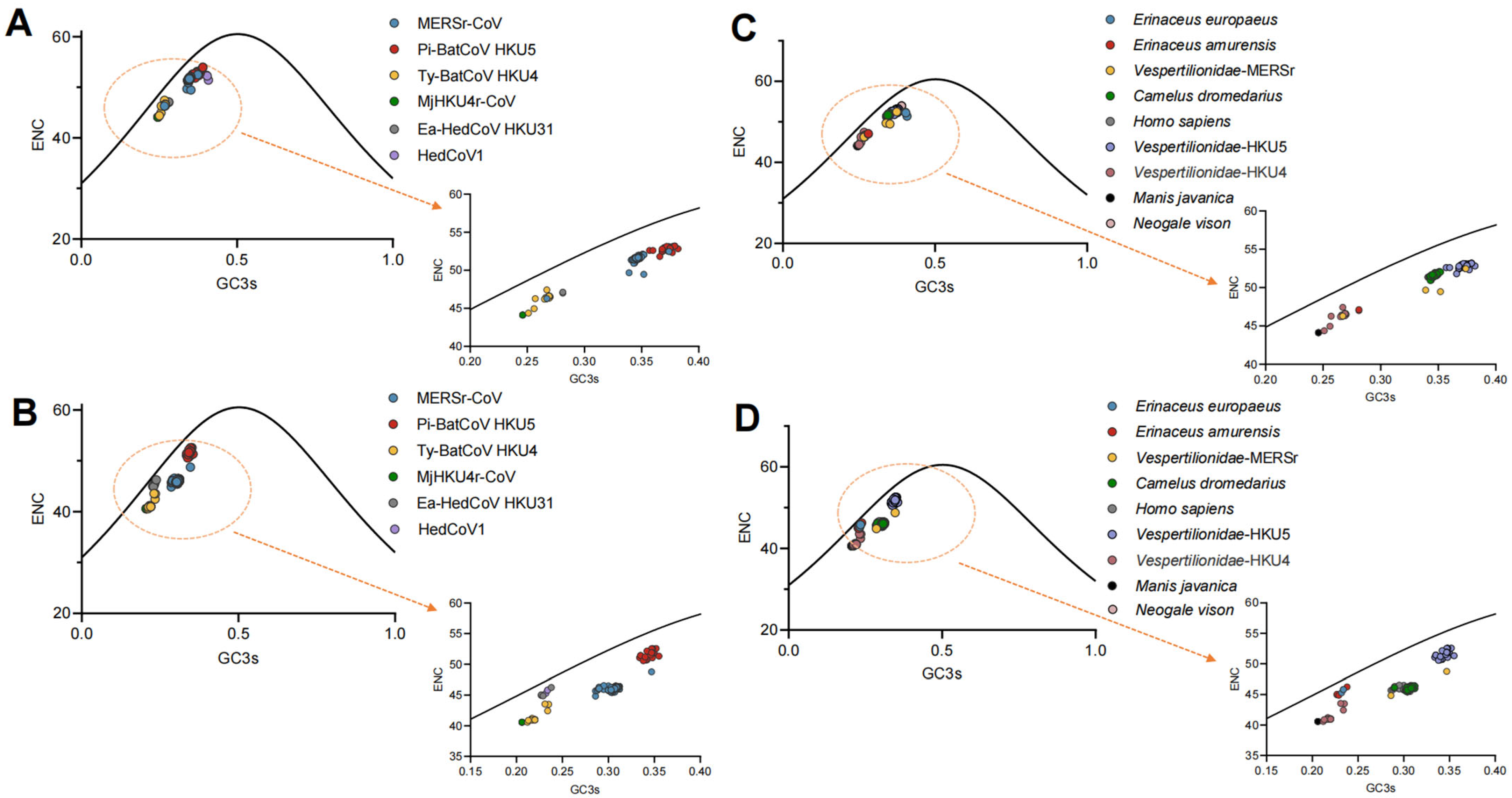
2.8. Analysis of Effective Number of Codons (ENC)
2.9. ENC-GC3s Plot Analysis
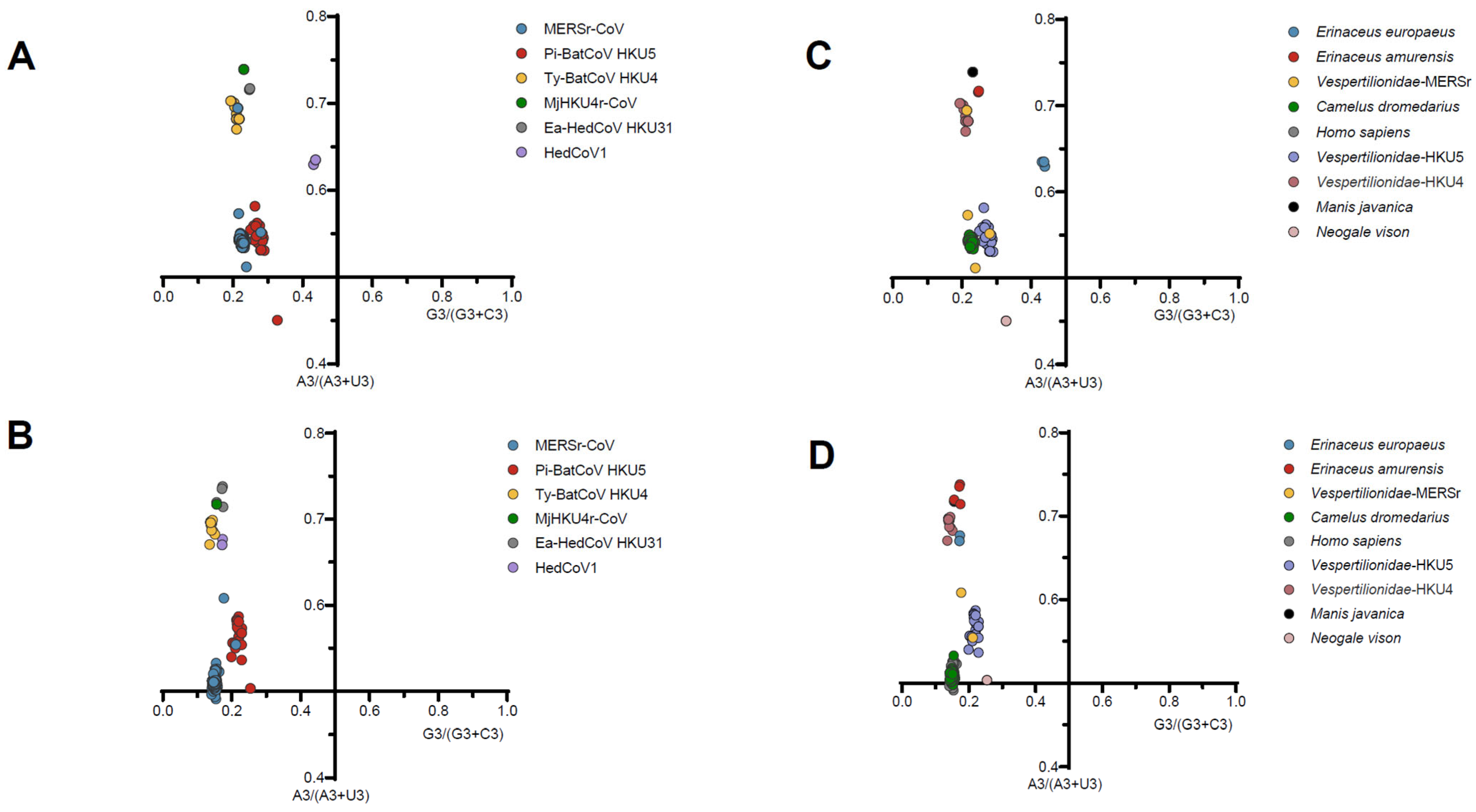
2.10. The Parity Rule 2 (PR2) Analysis
2.11. Neutrality Analysis
2.12. Correlation Analysis
2.13. Codon Adaptation Index (CAI) Analysis and Relative Codon Deoptimization Index (RCDI) Analysis
3. Results
3.1. The Phylogenetic Relationship of Merbecovirus Shows Clustering Patterns Similar to Those Seen in the PCA
3.2. Nucleotide Composition Analysis Indicated a High Abundance of AU
3.3. The RSCU of Merbecovirus Was A/U-End Biased and Opposite to the Hosts
3.4. Mutation Pressure and Natural Selection Have Both Influenced Codon Usage Patterns
3.5. Analysis of Dinucleotide Relative Abundance and Characterization
3.6. Codon Adaptation Index (CAI) and Relative Codon Deoptimization Index (RCDI) Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zaki, A.M.; Van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A.D.; Fouchier, R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization Regional Office for the Eastern Mediterranean (WHO-EMRO). MERS Situation Update, July 2025. 2025. Available online: https://applications.emro.who.int/docs/WHOEMCSR833E-eng.pdf (accessed on 1 July 2025).
