Multivalent Interactions Between the Picornavirus 3C(D) Main Protease and RNA Oligonucleotides Induce Liquid–Liquid Phase Separation
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Protein Expression and Isotopic Labeling
2.3. RNA Sample Preparation
2.4. Purification of PV-3C and PV-3CD
2.5. NMR Sample Preparation
2.6. Sedimentation Velocity Analytical Ultracentrifugation (AUC)
2.7. Differential Interference Contrast (DIC) Microscopy
2.8. Negative Staining Electron Microscopy
2.9. Molecular Dynamics (MD) Simulation: System Preparation
2.10. MD Simulation Runs and Analysis
3. Results
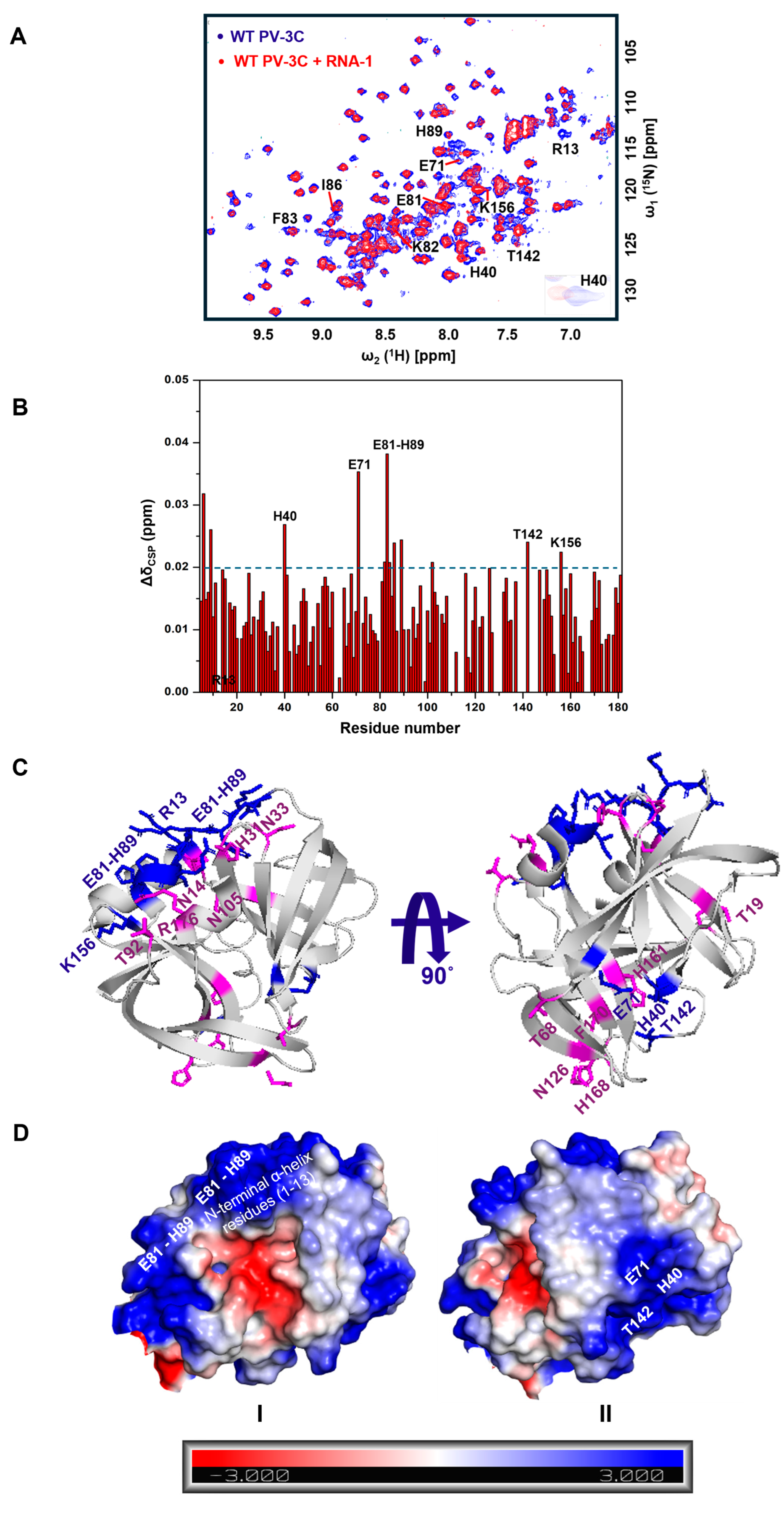
3.1. PV-3C Binds to a Diverse Set of RNA Oligonucleotides
3.2. AUC Reveals Multimeric Complex Formation Between PV-3C and RNA
3.3. Condensate Formation Observed from Differential Interference Contrast Microscopy
3.4. PV-3CD and RNA Interactions Also Lead to LLPS
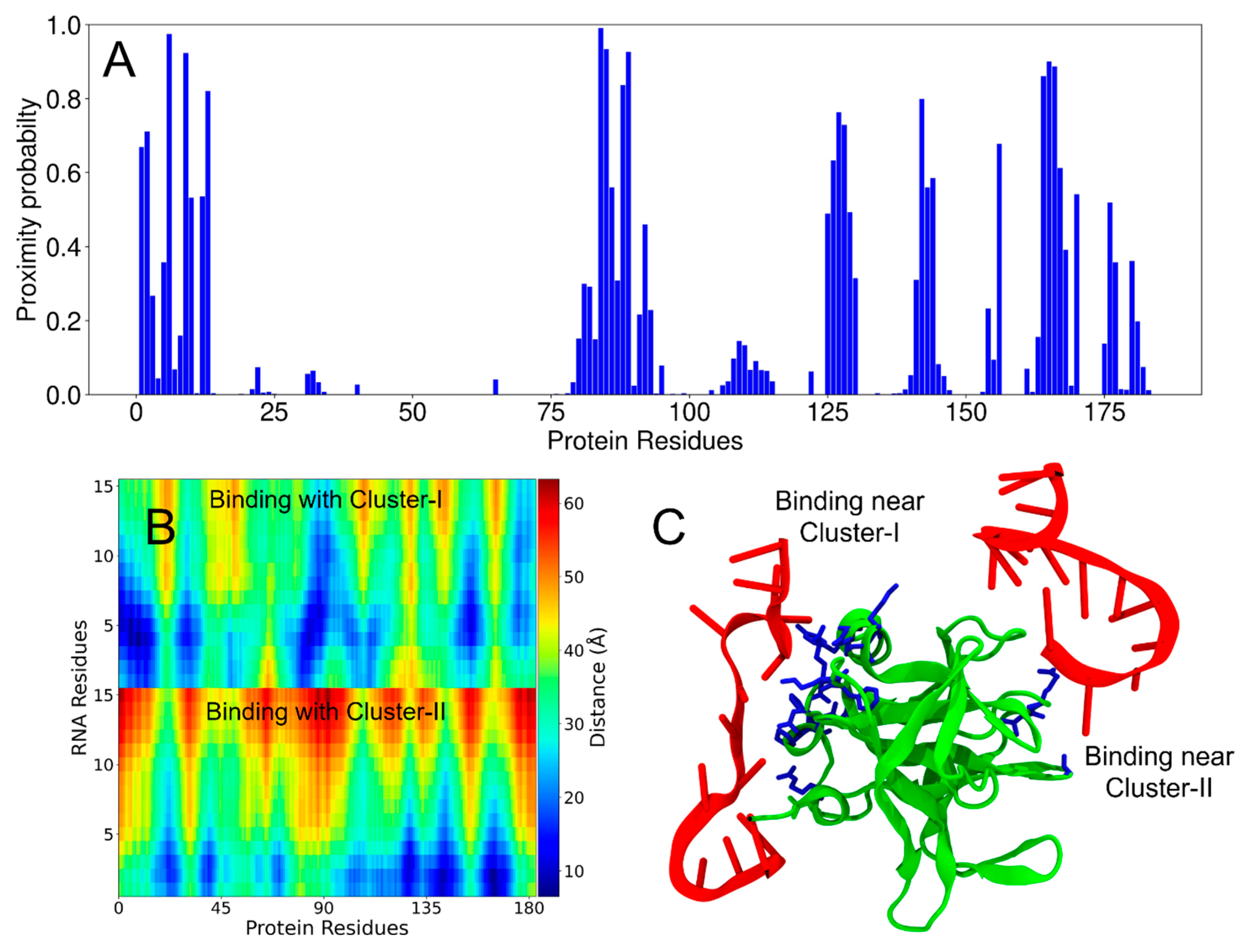
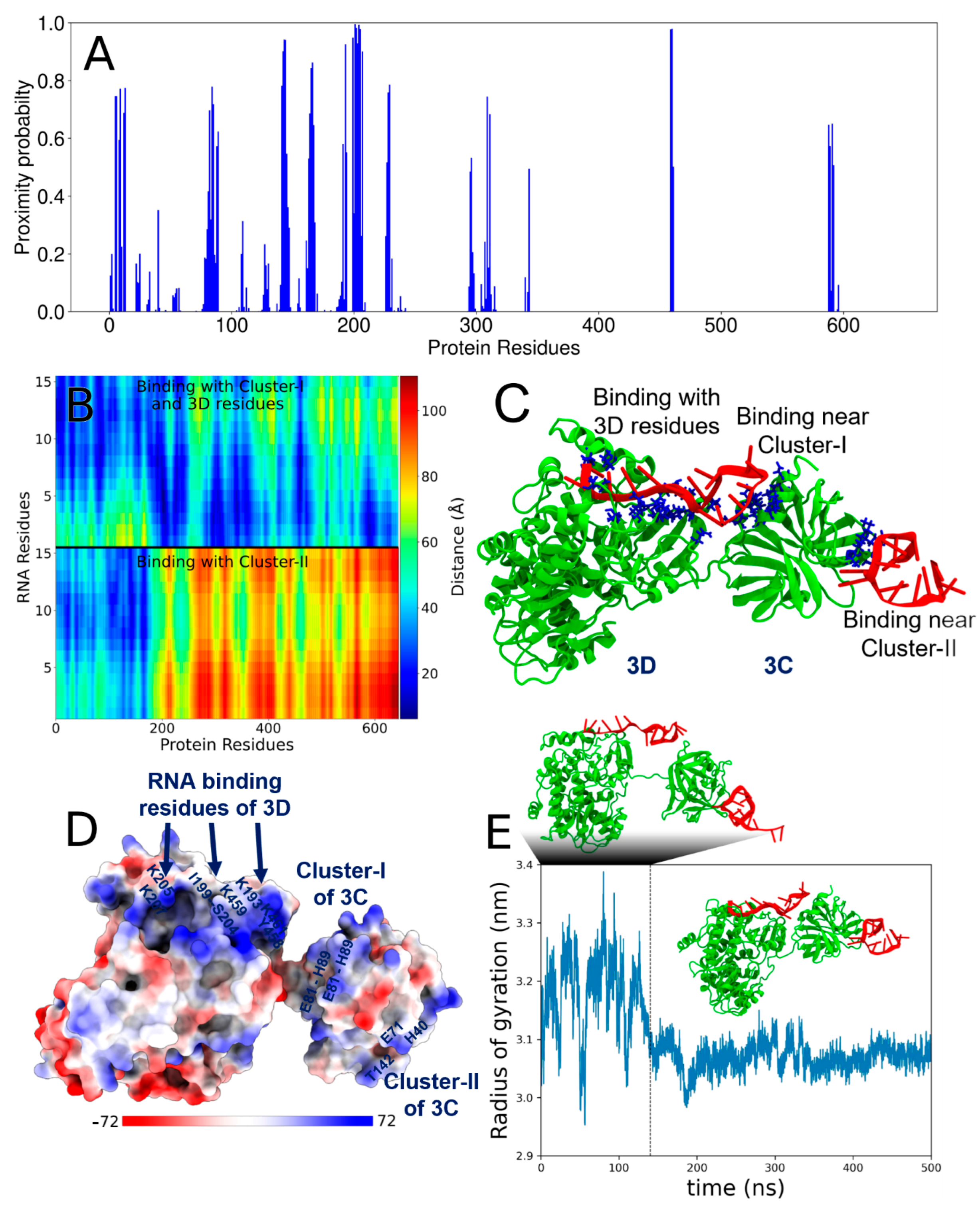
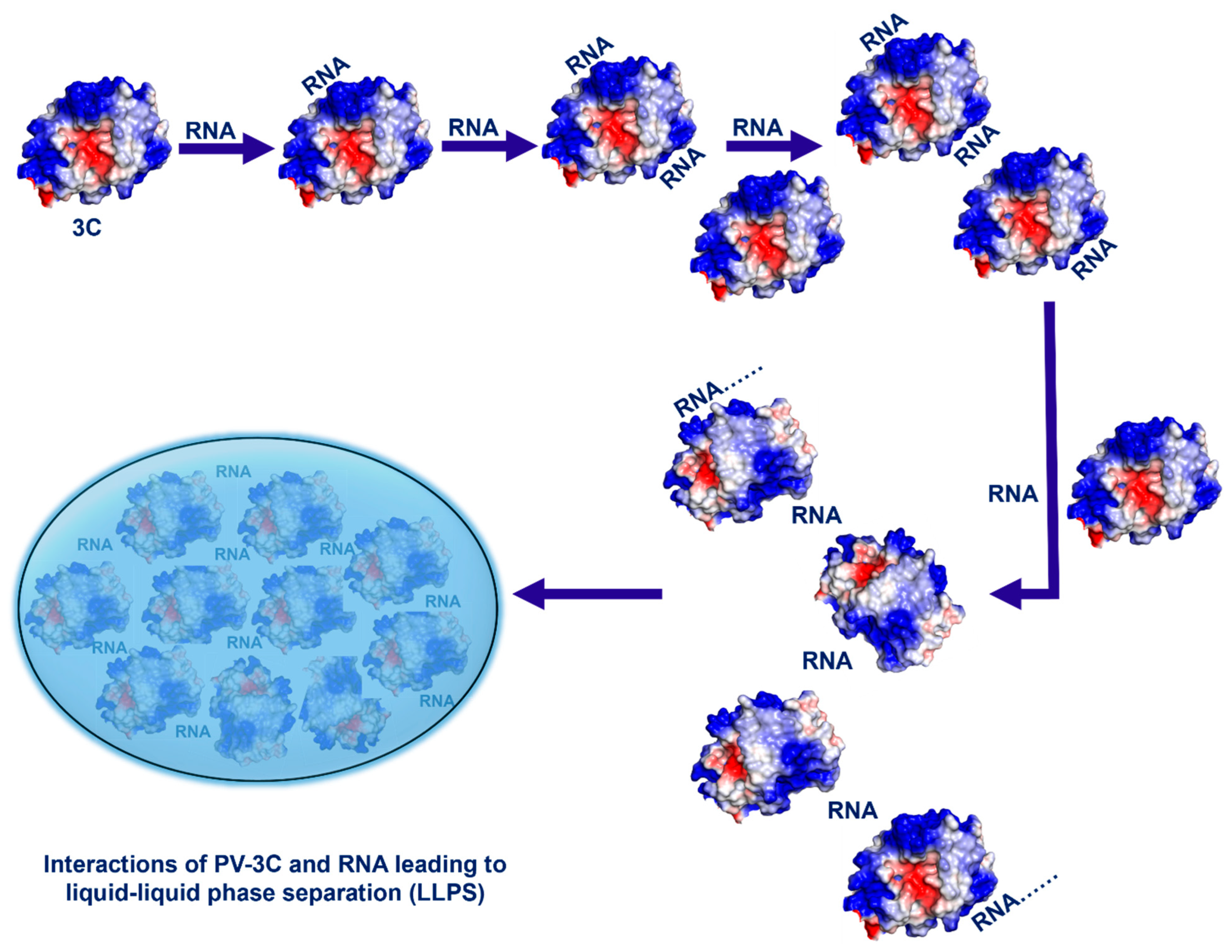
3.5. Molecular Model of the Multivalent Interactions Between PV-3C/3CD and RNA
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 3C | 3C main protease |
| 3D | 3D RNA-dependent RNA polymerase |
| CSP | Chemical shift perturbations |
| CREs | Cis-acting replication elements |
| HEPES | 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid |
| HMQC | Heteronuclear multiple quantum coherence |
| HPLC | High-performance liquid chromatography |
| IPTG | Isopropyl β-D-1-thiogalactopyranoside |
| LLPS | Liquid–liquid phase separation |
| MD | Molecular dynamics |
| MEM | Minimum essential medium |
| MWCO | Molecular weight cutoff |
| NaCl | Sodium chloride |
| NH4Cl | Ammonium chloride |
| Ni-NTA | Nickel–nitrilotriacetic acid |
| NMR | Nuclear magnetic resonance |
| NpT | Constant number of particles, pressure, and temperature |
| NVT | Constant number of particles, volume, and temperature |
| OD600 | Optical density at 600 nm |
| PDB | Protein Data Bank |
| PEI | Polyethyleneimine |
| PMSF | Phenylmethanesulfonyl fluoride |
| PV | Poliovirus |
| SARS-CoV | Severe acute respiratory syndrome-related coronavirus |
| SV-AUC | Sedimentation velocity analytical ultracentrifugation |
| TIA-1 | T-cell intracellular antigen 1 |
| VPg | Viral protein genome-linked |
References
- Flather, D.; Semler, B.L. Picornaviruses and nuclear functions: Targeting a cellular compartment distinct from the replication site of a positive-strand RNA virus. Front. Microbiol. 2015, 6, 594. [Google Scholar] [CrossRef]
