Global Surveillance and Biological Characterization of the SARS-CoV-2 NB.1.8.1 Variant: An Emerging VUM Lineage Under Scrutiny
Abstract
1. Introduction
2. The Backdrop of JN.1 Global Dominance and the Emergence of the Novel NB.1.8.1 Lineage
3. Methods
4. The Origin and Evolution of the NB.1.8.1 Lineage
5. Biological Characteristics and Functional Implications
6. Clinical and Epidemiological Perspectives
7. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154. [Google Scholar] [CrossRef]
- Dhama, K.; Khan, S.; Tiwari, R.; Sircar, S.; Bhat, S.; Malik, Y.S.; Singh, K.P.; Chaicumpa, W.; Bonilla-Aldana, D.K.; Rodriguez-Morales, A.J. Coronavirus Disease 2019-COVID-19. Clin. Microbiol. Rev. 2020, 33, e00028-20. [Google Scholar] [CrossRef]
- Rodriguez-Morales, A.J.; Bonilla-Aldana, D.K.; Balbin-Ramon, G.J.; Rabaan, A.A.; Sah, R.; Paniz-Mondolfi, A.; Pagliano, P.; Esposito, S. History is repeating itself: Probable zoonotic spillover as the cause of the 2019 novel Coronavirus Epidemic. Infez. Med. 2020, 28, 3–5. [Google Scholar]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef]
- The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [CrossRef] [PubMed]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef]
- Almehdi, A.M.; Khoder, G.; Alchakee, A.S.; Alsayyid, A.T.; Sarg, N.H.; Soliman, S.S.M. SARS-CoV-2 spike protein: Pathogenesis, vaccines, and potential therapies. Infection 2021, 49, 855–876. [Google Scholar] [CrossRef] [PubMed]
- Li, C.J.; Chang, S.C. SARS-CoV-2 spike S2-specific neutralizing antibodies. Emerg. Microbes Infect. 2023, 12, 2220582. [Google Scholar] [CrossRef] [PubMed]
- Souza, P.F.N.; Mesquita, F.P.; Amaral, J.L.; Landim, P.G.C.; Lima, K.R.P.; Costa, M.B.; Farias, I.R.; Belém, M.O.; Pinto, Y.O.; Moreira, H.H.T.; et al. The spike glycoprotein of SARS-CoV-2: A review of how mutations of spike glycoproteins have driven the emergence of variants with high transmissibility and immune escape. Int. J. Biol. Macromol. 2022, 208, 105–125. [Google Scholar] [CrossRef]
- Shehzadi, K.; Saba, A.; Yu, M.; Liang, J. Structure-Based Drug Design of RdRp Inhibitors against SARS-CoV-2. Top. Curr. Chem. 2023, 381, 22. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Chen, D.; Zhang, T.; Song, C.; Zhang, X.; Lin, L.; Huang, J.; Peng, X.; Liu, Y.; Wu, G.; et al. Recent advancements in the discovery of small-molecule non-nucleoside inhibitors targeting SARS-CoV-2 RdRp. Biomed. Pharmacother. 2024, 171, 116180. [Google Scholar] [CrossRef] [PubMed]
- Peyambari, M.; Guan, S.; Roossinck, M.J. RdRp or RT, That is the Question. Mol. Biol. Evol. 2021, 38, 5082–5091. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; Li, Y.; Song, X.; Lu, Z.; Zhai, H.; Qiu, H.J.; Sun, Y. Broad-spectrum vaccines against various and evolving viruses: From antigen design to nanoparticle delivery. J. Virol. 2025, 99, e0099725. [Google Scholar] [CrossRef]
- Malik, J.A.; Mulla, A.H.; Farooqi, T.; Pottoo, F.H.; Anwar, S.; Rengasamy, K.R.R. Targets and strategies for vaccine development against SARS-CoV-2. Biomed. Pharmacother. 2021, 137, 111254. [Google Scholar] [CrossRef]
- Zhou, Z.; Zhu, Y.; Chu, M. Role of COVID-19 Vaccines in SARS-CoV-2 Variants. Front. Immunol. 2022, 13, 898192. [Google Scholar] [CrossRef]
- Fiolet, T.; Kherabi, Y.; MacDonald, C.J.; Ghosn, J.; Peiffer-Smadja, N. Comparing COVID-19 vaccines for their characteristics, efficacy and effectiveness against SARS-CoV-2 and variants of concern: A narrative review. Clin. Microbiol. Infect. 2022, 28, 202–221. [Google Scholar] [CrossRef]
- Cao, Y.; Jian, F.; Wang, J.; Yu, Y.; Song, W.; Yisimayi, A.; Wang, J.; An, R.; Chen, X.; Zhang, N.; et al. Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. Nature 2023, 614, 521–529. [Google Scholar] [CrossRef]
- Focosi, D.; Quiroga, R.; McConnell, S.; Johnson, M.C.; Casadevall, A. Convergent Evolution in SARS-CoV-2 Spike Creates a Variant Soup from Which New COVID-19 Waves Emerge. Int. J. Mol. Sci. 2023, 24, 2264. [Google Scholar] [CrossRef]