- Woo, P.C.; Lau, S.K.; Li, K.S.; Poon, R.W.; Wong, B.H.; Tsoi, H.W.; Yip, B.C.; Huang, Y.; Chan, K.H.; Yuen, K.Y. Molecular diversity of coronaviruses in bats. Virology 2006, 351, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.; Wang, M.; Lau, S.K.; Xu, H.; Poon, R.W.; Guo, R.; Wong, B.H.; Gao, K.; Tsoi, H.W.; Huang, Y.; et al. Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features. J. Virol. 2007, 81, 1574–1585. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Wan, W.; Yu, K.; Lemey, P.; Pettersson, J.H.; Bi, Y.; Lu, M.; Li, X.; Chen, Z.; Zheng, M.; et al. Farmed fur animals harbour viruses with zoonotic spillover potential. Nature 2024, 634, 228–233. [Google Scholar] [CrossRef]
- Chen, J.; Yang, X.; Si, H.; Gong, Q.; Que, T.; Li, J.; Li, Y.; Wu, C.; Zhang, W.; Chen, Y.; et al. A bat MERS-like coronavirus circulates in pangolins and utilizes human DPP4 and host proteases for cell entry. Cell 2023, 186, 850–863.e16. [Google Scholar] [CrossRef]
- Lau, S.K.P.; Luk, H.K.H.; Wong, A.C.P.; Fan, R.Y.Y.; Lam, C.S.F.; Li, K.S.M.; Ahmed, S.S.; Chow, F.W.N.; Cai, J.P.; Zhu, X.; et al. Identification of a Novel Betacoronavirus (Merbecovirus) in Amur Hedgehogs from China. Viruses 2019, 11, 980. [Google Scholar] [CrossRef]
- Yao, Y.; Bao, L.; Deng, W.; Xu, L.; Li, F.; Lv, Q.; Yu, P.; Chen, T.; Xu, Y.; Zhu, H.; et al. An animal model of MERS produced by infection of rhesus macaques with MERS coronavirus. J. Infect. Dis. 2014, 209, 236–242. [Google Scholar] [CrossRef]
- Falzarano, D.; de Wit, E.; Feldmann, F.; Rasmussen, A.L.; Okumura, A.; Peng, X.; Thomas, M.J.; van Doremalen, N.; Haddock, E.; Nagy, L.; et al. Infection with MERS-CoV causes lethal pneumonia in the common marmoset. PLoS Pathog. 2014, 10, e1004250. [Google Scholar] [CrossRef]
- Kandeil, A.; Gomaa, M.; Shehata, M.; El-Taweel, A.; Kayed, A.E.; Abiadh, A.; Jrijer, J.; Moatasim, Y.; Kutkat, O.; Bagato, O.; et al. Middle East respiratory syndrome coronavirus infection in non-camelid domestic mammals. Emerg. Microbes Infect. 2019, 8, 103–108. [Google Scholar] [CrossRef]
- Meyer, B.; García-Bocanegra, I.; Wernery, U.; Wernery, R.; Sieberg, A.; Müller, M.A.; Drexler, J.F.; Drosten, C.; Eckerle, I. Serologic assessment of possibility for MERS-CoV infection in equids. Emerg. Infect. Dis. 2015, 21, 181–182. [Google Scholar] [CrossRef]
- Haagmans, B.L.; van den Brand, J.M.; Provacia, L.B.; Raj, V.S.; Stittelaar, K.J.; Getu, S.; de Waal, L.; Bestebroer, T.M.; van Amerongen, G.; Verjans, G.M.; et al. Asymptomatic Middle East respiratory syndrome coronavirus infection in rabbits. J. Virol. 2015, 89, 6131–6135. [Google Scholar] [CrossRef] [PubMed]
- Vergara-Alert, J.; van den Brand, J.M.; Widagdo, W.; Muñoz, M.t.; Raj, S.; Schipper, D.; Solanes, D.; Cordón, I.; Bensaid, A.; Haagmans, B.L.; et al. Livestock Susceptibility to Infection with Middle East Respiratory Syndrome Coronavirus. Emerg. Infect. Dis. 2017, 23, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Wang, Q.; Gao, G.F. Bat-to-human: Spike features determining ‘host jump’ of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 2015, 23, 468–478. [Google Scholar] [CrossRef] [PubMed]
