Development of a Highly Specific Monoclonal Antibody-Based Sandwich ELISA for Rapid Detection of Porcine Circovirus Type 3
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells, Vectors, and Animals
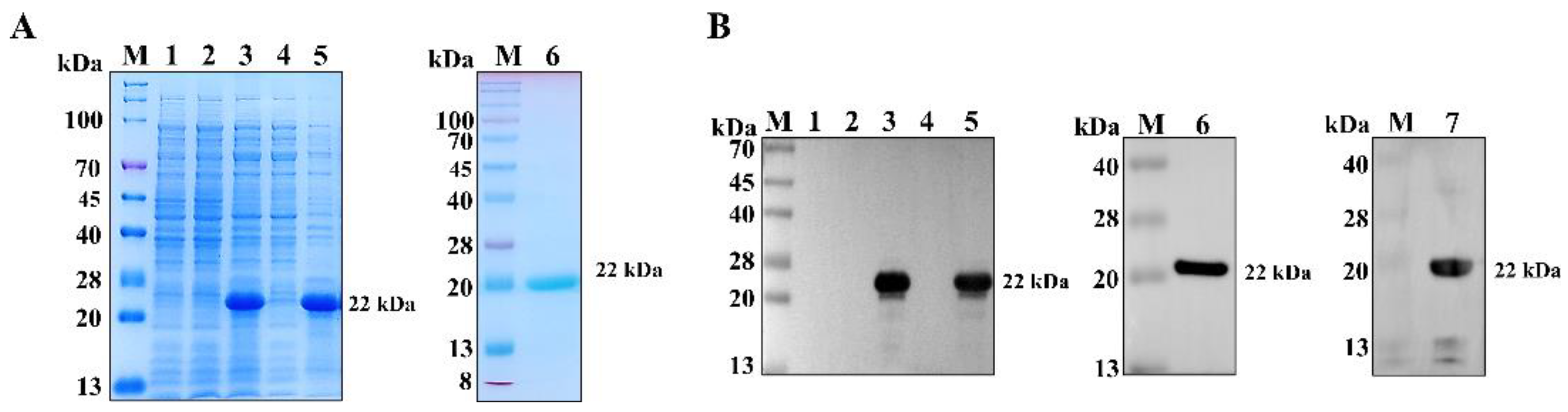
2.2. Expression and Purification of PCV3 Cap Protein