- Bouhaddou, M.; Reuschl, A.K.; Polacco, B.J.; Thorne, L.G.; Ummadi, M.R.; Ye, C.; Rosales, R.; Pelin, A.; Batra, J.; Jang, G.M.; et al. SARS-CoV-2 variants evolve convergent strategies to remodel the host response. Cell 2023, 186, 4597–4614.e26. [Google Scholar] [CrossRef]
- Focosi, D.; Spezia, P.G.; Maggi, F. Subsequent Waves of Convergent Evolution in SARS-CoV-2 Genes and Proteins. Vaccines 2024, 12, 887. [Google Scholar] [CrossRef] [PubMed]
- Tsai, K.C.; Lee, Y.C.; Tseng, T.S. Comprehensive Deep Mutational Scanning Reveals the Immune-Escaping Hotspots of SARS-CoV-2 Receptor-Binding Domain Targeting Neutralizing Antibodies. Front. Microbiol. 2021, 12, 698365. [Google Scholar] [CrossRef]
- Fung, K.M.; Lai, S.J.; Lin, T.L.; Tseng, T.S. Antigen-Antibody Complex-Guided Exploration of the Hotspots Conferring the Immune-Escaping Ability of the SARS-CoV-2 RBD. Front. Mol. Biosci. 2022, 9, 797132. [Google Scholar] [CrossRef]
- Yao, Z.; Zhang, L.; Duan, Y.; Tang, X.; Lu, J. Molecular insights into the adaptive evolution of SARS-CoV-2 spike protein. J. Infect. 2024, 88, 106121. [Google Scholar] [CrossRef]
- Pacchiarini, N.; Cronin, M.; Sawyer, C.; Williams, C.; Beazer, A.; Cottrell, S.; Morgan, M.; Saunders, V.; Moore, C.; Connor, T.R.; et al. Novel recombinant SARS-CoV-2 lineage detected through genomic surveillance in Wales, UK. Microb. Genom. 2023, 9, mgen000984. [Google Scholar] [CrossRef]
- Ryder, R.; Smith, E.; Borthwick, D.; Elder, J.; Panditrao, M.; Morales, C.; Wadford, D.A. Emergence of Recombinant SARS-CoV-2 Variants in California from 2020 to 2022. Viruses 2024, 16, 1209. [Google Scholar] [CrossRef]
- Dhawan, M.; Saied, A.A.; Mitra, S.; Alhumaydhi, F.A.; Emran, T.B.; Wilairatana, P. Omicron variant (B.1.1.529) and its sublineages: What do we know so far amid the emergence of recombinant variants of SARS-CoV-2? Biomed. Pharmacother. 2022, 154, 113522. [Google Scholar] [CrossRef]
- Parums, D.V. Editorial: World Health Organization (WHO) Variants of Concern Lineages Under Monitoring (VOC-LUM) in Response to the Global Spread of Lineages and Sublineages of Omicron, or B.1.1.529, SARS-CoV-2. Med. Sci. Monit. 2022, 28, e937676. [Google Scholar] [CrossRef] [PubMed]
- Tracking SARS-CoV-2 Variants. Available online: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants (accessed on 31 May 2021).
- Khare, S.; Gurry, C.; Freitas, L.; Schultz, M.B.; Bach, G.; Diallo, A.; Akite, N.; Ho, J.; Lee, R.T.; Yeo, W.; et al. GISAID’s Role in Pandemic Response. China CDC Wkly. 2021, 3, 1049–1051. [Google Scholar] [CrossRef] [PubMed]
- Cascella, M.; Rajnik, M.; Aleem, A.; Dulebohn, S.C.; Di Napoli, R. Features, Evaluation, and Treatment of Coronavirus (COVID-19). In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]
- Sarkar, M.; Madabhavi, I. COVID-19 mutations: An overview. World J. Methodol. 2024, 14, 89761. [Google Scholar] [CrossRef]
- Sanches, P.R.S.; Charlie-Silva, I.; Braz, H.L.B.; Bittar, C.; Freitas Calmon, M.; Rahal, P.; Cilli, E.M. Recent advances in SARS-CoV-2 Spike protein and RBD mutations comparison between new variants Alpha (B.1.1.7, United Kingdom), Beta (B.1.351, South Africa), Gamma (P.1, Brazil) and Delta (B.1.617.2, India). J. Virus Erad. 2021, 7, 100054. [Google Scholar] [CrossRef]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2013, 41, D36–D42. [Google Scholar] [CrossRef] [PubMed]
- Kaku, Y.; Okumura, K.; Padilla-Blanco, M.; Kosugi, Y.; Uriu, K.; Hinay, A.A., Jr.; Chen, L.; Plianchaisuk, A.; Kobiyama, K.; Ishii, K.J.; et al. Virological characteristics of the SARS-CoV-2 JN.1 variant. Lancet Infect. Dis. 2024, 24, e82. [Google Scholar] [CrossRef]
- Uriu, K.; Okumura, K.; Uwamino, Y.; Chen, L.; Tolentino, J.E.; Asakura, H.; Nagashima, M.; Sadamasu, K.; Yoshimura, K.; Ito, J.; et al. Virological characteristics of the SARS-CoV-2 NB.1.8.1 variant. Lancet Infect. Dis. 2025, 25, e443. [Google Scholar] [CrossRef]
- Eshraghi, R.; Bahrami, A.; Karimi Houyeh, M.; Nasr Azadani, M. JN.1 and the ongoing battle: Unpacking the characteristics of a new dominant COVID-19 variant. Pathog. Glob. Health 2024, 118, 453–458. [Google Scholar] [CrossRef]
- Hemo, M.K.; Islam, M.A. JN.1 as a new variant of COVID-19—Editorial. Ann. Med. Surg. 2024, 86, 1833–1835. [Google Scholar] [CrossRef]