- Gamarnik, A.V.; Andino, R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 1998, 12, 2293–2304. [Google Scholar] [CrossRef]
- Mondal, S.; Sarvari, G.; Boehr, D.D. Picornavirus 3C Proteins Intervene in Host Cell Processes through Proteolysis and Interactions with RNA. Viruses 2023, 15, 2413. [Google Scholar] [CrossRef]
- Winston, D.S.; Boehr, D.D. The Picornavirus Precursor 3CD Has Different Conformational Dynamics Compared to 3C(pro) and 3D(pol) in Functionally Relevant Regions. Viruses 2021, 13, 442. [Google Scholar] [CrossRef]
- Francisco-Velilla, R.; Embarc-Buh, A.; Abellan, S.; Martinez-Salas, E. Picornavirus translation strategies. FEBS Open Bio 2022, 12, 1125–1141. [Google Scholar] [CrossRef]
- Marcotte, L.L.; Wass, A.B.; Gohara, D.W.; Pathak, H.B.; Arnold, J.J.; Filman, D.J.; Cameron, C.E.; Hogle, J.M. Crystal Structure of Poliovirus 3CD Protein: Virally Encoded Protease and Precursor to the RNA-Dependent RNA Polymerase. J. Virol. 2007, 81, 3583–3596. [Google Scholar] [CrossRef]
- Parsley, T.B.; Towner, J.S.; Blyn, L.B.; Ehrenfeld, E.; Semler, B.L. Poly (rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA 1997, 3, 1124–1134. [Google Scholar]
- Gamarnik, A.V.; Andino, R. Interactions of Viral Protein 3CD and Poly(rC) Binding Protein with the 5′ Untranslated Region of the Poliovirus Genome. J. Virol. 2000, 74, 2219–2226. [Google Scholar] [CrossRef]
- Blair, W.S.; Parsley, T.B.; Bogerd, H.P.; Towner, J.S.; Semler, B.L.; Cullen, B.R. Utilization of a mammalian cell-based RNA binding assay to characterize the RNA binding properties of picornavirus 3C proteinases. RNA 1998, 4, 215. [Google Scholar]
- Harris, K.S.; Xiang, W.; Alexander, L.; Lane, W.S.; Paul, A.V.; Wimmer, E. Interaction of poliovirus polypeptide 3CDpro with the 5‘ and 3‘ termini of the poliovirus genome. Identification of viral and cellular cofactors needed for efficient binding. J. Biol. Chem. 1994, 269, 27004–27014. [Google Scholar] [CrossRef]
- Mosimann, S.C.; Cherney, M.M.; Sia, S.; Plotch, S.; James, M.N.G. Refined X-ray crystallographic structure of the poliovirus 3C gene product. J. Mol. Biol. 1997, 273, 1032–1047. [Google Scholar] [CrossRef]
- Melchers, W.J.G.; Zoll, J.; Tessari, M.; Bakhmutov, D.V.; Gmyl, A.P.; Agol, V.I.; Heus, H.A. A GCUA Tetranucleotide Loop Found in the Poliovirus OriL by in Vivo SELEX (Un)Expectedly Forms a YNMG-like Structure: Extending the YNMG Family with GYYA. RNA 2006, 12, 1671–1682. [Google Scholar] [CrossRef]