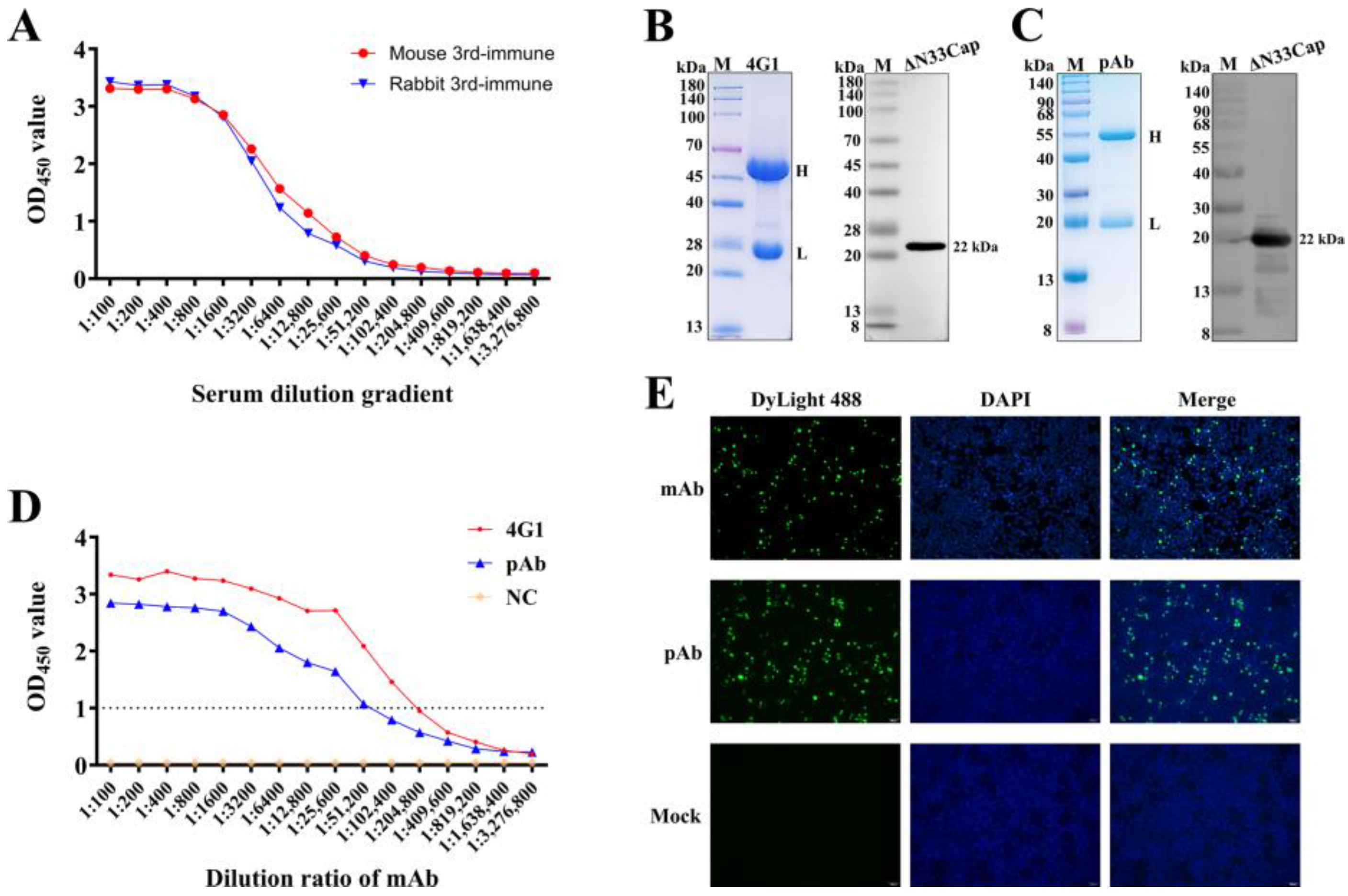
2.3. Production of Anti-PCV3 Cap Antibodies
2.4. Characterization of Anti-PCV3 Cap mAb and pAb
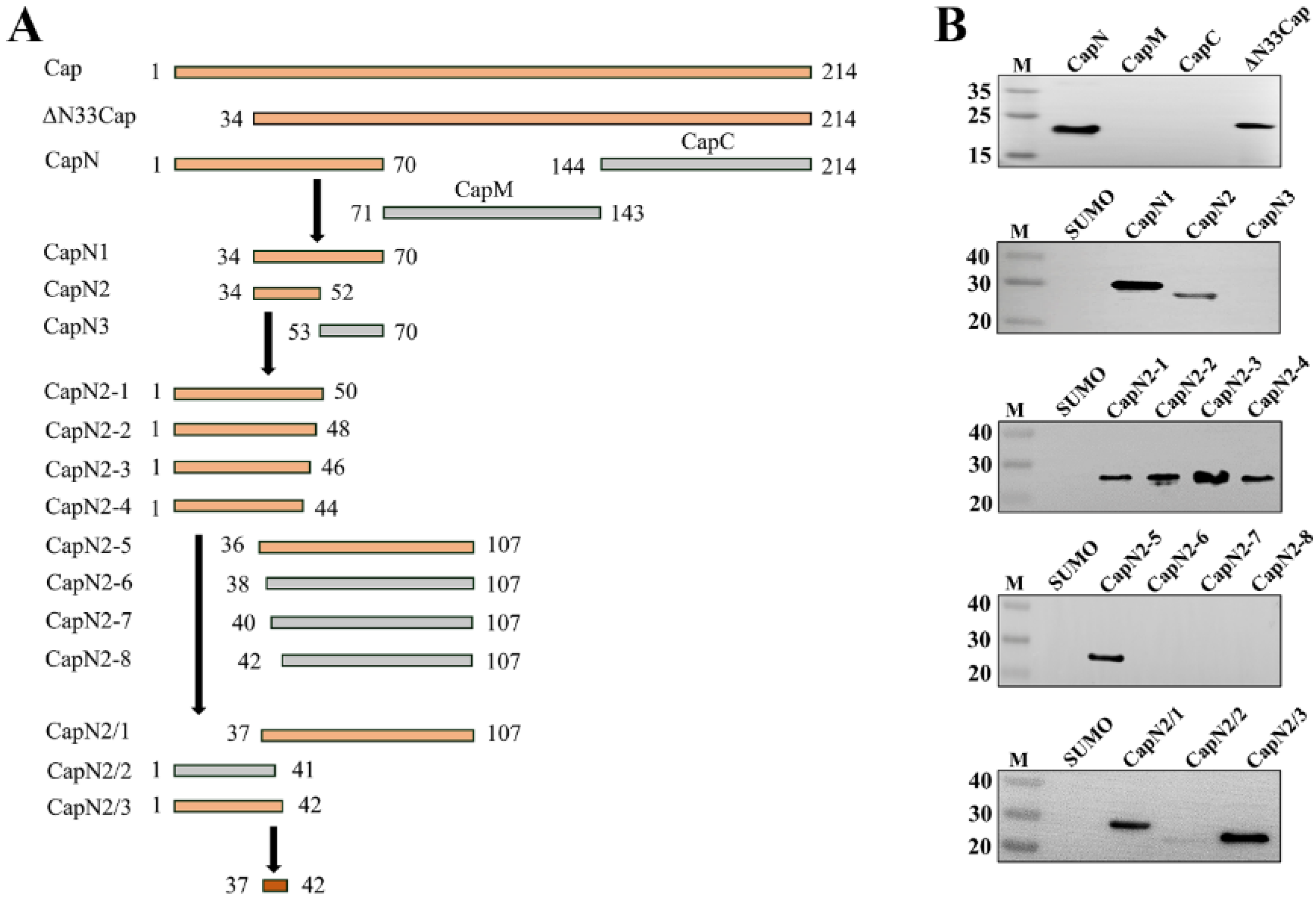
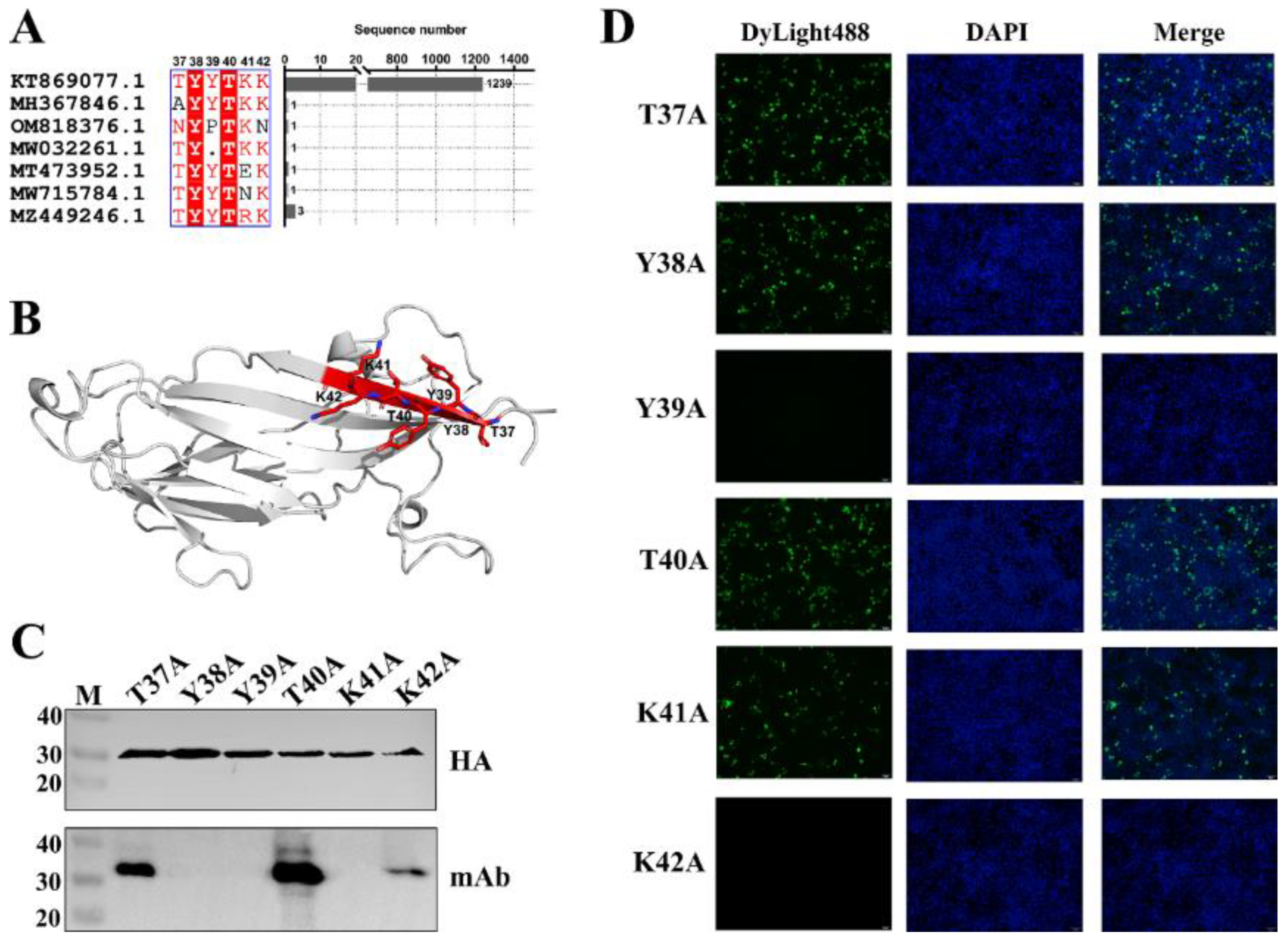
2.5. Fine Mapping of Epitope Targeted by mAb