- Yang, S.; Yu, Y.; Xu, Y.; Jian, F.; Song, W.; Yisimayi, A.; Wang, P.; Wang, J.; Liu, J.; Yu, L.; et al. Fast evolution of SARS-CoV-2 BA.2.86 to JN.1 under heavy immune pressure. Lancet Infect. Dis. 2024, 24, e70–e72. [Google Scholar] [CrossRef]
- Tamura, T.; Mizuma, K.; Nasser, H.; Deguchi, S.; Padilla-Blanco, M.; Oda, Y.; Uriu, K.; Tolentino, J.E.M.; Tsujino, S.; Suzuki, R.; et al. Virological characteristics of the SARS-CoV-2 BA.2.86 variant. Cell Host Microbe 2024, 32, 170–180.e12. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhao, X.; Shi, J.; Wang, Y.; Liu, H.; Hu, Y.F.; Hu, B.; Shuai, H.; Yuen, T.T.; Chai, Y.; et al. Lineage-specific pathogenicity, immune evasion, and virological features of SARS-CoV-2 BA.2.86/JN.1 and EG.5.1/HK.3. Nat. Commun. 2024, 15, 8728. [Google Scholar] [CrossRef] [PubMed]
- Satapathy, P.; Kumar, P.; Gupta, J.K.; Rabaan, A.A.; Al Kaabi, N.A.; Mohanty, D.; Naveen, P.; Khatib, M.N.; Gaidhane, S.; Zahiruddin, Q.S.; et al. The emergence and implications of SARS-CoV-2 omicron subvariant BA.2.86 on global health. Int. J. Surg. 2024, 110, 2498–2501. [Google Scholar] [CrossRef]
- Kamble, P.; Daulatabad, V.; Singhal, A.; Ahmed, Z.S.; Choubey, A.; Bhargava, S.; John, N.A. JN.1 variant in enduring COVID-19 pandemic: Is it a variety of interest (VoI) or variety of concern (VoC)? Horm. Mol. Biol. Clin. Investig. 2024, 45, 49–53. [Google Scholar] [CrossRef]
- Chakraborty, C.; Bhattacharya, M. FLip mutations (L455F + F456L) in newly emerging VOI, JN.1: Its antibody and immune escape. Int. Immunopharmacol. 2024, 133, 112146. [Google Scholar] [CrossRef]
- Yang, H.; Guo, H.; Wang, A.; Cao, L.; Fan, Q.; Jiang, J.; Wang, M.; Lin, L.; Ge, X.; Wang, H.; et al. Structural basis for the evolution and antibody evasion of SARS-CoV-2 BA.2.86 and JN.1 subvariants. Nat. Commun. 2024, 15, 7715. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Shi, K.; Gu, Y.; Xu, Z.; Shu, C.; Li, D.; Sun, J.; Cong, M.; Li, X.; Zhao, X.; et al. Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants. Structure 2024, 32, 1055–1067.e6. [Google Scholar] [CrossRef]
- Kaku, Y.; Uriu, K.; Kosugi, Y.; Okumura, K.; Yamasoba, D.; Uwamino, Y.; Kuramochi, J.; Sadamasu, K.; Yoshimura, K.; Asakura, H.; et al. Virological characteristics of the SARS-CoV-2 KP.2 variant. Lancet Infect. Dis. 2024, 24, e416. [Google Scholar] [CrossRef]
- Kaku, Y.; Uriu, K.; Okumura, K.; Ito, J.; Sato, K. Virological characteristics of the SARS-CoV-2 KP.3.1.1 variant. Lancet Infect. Dis. 2024, 24, e609. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, L.; Zamora, J.L.R.; Han, D.; Moshiri, J.; Peinovich, N.; Martinez, C.; Ho, P.Y.; Li, J.; Aeschbacher, T.; Martin, R.; et al. Remdesivir and Obeldesivir Retain Potent Antiviral Activity Against SARS-CoV-2 Omicron Variants. Viruses 2025, 17, 168. [Google Scholar] [CrossRef] [PubMed]
- Kaku, Y.; Yo, M.S.; Tolentino, J.E.; Uriu, K.; Okumura, K.; Ito, J.; Sato, K. Virological characteristics of the SARS-CoV-2 KP.3, LB.1, and KP.2.3 variants. Lancet Infect. Dis. 2024, 24, e482–e483. [Google Scholar] [CrossRef]
- Yang, J.; He, X.; Shi, H.; He, C.; Lei, H.; He, H.; Yang, L.; Wang, W.; Shen, G.; Yang, J.; et al. Recombinant XBB.1.5 boosters induce robust neutralization against KP.2- and KP.3-included JN.1 sublineages. Signal Transduct. Target. Ther. 2025, 10, 47. [Google Scholar] [CrossRef]
- Hansen, C.H.; Lassaunière, R.; Rasmussen, M.; Moustsen-Helms, I.R.; Valentiner-Branth, P. Effectiveness of the BNT162b2 and mRNA-1273 JN.1-adapted vaccines against COVID-19-associated hospitalisation and death: A Danish, nationwide, register-based, cohort study. Lancet Infect. Dis. 2025. [Google Scholar] [CrossRef]
- Lewnard, J.A.; Mahale, P.; Malden, D.; Hong, V.; Ackerson, B.K.; Lewin, B.J.; Link-Gelles, R.; Feldstein, L.R.; Lipsitch, M.; Tartof, S.Y. Immune escape and attenuated severity associated with the SARS-CoV-2 BA.2.86/JN.1 lineage. Nat. Commun. 2024, 15, 8550. [Google Scholar] [CrossRef]
- Naveed Siddiqui, A.; Musharaf, I.; Gulumbe, B.H. The JN.1 variant of COVID-19: Immune evasion, transmissibility, and implications for global health. Ther. Adv. Infect. Dis. 2025, 12, 20499361251314763. [Google Scholar] [CrossRef]