- Pilipenko, E.V.; Poperechny, K.V.; Maslova, S.V.; Melchers, W.J.; Slot, H.J.; Agol, V.I. Cis-Element, OriR, Involved in the Initiation of (-)Strand Poliovirus RNA: A Quasi-Globular Multi-Domain RNA Structure Maintained by Tertiary (‘kissing’) Interactions. EMBO J. 1996, 15, 5428–5436. [Google Scholar] [CrossRef]
- Amero, C.D.; Arnold, J.J.; Moustafa, I.M.; Cameron, C.E.; Foster, M.P. Identification of the oriI-Binding Site of Poliovirus 3C Protein by Nuclear Magnetic Resonance Spectroscopy. J. Virol. 2008, 82, 4363–4370. [Google Scholar] [CrossRef]
- Bergmann, E.M.; Mosimann, S.C.; Chernaia, M.M.; Malcolm, B.A.; James, M.N. The refined crystal structure of the 3C gene product from hepatitis A virus: Specific proteinase activity and RNA recognition. J. Virol. 1997, 71, 2436–2448. [Google Scholar] [CrossRef]
- Kim, Y.; Lovell, S.; Tiew, K.C.; Mandadapu, S.R.; Alliston, K.R.; Battaile, K.P.; Groutas, W.C.; Chang, K.O. Broad-Spectrum Antivirals against 3C or 3C-Like Proteases of Picornaviruses, Noroviruses, and Coronaviruses. J. Virol. 2012, 86, 11754–11762. [Google Scholar] [CrossRef]
- Roehl, H.H.; Parsley, T.B.; Ho, T.V.; Semler, B.L. Processing of a cellular polypeptide by 3CD proteinase is required for poliovirus ribonucleoprotein complex formation. J. Virol. 1997, 71, 578–585. [Google Scholar] [CrossRef]
- Erickson, J.W.; Frankenberger, E.A.; Rossmann, M.G.; Fout, G.S.; Medappa, K.C.; Rueckert, R.R. Crystallization of a common cold virus, human rhinovirus 14: ‘isomorphism’ with poliovirus crystals. Proc. Natl. Acad. Sci. USA 1983, 80, 931–934. [Google Scholar] [CrossRef]
- Zhang, X.; Zheng, R.; Li, Z.; Ma, J. Liquid-liquid Phase Separation in Viral Function. J. Mol. Biol. 2023, 435, 167955. [Google Scholar] [CrossRef]
- Ainani, H.; Bouchmaa, N.; Ben Mrid, R.; El Fatimy, R. Liquid-liquid phase separation of protein tau: An emerging process in Alzheimer’s disease pathogenesis. Neurobiol. Dis. 2023, 178, 106011. [Google Scholar] [CrossRef]
- Babinchak, W.M.; Surewicz, W.K. Liquid–Liquid Phase Separation and Its Mechanistic Role in Pathological Protein Aggregation. J. Mol. Biol. 2020, 432, 1910–1925. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.M.; Moustafa, I.M.; Arnold, J.J.; Cameron, C.E.; Boehr, D.D. Long-Range Communication between Different Functional Sites in the Picornaviral 3C Protein. Structure 2016, 24, 509–517. [Google Scholar] [CrossRef][Green Version]