- Pomorska-Mól, M.; Ruszkowski, J.J.; Gogulski, M.; Domanska-Blicharz, K. First detection of Hedgehog coronavirus 1 in Poland. Sci. Rep. 2022, 12, 2386. [Google Scholar] [CrossRef]
- He, W.T.; Hou, X.; Zhao, J.; Sun, J.; He, H.; Si, W.; Wang, J.; Jiang, Z.; Yan, Z.; Xing, G.; et al. Virome characterization of game animals in China reveals a spectrum of emerging pathogens. Cell 2022, 185, 1117–1129.e1118. [Google Scholar] [CrossRef]
- Aloor, A.; Aradhya, R.; Venugopal, P.; Gopalakrishnan Nair, B.; Suravajhala, R. Glycosylation in SARS-CoV-2 variants: A path to infection and recovery. Biochem. Pharmacol. 2022, 206, 115335. [Google Scholar] [CrossRef]
- Spencer, P.S.; Barral, J.M. Genetic code redundancy and its influence on the encoded polypeptides. Comput. Struct. Biotechnol. J. 2012, 1, e201204006. [Google Scholar] [CrossRef]
- Brule, C.E.; Grayhack, E.J. Synonymous Codons: Choose Wisely for Expression. Trends Genet. 2017, 33, 283–297. [Google Scholar] [CrossRef]
- Ingvarsson, P.K. Molecular evolution of synonymous codon usage in Populus. BMC Evol. Biol. 2008, 8, 307. [Google Scholar] [CrossRef]
- Bulmer, M. The selection-mutation-drift theory of synonymous codon usage. Genetics 1991, 129, 897–907. [Google Scholar] [CrossRef]
- Hershberg, R.; Petrov, D.A. Selection on codon bias. Annu. Rev. Genet. 2008, 42, 287–299. [Google Scholar] [CrossRef]
- Domingo, E.; Perales, C. Viral quasispecies. PLoS Genet. 2019, 15, e1008271. [Google Scholar] [CrossRef]
- Mordstein, C.; Cano, L.; Morales, A.C.; Young, B.; Ho, A.T.; Rice, A.M.; Liss, M.; Hurst, L.D.; Kudla, G. Transcription, mRNA Export, and Immune Evasion Shape the Codon Usage of Viruses. Genome Biol. Evol. 2021, 13, evab106. [Google Scholar] [CrossRef]
- Morgunov, A.S.; Babu, M.M. Optimizing membrane-protein biogenesis through nonoptimal-codon usage. Nat. Struct. Mol. Biol. 2014, 21, 1023–1025. [Google Scholar] [CrossRef]
- Victor, M.P.; Acharya, D.; Begum, T.; Ghosh, T.C. The optimization of mRNA expression level by its intrinsic properties-Insights from codon usage pattern and structural stability of mRNA. Genomics 2019, 111, 1292–1297. [Google Scholar] [CrossRef] [PubMed]
- Bu, C.; Zheng, X.; Zhao, X.; Xu, T.; Bai, X.; Jia, Y.; Chen, M.; Hao, L.; Xiao, J.; Zhang, Z.; et al. GenBase: A Nucleotide Sequence Database. Genom. Proteom. Bioinform. 2024, 22, qzae047. [Google Scholar] [CrossRef]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015, 1, vev003. [Google Scholar] [CrossRef]
- Martin, D.; Rybicki, E. RDP: Detection of recombination amongst aligned sequences. Bioinformatics 2000, 16, 562–563. [Google Scholar] [CrossRef]
- Padidam, M.; Sawyer, S.; Fauquet, C.M. Possible emergence of new geminiviruses by frequent recombination. Virology 1999, 265, 218–225. [Google Scholar] [CrossRef]
- Boni, M.F.; Posada, D.; Feldman, M.W. An exact nonparametric method for inferring mosaic structure in sequence triplets. Genetics 2007, 176, 1035–1047. [Google Scholar] [CrossRef]