- Markov, P.V.; Ghafari, M.; Beer, M.; Lythgoe, K.; Simmonds, P.; Stilianakis, N.I.; Katzourakis, A. The evolution of SARS-CoV-2. Nat. Rev. Microbiol. 2023, 21, 361–379. [Google Scholar] [CrossRef]
- Patterson, L.D.; Elsharkawy, A.; Jahantigh, H.R.; Nabi, Z.; Stone, S.; Kumar, M. Pathogenicity of SARS-CoV-2 Omicron Subvariants JN.1, KP.2, and EG.5.1 in K18-hACE2 Transgenic Mice. Viruses 2025, 17, 1177. [Google Scholar] [CrossRef]
- Jian, F.; Wang, J.; Yisimayi, A.; Song, W.; Xu, Y.; Chen, X.; Niu, X.; Yang, S.; Yu, Y.; Wang, P.; et al. Evolving antibody response to SARS-CoV-2 antigenic shift from XBB to JN.1. Nature 2025, 637, 921–929. [Google Scholar] [CrossRef]
- Malay, S.; Madabhavi, I.V.; Tripathi, A. SARS-CoV-2 JN.1 variant: A short review. Monaldi Arch. Chest Dis. 2024. [Google Scholar] [CrossRef]
- Hu, Y.; Zou, J.; Nguyen, M.D.; Chang, H.C.; Yeung, J.; Hao, H.; Shi, P.Y.; Ren, P.; Xie, X. Comparative analysis of replication and immune evasion among SARS-CoV-2 subvariants BA.2.86, JN.1, KP.2, and KP.3. mBio 2025, 16, e0350324. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Mellis, I.A.; Ho, J.; Bowen, A.; Kowalski-Dobson, T.; Valdez, R.; Katsamba, P.S.; Wu, M.; Lee, C.; Shapiro, L.; et al. Recurrent SARS-CoV-2 spike mutations confer growth advantages to select JN.1 sublineages. Emerg. Microbes Infect. 2024, 13, 2402880. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Huang, J.; Baboo, S.; Diedrich, J.K.; Bangaru, S.; Paulson, J.C.; Yates, J.R., 3rd; Yuan, M.; Wilson, I.A.; Ward, A.B. Structural and functional insights into the evolution of SARS-CoV-2 KP.3.1.1 spike protein. Cell Rep. 2025, 44, 115941. [Google Scholar] [CrossRef]
- Scarpa, F.; Branda, F.; Ceccarelli, G.; Romano, C.; Locci, C.; Pascale, N.; Azzena, I.; Fiori, P.L.; Casu, M.; Pascarella, S.; et al. SARS-CoV-2 XEC: A Genome-Based Survey. Microorganisms 2025, 13, 253. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Kempf, A.; Nehlmeier, I.; Chen, N.; Graichen, L.; Moldenhauer, A.S.; Stankov, M.V.; Happle, C.; Schulz, S.R.; Dopfer-Jablonka, A.; et al. Host cell entry efficiency and neutralization sensitivity of the SARS-CoV-2 MC.10.1 variant. Virology 2025, 612, 110675. [Google Scholar] [CrossRef]
- Zhang, Y.; He, X.; Zhai, J.; Ji, B.; Man, V.H.; Wang, J. In silico binding profile characterization of SARS-CoV-2 spike protein and its mutants bound to human ACE2 receptor. Brief. Bioinform. 2021, 22, bbab188. [Google Scholar] [CrossRef]
- Qu, P.; Xu, K.; Faraone, J.N.; Goodarzi, N.; Zheng, Y.M.; Carlin, C.; Bednash, J.S.; Horowitz, J.C.; Mallampalli, R.K.; Saif, L.J.; et al. Immune evasion, infectivity, and fusogenicity of SARS-CoV-2 BA.2.86 and FLip variants. Cell 2024, 187, 585–595.e6. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Yu, Y.; Liu, J.; Jian, F.; Yang, S.; Song, W.; Yu, L.; Shao, F.; Cao, Y. Antigenic and virological characteristics of SARS-CoV-2 variants BA.3.2, XFG, and NB.1.8.1. Lancet Infect. Dis. 2025, 25, e374–e377. [Google Scholar] [CrossRef] [PubMed]
- Mellis, I.A.; Wu, M.; Hong, H.; Tzang, C.C.; Bowen, A.; Wang, Q.; Gherasim, C.; Pierce, V.M.; Shah, J.G.; Purpura, L.J.; et al. Antibody evasion and receptor binding of SARS-CoV-2 LP.8.1.1, NB.1.8.1, XFG, and related subvariants. Cell Rep. 2025, 44, 116440. [Google Scholar] [CrossRef]
- Abbad, A.; Lerman, B.; Ehrenhaus, J.; Monahan, B.; Singh, G.; Wilson, A.; Slamanig, S.; Aracena, A.; Lyttle, N.; Nardulli, J.; et al. Antibody responses to SARS-CoV-2 variants LP.8.1, LF.7.1, NB.1.8.1, XFG and BA.3.2 following KP.2 monovalent mRNA vaccination. medRxiv 2025. [Google Scholar] [CrossRef]
- Gong, T.; Zhang, X.; Lin, H.; Li, J.; Tao, J.; Zeng, T.; Ren, X.; Xie, Z.; Lei, X.; Zhang, S.; et al. Mutation profiling, evolution analysis, molecular dynamics simulation, and functional characterization of Omicron sub-strains. Virus Res. 2025, 360, 199626. [Google Scholar] [CrossRef] [PubMed]