- Mondal, S.; Shet, D.; Prasanna, C.; Atreya, H.S. High yield expression of proteins in E. coli for NMR studies. Adv. Biosci. Biotechnol. 2013, 04, 751–767. [Google Scholar] [CrossRef]
- Lee, W.; Tonelli, M.; Markley, J.L. NMRFAM-SPARKY: Enhanced software for biomolecular NMR spectroscopy. Bioinformatics 2015, 31, 1325–1327. [Google Scholar] [CrossRef]
- Mondal, S.; Ravula, T.; Rao, P.L.; Atreya, H.S. Unraveling the Dynamic Nature of Protein–Graphene Oxide Interactions. RSC Adv. 2016, 6, 52539–52548. [Google Scholar] [CrossRef]
- Jaipuria, G.; Shet, D.; Malik, S.; Swain, M.; Atreya, H.S.; Galea, C.A.; Slomiany, M.G.; Rosenzweig, S.A.; Forbes, B.E.; Norton, R.S.; et al. IGF-dependent dynamic modulation of a protease cleavage site in the intrinsically disordered linker domain of human IGFBP2. Proteins 2022, 90, 1732–1743. [Google Scholar] [CrossRef]
- Werbeck, N.D.; Shukla, V.K.; Kunze, M.B.A.; Yalinca, H.; Pritchard, R.B.; Siemons, L.; Mondal, S.; Greenwood, S.O.R.; Kirkpatrick, J.; Marson, C.M.; et al. A distal regulatory region of a class I human histone deacetylase. Nat. Commun. 2020, 11, 3841. [Google Scholar] [CrossRef]
- Malik, S.A.; Mondal, S.; Atreya, H.S. Alpha-Synuclein Aggregation Mechanism in the Presence of Nanomaterials. Biochemistry 2024, 63, 1162–1169. [Google Scholar] [CrossRef]
- Malik, S.A.; Mondal, S.; Atreya, H.S. Enhanced stability of an intrinsically disordered protein against proteolytic cleavage through interactions with silver nanoparticles. RSC Adv. 2019, 9, 28746. [Google Scholar] [CrossRef]
- Namitz, K.E.W.; Showalter, S.A.; Cosgrove, M.S. Phase separation promotes a highly active oligomeric scaffold of the MLL1 core complex for regulation of histone H3K4 methylation. J. Biol. Chem. 2023, 299, 105204. [Google Scholar] [CrossRef]
- Preza, C.; Snyder, D.L. Theoretical development and experimental evaluation of imaging models for differential-interference-contrast microscopy. J. Opt. Soc. Am. A Opt. Image Sci. Vis. 1999, 16, 2185–21999. [Google Scholar] [CrossRef]
- Obara, B.; Roberts, M.A.J.; Armitage, J.P.; Grau, V. Bacterial cell identification in differential interference contrast microscopy images. BMC Bioinform. 2013, 14, 134. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, H.; Wang, J.; Liu, X.; Hao, H.; Tan, Y.S.; Zhang, Y.; Zhang, H.; Ding, X.; Zhao, W.; et al. Single-shot isotropic differential interference contrast microscopy. Nat. Commun. 2023, 14, 2063. [Google Scholar] [CrossRef] [PubMed]