- Gibbs, M.J.; Armstrong, J.S.; Gibbs, A.J. Sister-scanning: A Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics 2000, 16, 573–582. [Google Scholar] [CrossRef]
- Smith, J.M. Analyzing the mosaic structure of genes. J. Mol. Evol. 1992, 34, 126–129. [Google Scholar] [CrossRef] [PubMed]
- Holmes, E.C.; Worobey, M.; Rambaut, A. Phylogenetic evidence for recombination in dengue virus. Mol. Biol. Evol. 1999, 16, 405–409. [Google Scholar] [CrossRef] [PubMed]
- Sabir, J.S.; Lam, T.T.; Ahmed, M.M.; Li, L.; Shen, Y.; Abo-Aba, S.E.; Qureshi, M.I.; Abu-Zeid, M.; Zhang, Y.; Khiyami, M.A.; et al. Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science 2016, 351, 81–84. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- Wang, M.; Liu, Y.-S.; Zhou, J.-H.; Chen, H.-T.; Ma, L.-N.; Ding, Y.-Z.; Liu, W.-Q.; Gu, Y.-X.; Zhang, J. Analysis of codon usage in Newcastle disease virus. Virus Genes 2011, 42, 245–253. [Google Scholar] [CrossRef]
- Subramanian, K.; Payne, B.; Feyertag, F.; Alvarez-Ponce, D. The codon statistics database: A database of codon usage bias. Mol. Biol. Evol. 2022, 39, msac157. [Google Scholar] [CrossRef]
- Sharp, P.M.; Tuohy, T.M.; Mosurski, K.R. Codon usage in yeast: Cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 1986, 14, 5125–5143. [Google Scholar] [CrossRef] [PubMed]
- Wright, F. The ‘effective number of codons’ used in a gene. Gene 1990, 87, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Puigbò, P.; Aragonès, L.; Garcia-Vallvé, S. RCDI/eRCDI: A web-server to estimate codon usage deoptimization. BMC Res. Notes 2010, 3, 87. [Google Scholar] [CrossRef]
- Cotten, M.; Watson, S.J.; Kellam, P.; Al-Rabeeah, A.A.; Makhdoom, H.Q.; Assiri, A.; Al-Tawfiq, J.A.; Alhakeem, R.F.; Madani, H.; AlRabiah, F.A. Transmission and evolution of the Middle East respiratory syndrome coronavirus in Saudi Arabia: A descriptive genomic study. Lancet 2013, 382, 1993–2002. [Google Scholar] [CrossRef]
- Bellare, P.; Dufresne, A.; Ganem, D. Inefficient Codon Usage Impairs mRNA Accumulation: The Case of the v-FLIP Gene of Kaposi’s Sarcoma-Associated Herpesvirus. J. Virol. 2015, 89, 7097–7107. [Google Scholar] [CrossRef]
- Hussain, S.; Shinu, P.; Islam, M.M.; Chohan, M.S.; Rasool, S.T. Analysis of Codon Usage and Nucleotide Bias in Middle East Respiratory Syndrome Coronavirus Genes. Evol. Bioinform. 2020, 16, 1176934320918861. [Google Scholar] [CrossRef]
- Wang, W.; Zhou, L.; Ge, X.; Han, J.; Guo, X.; Zhang, Y.; Yang, H. Analysis of codon usage patterns of porcine enteric alphacoronavirus and its host adaptability. Virology 2023, 587, 109879. [Google Scholar] [CrossRef]
- Peng, Q.; Zhang, X.; Li, J.; He, W.; Fan, B.; Ni, Y.; Liu, M.; Li, B. Comprehensive analysis of codon usage patterns of porcine deltacoronavirus and its host adaptability. Transbound. Emerg. Dis. 2022, 69, e2443–e2455. [Google Scholar] [CrossRef]