- Aljabali, A.A.A.; Lundstrom, K.; Hromić-Jahjefendić, A.; El-Baky, N.A.; Nawn, D.; Hassan, S.S.; Rubio-Casillas, A.; Redwan, E.M.; Uversky, V.N. The XEC Variant: Genomic Evolution, Immune Evasion, and Public Health Implications. Viruses 2025, 17, 985. [Google Scholar] [CrossRef]
- Malone, B.; Urakova, N.; Snijder, E.J.; Campbell, E.A. Structures and functions of coronavirus replication-transcription complexes and their relevance for SARS-CoV-2 drug design. Nat. Rev. Mol. Cell Biol. 2022, 23, 21–39. [Google Scholar] [CrossRef]
- Sáenz Altamirano, E.E.; Huang, C.H.; Tsai, Y.L.; Tzeng, T.J.; Borges Teruel, N.F.; Pandey, I.; Oswal, N.; Chen, Y.G.; Chang, C.W.; Rajsbaum, R.; et al. Coronavirus NSP14 Drives Internal m 7G Modification to Rewire Host Splicing and Promote Viral Replication. bioRxiv 2025. [Google Scholar] [CrossRef]
- Ru, Y.; Ma-Lauer, Y.; Xiang, C.; Li, P.; von Brunn, B.; Richter, A.; Drosten, C.; Pichlmair, A.; Pfefferle, S.; Klein, M.; et al. LMP7 as a Target for Coronavirus Therapy: Inhibition by Ixazomib and Interaction with SARS-CoV-2 Proteins Nsp13 and Nsp16. Pathogens 2025, 14, 871. [Google Scholar] [CrossRef] [PubMed]
- Lei, F.; Shu, J.; Xia, C.; Lei, Y.; Zhang, Y.; Zhang, Y.; Yuan, Z.; Yi, Z. Viral determinants of cis- and trans-cleavage by SARS-CoV-2 Nsp3 and an on-off reporter for monitoring intracellular protease activity. Antivir. Res. 2025, 242, 106262. [Google Scholar] [CrossRef] [PubMed]
- Arya, R.; Ganesh, J.; Prashar, V.; Kumar, M. Structural and functional insights into Ubl domain-mediated regulation of SARS-CoV-2 PLpro. Biol. Direct 2025, 20, 102. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Pan, Q.; Xiao, H.; Pan, Q. Therapeutic potential of targeting ORF6 in persistent SARS-CoV-2 infections. Lancet Microbe 2025, 101253. [Google Scholar] [CrossRef]
- González-Vázquez, L.D.; Iglesias-Rivas, P.; Ferreiro, D.; Arenas, M. Heterogeneous Evolution Among SARS-CoV-2 Genes and Variants of Concern. J. Med. Virol. 2025, 97, e70604. [Google Scholar] [CrossRef]
- Lokugamage, K.G.; Zhou, Y.; Alvarado, R.E.; Plante, J.A.; Ahearn, Y.; Chen, J.; Estes, L.; Meyers, W.; Nilsson, J.; Routh, A.L.; et al. Convergent evolution in nucleocapsid facilitated SARS-CoV-2 adaptation for human infection. J. Virol. 2025, 99, e0209124. [Google Scholar] [CrossRef]
- Ferdoush, J.; Abdul Kadir, R.; Simay Kaplanoglu, S.; Osborn, M. SARS-CoV-2 and UPS with potentials for therapeutic interventions. Gene 2024, 912, 148377. [Google Scholar] [CrossRef]
- Tsujino, S.; Tsuda, M.; Nao, N.; Okumura, K.; Wang, L.; Oda, Y.; Mimura, Y.; Li, J.; Hashimoto, R.; Matsumura, Y.; et al. Evolution of BA.2.86 to JN.1 reveals that functional changes in non-structural viral proteins are required for fitness of SARS-CoV-2. J. Virol. 2025, 99, e0090825. [Google Scholar] [CrossRef]
- Tsujino, S.; Tsuda, M.; Ito, J.; Deguchi, S.; Taha, T.Y.; Nasser, H.; Wang, L.; Rosecrans, J.; Suzuki, R.; Suzuki, S.; et al. A non-spike nucleocapsid R204P mutation in SARS-CoV-2 Omicron XEC enhances inflammation and pathogenicity. bioRxiv 2025. [Google Scholar] [CrossRef]
- Hong Kong China News Agency. Taiwanese Experts are Concerned About the Increased Transmissibility of New COVID-19 Variants. Available online: https://hkcna.hk/docDetail.jsp?channel=2803&id=101013530 (accessed on 31 May 2025).
- Branda, F.; Ciccozzi, M.; Scarpa, F. Genome-based analyses of SARS-CoV-2 NB.1.8.1 variant reveals its low potential. Infect. Dis. 2025, 57, 805–808. [Google Scholar] [CrossRef]
- Shu, Y.; McCauley, J. GISAID: Global initiative on sharing all influenza data—From vision to reality. Eurosurveillance 2017, 22, 30494. [Google Scholar] [CrossRef] [PubMed]
- WHO. Risk Evaluation for SARS-CoV-2 Variant Under Monitoring: NB.1.8.1. Available online: https://www.who.int/publications/m/item/risk-evaluation-for-sars-cov-2-variant-under-monitoring-nb.1.8.1 (accessed on 23 May 2025).
- Chinese Center for Disease Control and Prevention. National COVID-19 Infection Status. Available online: https://www.chinacdc.cn/jksj/xgbdyq/ (accessed on 29 September 2025).
- Chinese Center for Disease Control and Prevention. Symptoms of COVID-19. Available online: https://www.cdc.gov/covid/signs-symptoms/index.html (accessed on 29 September 2025).