- Weissenberger, G.; Henderikx, R.J.M.; Peters, P.J. Understanding the invisible hands of sample preparation for cryo-EM. Nat. Methods 2021, 18, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Schrödinger, LLC. The PyMOL Molecular Graphics System, Version 2.5.0; Schrödinger, LLC: New York, NY, USA, 2015. [Google Scholar]
- Mukherjee, S.; Schäfer, L.V. Thermodynamic forces from protein and water govern condensate formation of an intrinsically disordered protein domain. Nat. Commun. 2023, 14, 5892. [Google Scholar] [CrossRef]
- Fiser, A.; Sali, A. ModLoop: Automated modeling of loops in protein structures. Bioinformatics 2003, 19, 2500–2501. [Google Scholar] [CrossRef]
- Hospital, A.; Faustino, I.; Collepardo-Guevara, R.; González, C.; Gelpí, J.L.; Orozco, M. NAFlex: A web server for the study of nucleic acid flexibility. Nucleic Acids Res. 2013, 41, W47–W55. [Google Scholar] [CrossRef]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B.L.; Grubmüller, H.; MacKerell, A.D. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar] [CrossRef]
- Hess, B.; Kutzner, C.; van der Spoel, D.; Lindahl, E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J. Chem. Theory Comput. 2008, 4, 435–447. [Google Scholar] [CrossRef]
- Bernetti, M.; Bussi, G. Pressure control using stochastic cell rescaling. J. Chem. Phys. 2020, 153, 114107. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Claridge, J.K.; Headey, S.J.; Chow, J.Y.H.; Schwalbe, M.; Edwards, P.J.; Jeffries, C.M.; Venugopal, H.; Trewhella, J.; Pascal, S.M. A picornaviral loop-to-loop replication complex. J. Struct. Biol. 2009, 166, 251–262. [Google Scholar] [CrossRef]
- Nayak, A.; Goodfellow, I.G.; Woolaway, K.E.; Birtley, J.; Curry, S.; Belsham, G.J. Role of RNA Structure and RNA Binding Activity of Foot-and-Mouth Disease Virus 3C Protein in VPg Uridylylation and Virus Replication. J. Virol. 2006, 80, 9865–9875. [Google Scholar] [CrossRef][Green Version]
- Contursi, P.; Farina, B.; Pirone, L.; Fusco, S.; Russo, L.; Bartolucci, S.; Fattorusso, R.; Pedone, E. Structural and functional studies of Stf76 from the Sulfolobus islandicus plasmid–virus pSSVx: A novel peculiar member of the winged helix–turn–helix transcription factor family. Nucleic Acids Res. 2014, 42, 5993–6011. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.; Li, Q.; Xing, R.; Li, J.; Yan, X. Peptide self-assembly through liquid-liquid phase separation. Chem 2023, 9, 2425–2445. [Google Scholar] [CrossRef]