- Kustin, T.; Stern, A. Biased Mutation and Selection in RNA Viruses. Mol. Biol. Evol. 2021, 38, 575–588. [Google Scholar] [CrossRef]
- Ventoso, I. Codon Usage Bias in Human RNA Viruses and Its Impact on Viral Translation, Fitness, and Evolution. Viruses 2025, 17, 1218. [Google Scholar] [CrossRef]
- Cheng, S.; Wu, H.; Chen, Z. Evolution of Transmissible Gastroenteritis Virus (TGEV): A Codon Usage Perspective. Int. J. Mol. Sci. 2020, 21, 7898. [Google Scholar] [CrossRef]
- Rima, B.K.; McFerran, N.V. Dinucleotide and stop codon frequencies in single-stranded RNA viruses. J. Gen. Virol. 1997, 78, 2859–2870. [Google Scholar] [CrossRef] [PubMed]
- Piovesan, A.; Pelleri, M.C.; Antonaros, F.; Strippoli, P.; Caracausi, M.; Vitale, L. On the length, weight and GC content of the human genome. BMC Res. Notes 2019, 12, 106. [Google Scholar] [CrossRef] [PubMed]
- Dutta, R.; Buragohain, L.; Borah, P. Analysis of codon usage of severe acute respiratory syndrome corona virus 2 (SARS-CoV-2) and its adaptability in dog. Virus Res. 2020, 288, 198113. [Google Scholar] [CrossRef] [PubMed]
- Takata, M.A.; Gonçalves-Carneiro, D.; Zang, T.M.; Soll, S.J.; York, A.; Blanco-Melo, D.; Bieniasz, P.D. CG dinucleotide suppression enables antiviral defence targeting non-self RNA. Nature 2017, 550, 124–127. [Google Scholar] [CrossRef]
- Franzo, G. SARS-CoV-2 and other human coronavirus show genome patterns previously associated to reduced viral recognition and altered immune response. Sci. Rep. 2021, 11, 10696. [Google Scholar] [CrossRef]
- Hu, J.S.; Wang, Q.Q.; Zhang, J.; Chen, H.T.; Xu, Z.W.; Zhu, L.; Ding, Y.Z.; Ma, L.N.; Xu, K.; Gu, Y.X.; et al. The characteristic of codon usage pattern and its evolution of hepatitis C virus. Infect. Genet. Evol. 2011, 11, 2098–2102. [Google Scholar] [CrossRef]
- Kumar, N.; Kulkarni, D.D.; Lee, B.; Kaushik, R.; Bhatia, S.; Sood, R.; Pateriya, A.K.; Bhat, S.; Singh, V.P. Evolution of Codon Usage Bias in Henipaviruses Is Governed by Natural Selection and Is Host-Specific. Viruses 2018, 10, 604. [Google Scholar] [CrossRef]
- Huang, W.; Guo, Y.; Li, N.; Feng, Y.; Xiao, L. Codon usage analysis of zoonotic coronaviruses reveals lower adaptation to humans by SARS-CoV-2. Infect. Genet. Evol. 2021, 89, 104736. [Google Scholar] [CrossRef]
- Maldonado, L.L.; Bertelli, A.M.; Kamenetzky, L. Molecular features similarities between SARS-CoV-2, SARS, MERS and key human genes could favour the viral infections and trigger collateral effects. Sci. Rep. 2021, 11, 4108. [Google Scholar] [CrossRef]
- Kumar, N.; Kaushik, R.; Tennakoon, C.; Uversky, V.N.; Mishra, A.; Sood, R.; Srivastava, P.; Tripathi, M.; Zhang, K.Y.J.; Bhatia, S. Evolutionary Signatures Governing the Codon Usage Bias in Coronaviruses and Their Implications for Viruses Infecting Various Bat Species. Viruses 2021, 13, 1847. [Google Scholar] [CrossRef]
- Ramazzotti, D.; Angaroni, F.; Maspero, D.; Mauri, M.; D’Aliberti, D.; Fontana, D.; Antoniotti, M.; Elli, E.M.; Graudenzi, A.; Piazza, R. Large-scale analysis of SARS-CoV-2 synonymous mutations reveals the adaptation to the human codon usage during the virus evolution. Virus Evol. 2022, 8, veac026. [Google Scholar] [CrossRef]