- Ochani, R.; Asad, A.; Yasmin, F.; Shaikh, S.; Khalid, H.; Batra, S.; Sohail, M.R.; Mahmood, S.F.; Ochani, R.; Hussham Arshad, M.; et al. COVID-19 pandemic: From origins to outcomes. A comprehensive review of viral pathogenesis, clinical manifestations, diagnostic evaluation, and management. Infez. Med. 2021, 29, 20–36. [Google Scholar]
- Focosi, D.; Franchini, M.; Maggi, F.; Shoham, S. COVID-19 therapeutics. Clin. Microbiol. Rev. 2024, 37, e0011923. [Google Scholar] [CrossRef]
- Ahmad, F.; Basharat, Z.; Janjua, A.; Najmi, M.H.; Almajhdi, F.N.; Hussain, T.; Ozsahin, D.U.; Waheed, Y. An exploratory binding study of molnupiravir efficacy against emerging Omicron SARS-CoV-2 variants. Sci. Rep. 2025, 15, 36549. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.H.; Lee, E.J.; No, J.S.; Noh, J.Y.; Lee, C.Y.; O, S.W.; Choi, Y.J.; Kim, J.A.; An, B.M.; Nam, J.H.; et al. Long-Term Genomic Surveillance and Immune Escape of SARS-CoV-2 in the Republic of Korea, with a Focus on JN.1-Derived Variants. Viruses 2025, 17, 1202. [Google Scholar] [CrossRef]
- Kobayashi, M.; Kakimoto, K.; Yahata, Y.; Kobayashi, Y.; Nagai, H.; Tanikake, C.; Fukumura, K.; Date, K.; Murata, H.; Kitagawa, S.; et al. Enhanced case finding and self-isolation measures in the early phase of SARS-CoV-2 Omicron transmission, Osaka, Japan, December 2021–January 2022. West. Pac. Surveill. Response J. 2025, 16, 1–10. [Google Scholar] [CrossRef]
- Correia, T.; Buissonnière, M.; McKee, M. The Pandemic Agreement: What’s Next? Int. J. Health Plan. Manag. 2025, 40, 1029–1032. [Google Scholar] [CrossRef]
- Mura, M.; Trignol, A.; Le Dault, E.; Tournier, J.N. Lessons for medical countermeasure development from unforeseen outbreaks. Emerg. Microbes Infect. 2025, 14, 2471035. [Google Scholar] [CrossRef]