- Cabral, S.E.; Otis, J.P.; Mowry, K.L. Multivalent interactions with RNA drive recruitment and dynamics in biomolecular condensates in Xenopus oocytes. iScience 2022, 25, 104811. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Jia, H.; Nakamura, Y.; Kanekura, K.; Hayamizu, Y. Effect of Multivalency on Phase-Separated Droplets Consisting of Poly(PR) Dipeptide Repeats and RNA at the Solid/Liquid Interface. ACS Omega 2022, 7, 19280–19287. [Google Scholar] [CrossRef]
- Dias-Solange, D.; Le, M.T.; Gottipati, K.; Choi, K.H. Structure of Coxsackievirus Cloverleaf RNA and 3C pro Dimer Establishes the RNA-Binding Mechanism of Enterovirus Protease 3C Pro. Sci. Adv. 2025, 11, eads6862. [Google Scholar] [CrossRef]
- Weidman, M.K.; Yalamanchili, P.; Ng, B.; Tsai, W.; Dasgupta, A. Poliovirus 3C protease-mediated degradation of transcriptional activator p53 requires a cellular activity. Virology 2001, 291, 260–271. [Google Scholar] [CrossRef] [PubMed]
- Kundu, P.; Raychaudhuri, S.; Tsai, W.; Dasgupta, A. Shutoff of RNA polymerase II transcription by poliovirus involves 3C protease-mediated cleavage of the TATA-binding protein at an alternative site: Incomplete shutoff of transcription interferes with efficient viral replication. J. Virol. 2005, 79, 9702–9713. [Google Scholar] [CrossRef] [PubMed]
- Shengjuler, D.; Chan, Y.M.; Sun, S.; Moustafa, I.M.; Li, Z.L.; Gohara, D.W.; Buck, M.; Cremer, P.S.; Boehr, D.D.; Cameron, C.E. The RNA-Binding Site of Poliovirus 3C Protein Doubles as a Phosphoinositide-Binding Domain. Structure 2017, 25, 1875–1886. [Google Scholar] [CrossRef]
- Sun, D.; Chen, S.; Cheng, A.; Wang, M. Roles of the Picornaviral 3C Proteinase in the Viral Life Cycle and Host Cells. Viruses 2016, 8, 82. [Google Scholar] [CrossRef]
- Wei, W.; Bai, L.; Yan, B.; Meng, W.; Wang, H.; Zhai, J.; Si, F.; Zheng, C. When liquid-liquid phase separation meets viral infections. Front. Immunol. 2022, 13, 985622. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Yang, Y.; Zhang, S.; Zhang, T.; Lü, P.; Chen, K. Liquid-liquid phase separation in viral infection: From the occurrence and function to treatment potentials. Colloids Surf. B Biointerfaces 2025, 246, 114385. [Google Scholar] [CrossRef]