- Gu, H.; Chu, D.K.W.; Peiris, M.; Poon, L.L.M. Multivariate analyses of codon usage of SARS-CoV-2 and other betacoronaviruses. Virus Evol. 2020, 6, veaa032. [Google Scholar] [CrossRef]
- Lu, G.; Hu, Y.; Wang, Q.; Qi, J.; Gao, F.; Li, Y.; Zhang, Y.; Zhang, W.; Yuan, Y.; Bao, J.; et al. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature 2013, 500, 227–231. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Q.; Cao, L.; Ma, C.; Tortorici, M.A.; Liu, C.; Si, J.; Liu, P.; Gu, M.; Walls, A.C.; Wang, C.; et al. Close relatives of MERS-CoV in bats use ACE2 as their functional receptors. Nature 2022, 612, 748–757. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.B.; Liu, C.; Park, Y.J.; Tang, J.; Chen, J.; Xiong, Q.; Lee, J.; Stewart, C.; Asarnow, D.; Brown, J.; et al. Multiple independent acquisitions of ACE2 usage in MERS-related coronaviruses. Cell 2025, 188, 1693–1710.e18. [Google Scholar] [CrossRef] [PubMed]
- Madel Alfajaro, M.; Keeler, E.L.; Li, N.; Catanzaro, N.J.; Teng, I.T.; Zhao, Z.; Grunst, M.W.; Yount, B.; Schäfer, A.; Wang, D.; et al. HKU5 bat merbecoviruses engage bat and mink ACE2 as entry receptors. Nat. Commun. 2025, 16, 6822. [Google Scholar] [CrossRef]
- Park, Y.J.; Liu, C.; Lee, J.; Brown, J.T.; Ma, C.B.; Liu, P.; Gen, R.; Xiong, Q.; Zepeda, S.K.; Stewart, C.; et al. Molecular basis of convergent evolution of ACE2 receptor utilization among HKU5 coronaviruses. Cell 2025, 188, 1711–1728.e1721. [Google Scholar] [CrossRef]
- Ma, C.; Liu, C.; Xiong, Q.; Gu, M.; Shi, L.; Wang, C.; Si, J.; Tong, F.; Liu, P.; Huang, M.; et al. Broad host tropism of ACE2-using MERS-related coronaviruses and determinants restricting viral recognition. Cell Discov. 2023, 9, 57. [Google Scholar] [CrossRef]
- Wu, K.; Li, W.; Peng, G.; Li, F. Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor. Proc. Natl. Acad. Sci. USA 2009, 106, 19970–19974. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef]
- Zhao, J.; Kang, M.; Wu, H.; Sun, B.; Baele, G.; He, W.T.; Lu, M.; Suchard, M.A.; Ji, X.; He, N.; et al. Risk assessment of SARS-CoV-2 replicating and evolving in animals. Trends Microbiol. 2024, 32, 79–92. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, W.; Li, Y.; Liu, C.; Dong, T.; Chen, H.; Wu, C.; Su, J.; Li, B.; Zhang, W.; et al. Bat-infecting merbecovirus HKU5-CoV lineage 2 can use human ACE2 as a cell entry receptor. Cell 2025, 188, 1729–1742.e1716. [Google Scholar] [CrossRef]
- Hemida, M.G.; Chu, D.K.W.; Perera, R.; Ko, R.L.W.; So, R.T.Y.; Ng, B.C.Y.; Chan, S.M.S.; Chu, S.; Alnaeem, A.A.; Alhammadi, M.A.; et al. Coronavirus infections in horses in Saudi Arabia and Oman. Transbound. Emerg. Dis. 2017, 64, 2093–2103. [Google Scholar] [CrossRef]

| Species | ENC | |||||
|---|---|---|---|---|---|---|
| RDRP | S | E | M | N | AVERAGE | |
| HedCoV1 | 52 | 45.65 | 53.42 | 42.69 | 49.71 | 48.69 ± 3.99 |
| Ea-HedCoV HKU31 | 47.06 | 45.29 | 50.15 | 48.56 | 51.85 | 48.58 ± 2.29 |