- Seki, N.; Mikami, A.; Kokufu, T.; Kusaka, T.; Yamaguchi, K.; Takahashi, C.; Izuno, T.; Saito, T. Impact of border control measures on public health center operations and staffing at international airports during the emergence of the SARS-CoV-2 B.1.1.529 (Omicron) variant of concern. Jpn. J. Public Health 2025, 71, 775–786. [Google Scholar]
- El-Haddad, K.; Liu, W.; Burke, P.; Wang, H.; Esper, F.P. Changes to Endemic Respiratory Virus Circulation and Testing Before, During, and After the COVID-19 Pandemic. Open Forum Infect. Dis. 2025, 12, ofaf493. [Google Scholar] [CrossRef]
- Mohamadian, M.; Chiti, H.; Shoghli, A.; Biglari, S.; Parsamanesh, N.; Esmaeilzadeh, A. COVID-19: Virology, biology and novel laboratory diagnosis. J. Gene Med. 2021, 23, e3303. [Google Scholar] [CrossRef] [PubMed]
- Xie, N.N.; Zhang, W.C.; Chen, J.; Tian, F.B.; Song, J.X. Clinical Characteristics, Diagnosis, and Therapeutics of COVID-19: A Review. Curr. Med. Sci. 2023, 43, 1066–1074. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.C.; Webber, A.; Lauring, A.S.; Bendall, E.; Papalambros, L.K.; Safdar, B.; Ginde, A.A.; Peltan, I.D.; Brown, S.M.; Gaglani, M.; et al. Effectiveness of 2024–2025 COVID-19 Vaccination Against COVID-19 Hospitalization and Severe In-Hospital Outcomes—IVY Network, 26 Hospitals, 1 September 2024–30 April 2025. medRxiv 2025. [Google Scholar] [CrossRef]
- Kim, A.H.J.; Nakamura, M.C. COVID-19 Breakthrough Infection Among Immunocompromised Persons. JAMA Intern. Med. 2022, 182, 163–164. [Google Scholar] [CrossRef]
- Smits, P.D.; Gratzl, S.; Simonov, M.; Nachimuthu, S.K.; Goodwin Cartwright, B.M.; Wang, M.D.; Baker, C.; Rodriguez, P.; Bogiages, M.; Althouse, B.M.; et al. Risk of COVID-19 breakthrough infection and hospitalization in individuals with comorbidities. Vaccine 2023, 41, 2447–2455. [Google Scholar] [CrossRef]
- Miyamoto, S.; Suzuki, T. Infection-mediated immune response in SARS-CoV-2 breakthrough infection and implications for next-generation COVID-19 vaccine development. Vaccine 2024, 42, 1401–1406. [Google Scholar] [CrossRef] [PubMed]