- Glon, D.; Léonardon, B.; Guillemot, A.; Albertini, A.; Lagaudrière-Gesbert, C.; Gaudin, Y. Biomolecular condensates with liquid properties formed during viral infections. Microbes Infect. 2024, 26, 105402. [Google Scholar] [CrossRef]
- Etibor, T.A.; Yamauchi, Y.; Amorim, M.J. Liquid biomolecular condensates and viral lifecycles: Review and perspectives. Viruses 2021, 13, 366. [Google Scholar] [CrossRef]
- Yan, Y.; Gan, J.; Tao, Y.; Okita, T.W.; Tian, L. RNA-Binding Proteins: The Key Modulator in Stress Granule Formation and Abiotic Stress Response. Front. Plant Sci. 2022, 13, 882596. [Google Scholar] [CrossRef]
- Guan, Y.; Wang, Y.; Fu, X.; Bai, G.; Li, X.; Mao, J.; Yan, Y.; Hu, L. Multiple functions of stress granules in viral infection at a glance. Front. Microbiol. 2023, 14, 1138864. [Google Scholar] [CrossRef]
- Dougherty, J.D.; Tsai, W.C.; Lloyd, R.E. Multiple poliovirus proteins repress cytoplasmic RNA granules. Viruses 2015, 7, 6127–6140. [Google Scholar] [CrossRef] [PubMed]
- Amineva, S.P.; Aminev, A.G.; Palmenberg, A.C.; Gern, J.E. Rhinovirus 3C protease precursors 3CD and 3CD′ localize to the nuclei of infected cells. J. Gen. Virol. 2004, 85, 2969–2979. [Google Scholar] [CrossRef]
- Sharma, R.; Raychaudhuri, S.; Dasgupta, A. Nuclear entry of poliovirus protease-polymerase precursor 3CD: Implications for host cell transcription shut-off. Virology 2004, 320, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, K.D.; Semler, B.L. Poliovirus infection induces the co-localization of cellular protein SRp20 with TIA-1, a cytoplasmic stress granule protein. Virus Res. 2013, 176, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Visser, L.J.; Medina, G.N.; Rabouw, H.H.; de Groot, R.J.; Langereis, M.A.; de los Santos, T.; van Kuppeveld, F.J.M. Foot-and-Mouth Disease Virus Leader Protease Cleaves G3BP1 and G3BP2 and Inhibits Stress Granule Formation. J. Virol. 2019, 93, e00922-18. [Google Scholar] [CrossRef]
- Saiz, M.; Martinez-Salas, E. Uncovering Targets of the Leader Protease: Linking RNA-Mediated Pathways and Antiviral Defense. Wiley Interdiscip. Rev. RNA 2021, 12, e1645. [Google Scholar] [CrossRef]
- Galan, A.; Lozano, G.; Piñeiro, D.; Martinez-Salas, E. G3BP1 Interacts Directly with the FMDV IRES and Negatively Regulates Translation. FEBS J. 2017, 284, 3202–3217. [Google Scholar] [CrossRef]