| Ty-BatCoV HKU4 | 46.36 | 41.26 | 51.09 | 47.17 | 48.81 | 46.94 ± 3.27 |
| Pi-BatCoV HKU5 | 52.80 | 51.62 | 47.90 | 53.34 | 52.00 | 51.53 ± 1.91 |
| MERSr-CoV | 51.60 | 46.04 | 55.91 | 59.85 | 49.50 | 52.58 ± 4.84 |
| MjHKU4r-CoV | 44.12 | 40.60 | 57.90 | 41.11 | 47.77 | 46.30 ± 6.34 |
| Mink-derived HKU5-like | 53.93 | 51.62 | 61.00 | 58.47 | 51.22 | 55.24 ± 4.31 |
| (a) | |||||||||||
| Bos taurus | Camelus dromedarius | Equus asinus | Erinaceus europaeus | Capra hircus | Homo sapiens | Ovis aries | Rhinolophus ferrumequinum | Sus scrofa | Equus caballus | Oryctolagus cuniculus | |
| RdRp | 0.65 | 0.59 | 0.64 | 0.67 | 0.66 | 0.73 | 0.61 | 0.52 | 0.61 | 0.65 | 0.56 |
| S | 0.63 | 0.57 | 0.61 | 0.65 | 0.64 | 0.71 | 0.59 | 0.49 | 0.58 | 0.63 | 0.53 |
| E | 0.61 | 0.57 | 0.58 | 0.66 | 0.61 | 0.68 | 0.57 | 0.47 | 0.57 | 0.61 | 0.53 |
| M | 0.62 | 0.57 | 0.59 | 0.61 | 0.62 | 0.69 | 0.58 | 0.48 | 0.58 | 0.62 | 0.54 |
| N | 0.66 | 0.61 | 0.66 | 0.71 | 0.67 | 0.73 | 0.64 | 0.51 | 0.62 | 0.67 | 0.57 |
| AVERAGE | 0.64 ± 0.02 | 0.58 ± 0.02 | 0.62 ± 0.03 | 0.66 ± 0.03 | 0.64 ± 0.02 | 0.71 ± 0.02 | 0.6 ± 0.02 | 0.49 ± 0.02 | 0.59 ± 0.02 | 0.64 ± 0.02 | 0.55 ± 0.02 |
| (b) | |||||||||||
| Bos taurus | Camelus dromedarius | Equus asinus | Erinaceus europaeus | Capra hircus | Homo sapiens | Ovis aries | Rhinolophus ferrumequinum | Sus scrofa | Equus caballus | Oryctolagus cuniculus | |
| RdRp | 1.45 | 1.56 | 1.49 | 1.51 | 1.42 | 1.36 | 1.54 | 1.82 | 1.54 | 1.45 | 1.65 |
| S | 1.65 | 1.78 | 1.72 | 1.73 | 1.61 | 1.53 | 1.8 | 2.06 | 1.77 | 1.64 | 1.89 |
| E | 1.94 | 2.18 | 2.2 | 1.89 | 1.99 | 1.79 | 2.19 | 3.03 | 2.1 | 2.01 | 2.24 |
| M | 1.44 | 1.52 | 1.62 | 1.56 | 1.43 | 1.35 | 1.61 | 1.79 | 1.52 | 1.45 | 1.61 |
| N | 1.48 | 1.53 | 1.46 | 1.38 | 1.43 | 1.39 | 1.53 | 1.8 | 1.55 | 1.45 | 1.65 |
| AVERAGE | 1.59 ± 0.19 | 1.71 ± 0.25 | 1.7 ± 0.27 | 1.61 ± 0.18 | 1.58 ± 0.22 | 1.48 ± 0.17 | 1.73 ± 0.25 | 2.1 ± 0.48 | 1.7 ± 0.22 | 1.6 ± 0.22 | 1.81 ± 0.24 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, G.; Li, Y.; Zhou, H.; Franzo, G.; Zheng, M.; Liu, H.; Chen, X.; Dai, J.; He, W.-T. Comparative Analysis of Codon Usage Patterns and Host Adaptation in Merbecoviruses. Viruses 2025, 17, 1479. https://doi.org/10.3390/v17111479
Yan G, Li Y, Zhou H, Franzo G, Zheng M, Liu H, Chen X, Dai J, He W-T. Comparative Analysis of Codon Usage Patterns and Host Adaptation in Merbecoviruses. Viruses. 2025; 17(11):1479. https://doi.org/10.3390/v17111479
Chicago/Turabian StyleYan, Ge, Yue Li, Huimin Zhou, Giovanni Franzo, Mengdi Zheng, Hao Liu, Xiang Chen, Jianjun Dai, and Wan-Ting He. 2025. "Comparative Analysis of Codon Usage Patterns and Host Adaptation in Merbecoviruses" Viruses 17, no. 11: 1479. https://doi.org/10.3390/v17111479
APA StyleYan, G., Li, Y., Zhou, H., Franzo, G., Zheng, M., Liu, H., Chen, X., Dai, J., & He, W.-T. (2025). Comparative Analysis of Codon Usage Patterns and Host Adaptation in Merbecoviruses. Viruses, 17(11), 1479. https://doi.org/10.3390/v17111479