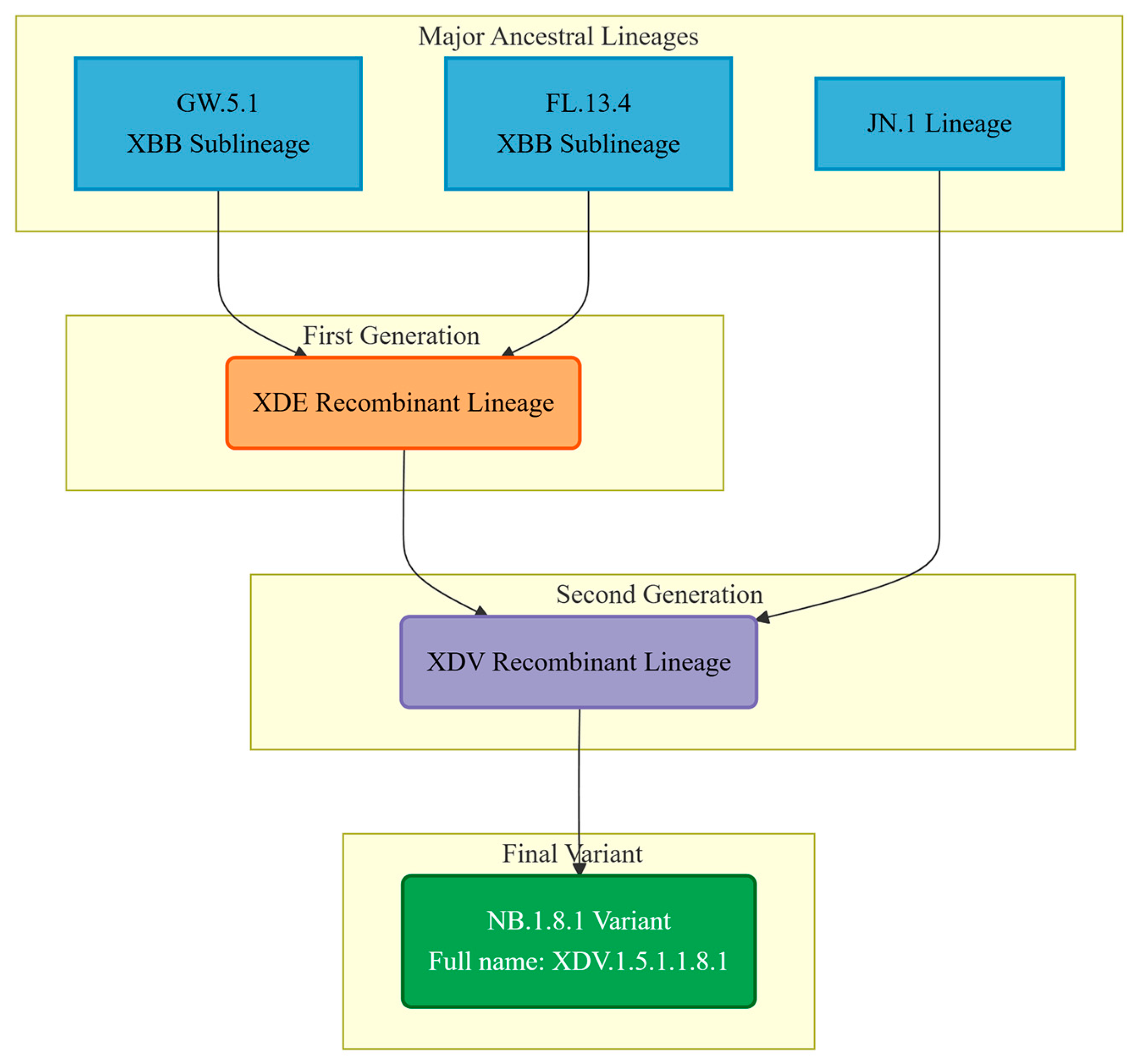
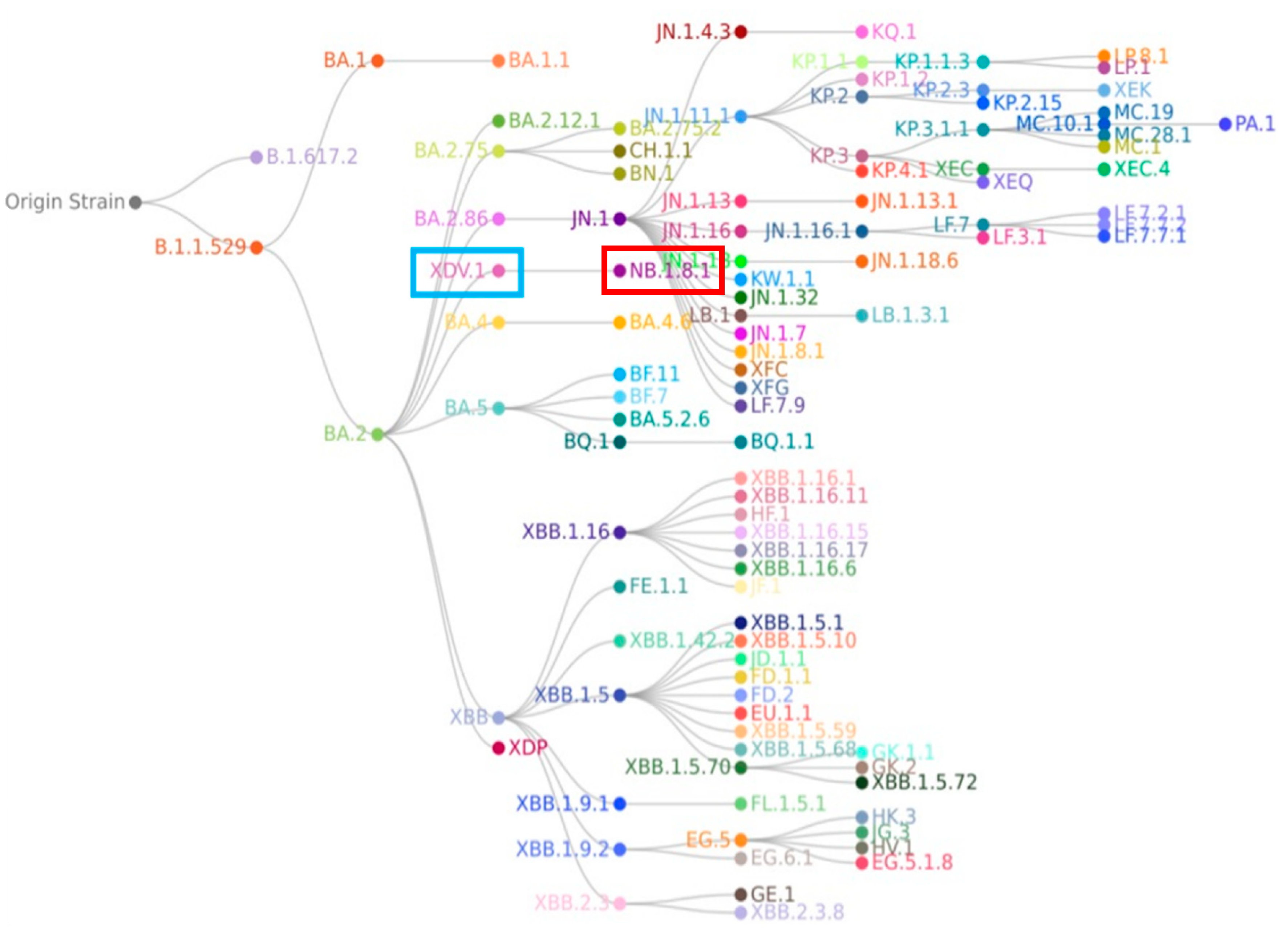
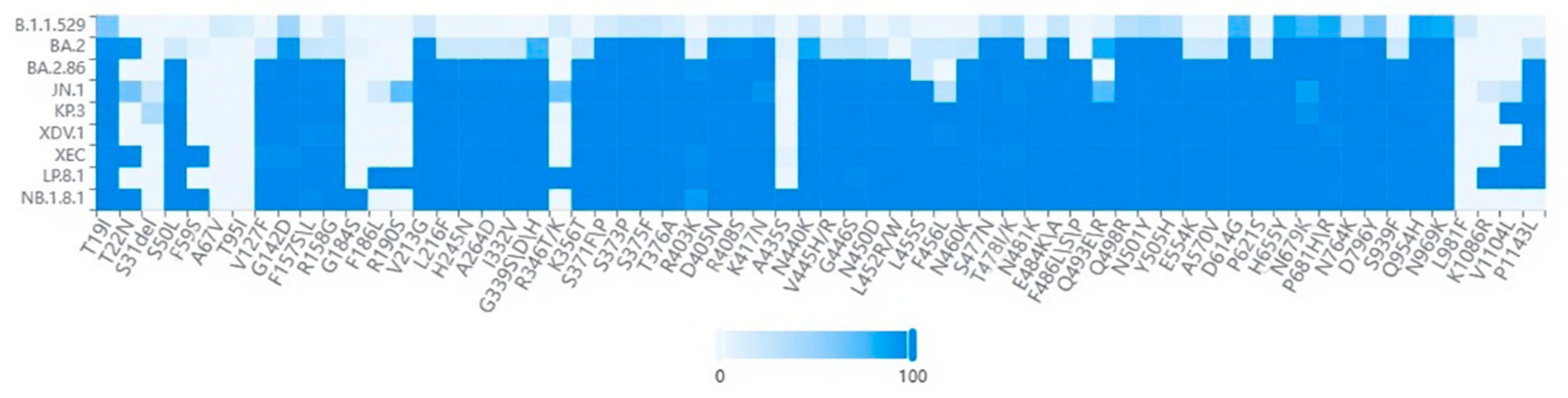
| Risk Assessment Indicators | Assessment Results |
|---|---|
| Disease severity | No substantial change; similar to previously circulating Omicron sub-variants [83,85] |
| ICU admission/hospitalization rate | No significant increase; remained below all preceding peaks [86] |
| Predominant clinical manifestation | Sore throat, fatigue, fever, mild cough, myalgia, and nasal congestion [87,88] |
| Vaccine effectiveness | Current vaccines retain high effectiveness against severe disease [67,68] |
| Antiviral drug efficacy | No well-defined resistance-associated mutations detected [89,90] |
| Transmissibility | Moderate to high [36,66] |
| Immune escape | Moderate [67,91] |
| Health-system impact | Low to moderate [86] |
| Effectiveness of countermeasures | High [92,93,94,95,96] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, G.; Xu, C.; Wang, L.; Chai, K.; Wu, B. Global Surveillance and Biological Characterization of the SARS-CoV-2 NB.1.8.1 Variant: An Emerging VUM Lineage Under Scrutiny. Viruses 2025, 17, 1457. https://doi.org/10.3390/v17111457
Cao G, Xu C, Wang L, Chai K, Wu B. Global Surveillance and Biological Characterization of the SARS-CoV-2 NB.1.8.1 Variant: An Emerging VUM Lineage Under Scrutiny. Viruses. 2025; 17(11):1457. https://doi.org/10.3390/v17111457
Chicago/Turabian StyleCao, Gaojie, Chenhui Xu, Linxi Wang, Keikei Chai, and Beibei Wu. 2025. "Global Surveillance and Biological Characterization of the SARS-CoV-2 NB.1.8.1 Variant: An Emerging VUM Lineage Under Scrutiny" Viruses 17, no. 11: 1457. https://doi.org/10.3390/v17111457
APA StyleCao, G., Xu, C., Wang, L., Chai, K., & Wu, B. (2025). Global Surveillance and Biological Characterization of the SARS-CoV-2 NB.1.8.1 Variant: An Emerging VUM Lineage Under Scrutiny. Viruses, 17(11), 1457. https://doi.org/10.3390/v17111457