- Ye, X.; Pan, T.; Wang, D.; Fang, L.; Ma, J.; Zhu, X.; Shi, Y.; Zhang, K.; Zheng, H.; Chen, H.; et al. Foot-and-Mouth Disease Virus Counteracts on Internal Ribosome Entry Site Suppression by G3BP1 and Inhibits G3BP1-Mediated Stress Granule Assembly via Post-Translational Mechanisms. Front. Immunol. 2018, 9, 1142. [Google Scholar] [CrossRef]
- White, J.P.; Cardenas, A.M.; Marissen, W.E.; Lloyd, R.E. Inhibition of Cytoplasmic MRNA Stress Granule Formation by a Viral Proteinase. Cell Host Microbe 2007, 2, 295–305. [Google Scholar] [CrossRef]
- Perera, R.; Daijogo, S.; Walter, B.L.; Nguyen, J.H.C.; Semler, B.L. Cellular protein modification by poliovirus: The two faces of poly(rC)-binding protein. J. Virol. 2007, 81, 8919–8932. [Google Scholar] [CrossRef] [PubMed]
- Kafasla, P.; Morgner, N.; Robinson, C.V.; Jackson, R.J. Polypyrimidine tract-binding protein stimulates the poliovirus IRES by modulating eIF4G binding. EMBO J. 2010, 29, 3710–3722. [Google Scholar] [CrossRef] [PubMed]
- McCormick, C.; Khaperskyy, D.A. Translation inhibition and stress granules in the antiviral immune response. Nat. Rev. Immunol. 2017, 17, 647–660. [Google Scholar] [CrossRef]
- Campos-Melo, D.; Hawley, Z.C.E.; Droppelmann, C.A.; Strong, M.J. The Integral Role of RNA in Stress Granule Formation and Function. Front. Cell Dev. Biol. 2021, 9, 621779. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mondal, S.; Mukherjee, S.; Namitz, K.E.W.; Yennawar, N.H.; Boehr, D.D. Multivalent Interactions Between the Picornavirus 3C(D) Main Protease and RNA Oligonucleotides Induce Liquid–Liquid Phase Separation. Viruses 2025, 17, 1473. https://doi.org/10.3390/v17111473
Mondal S, Mukherjee S, Namitz KEW, Yennawar NH, Boehr DD. Multivalent Interactions Between the Picornavirus 3C(D) Main Protease and RNA Oligonucleotides Induce Liquid–Liquid Phase Separation. Viruses. 2025; 17(11):1473. https://doi.org/10.3390/v17111473
Chicago/Turabian StyleMondal, Somnath, Saumyak Mukherjee, Kevin E. W. Namitz, Neela H. Yennawar, and David D. Boehr. 2025. "Multivalent Interactions Between the Picornavirus 3C(D) Main Protease and RNA Oligonucleotides Induce Liquid–Liquid Phase Separation" Viruses 17, no. 11: 1473. https://doi.org/10.3390/v17111473
APA StyleMondal, S., Mukherjee, S., Namitz, K. E. W., Yennawar, N. H., & Boehr, D. D. (2025). Multivalent Interactions Between the Picornavirus 3C(D) Main Protease and RNA Oligonucleotides Induce Liquid–Liquid Phase Separation. Viruses, 17(11), 1473. https://doi.org/10.3390/v17111473