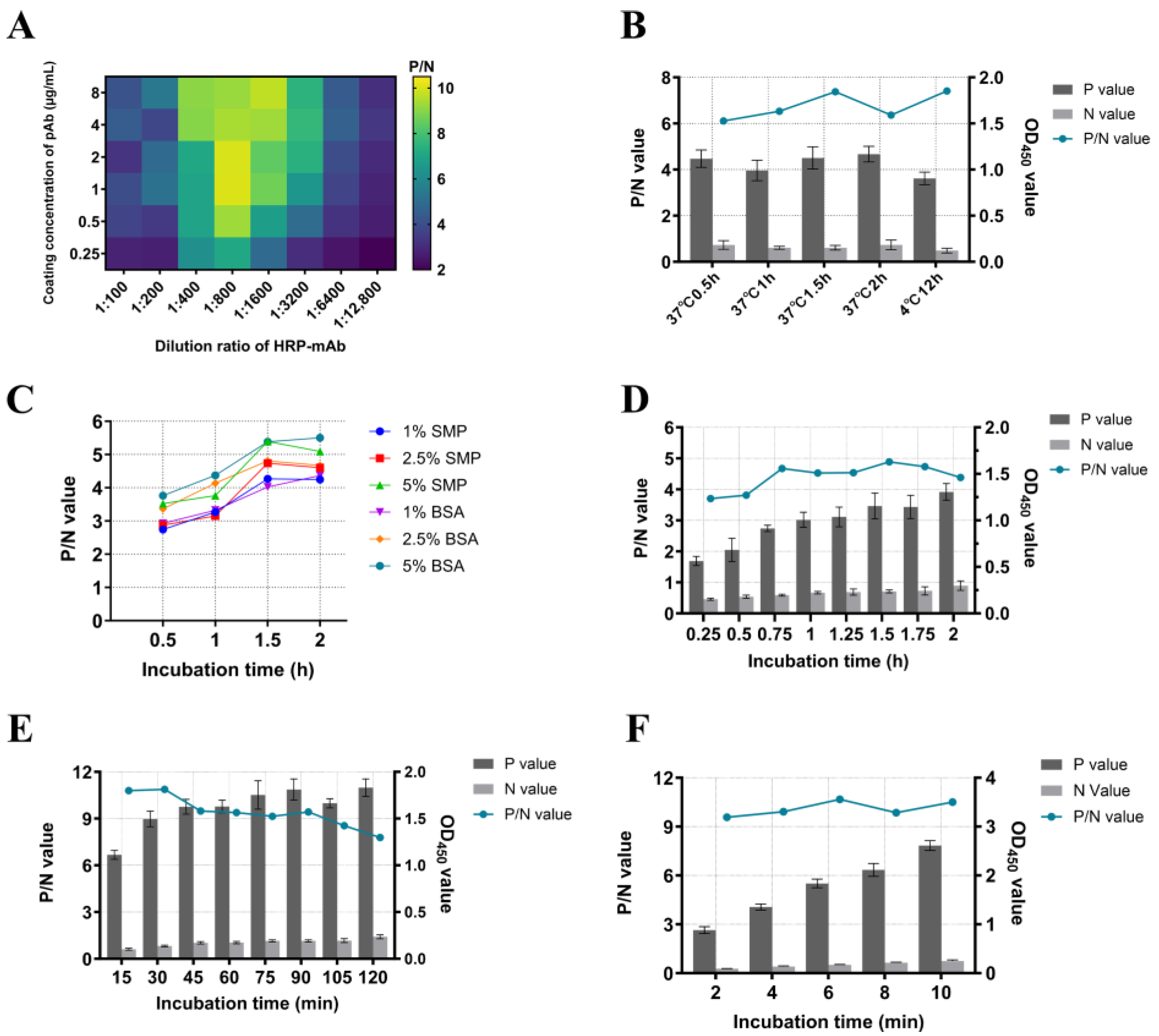
2.6. Optimization of DAS-ELISA Procedure
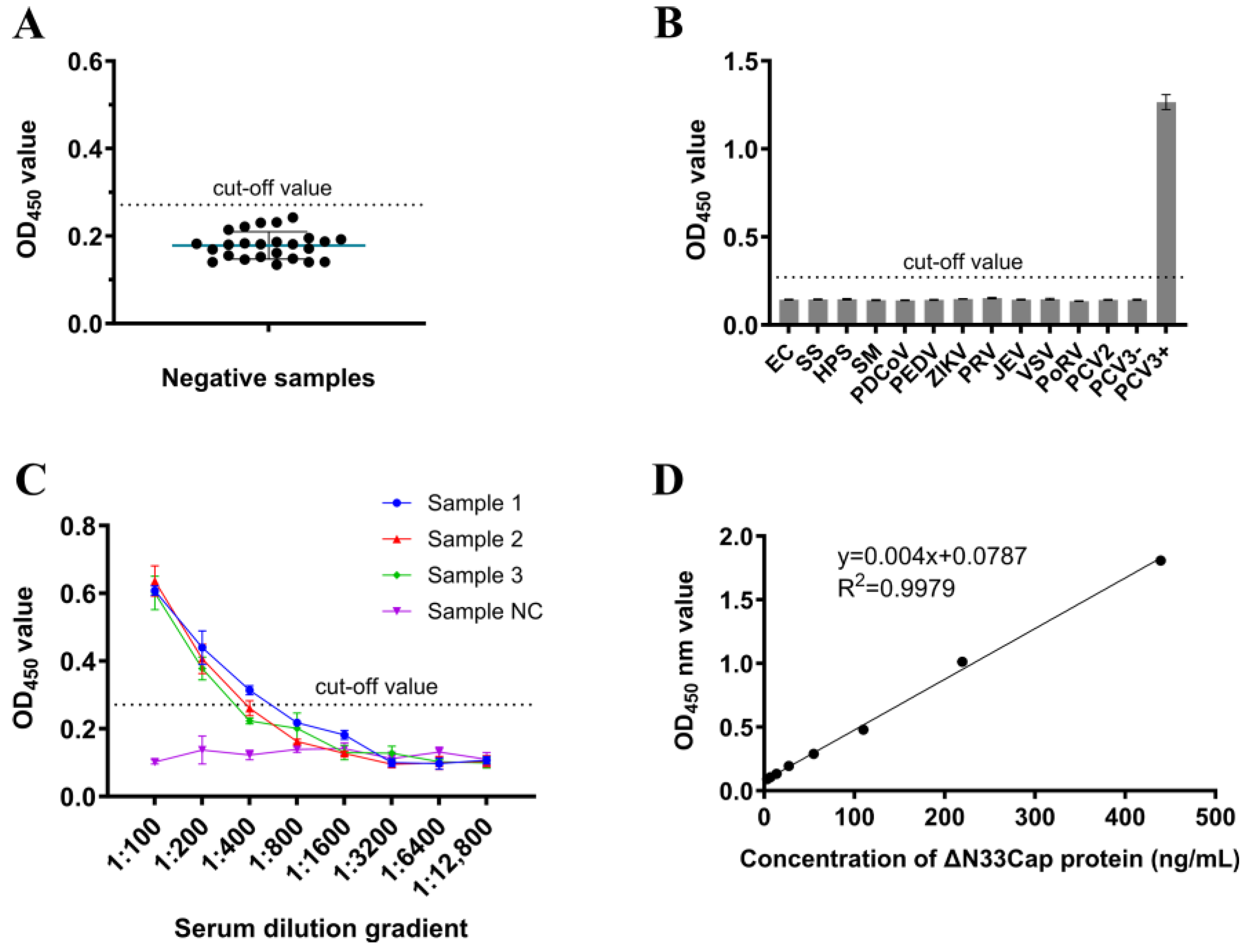
2.7. Performance Verification of DAS-ELISA
2.8. Statistical Analysis
3. Results
3.1. PCV3 Cap Antigen Preparation
3.2. Preparation and Characterization of mAb and pAb Against PCV3 Cap
3.3. Fine Mapping of Epitope Targeted by mAb Against PCV3 Cap
3.4. Optimal DAS-ELISA Procedure
3.5. Evaluation of DAS-ELISA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- da Silva, R.R.; da Silva, D.F.; da Silva, V.H.; de Castro, A. Porcine circovirus 3: A new challenge to explore. Front. Vet. Sci. 2023, 10, 1266499. [Google Scholar] [CrossRef]
- Varsani, A.; Harrach, B.; Roumagnac, P.; Benko, M.; Breitbart, M.; Delwart, E.; Franzo, G.; Kazlauskas, D.; Rosario, K.; Segalés, J.; et al. 2024 taxonomy update for the family Circoviridae. Arch. Virol. 2024, 169, 176. [Google Scholar] [CrossRef]
- Phan, T.G.; Giannitti, F.; Rossow, S.; Marthaler, D.; Knutson, T.P.; Li, L.; Deng, X.; Resende, T.; Vannucci, F.; Delwart, E. Detection of a novel circovirus PCV3 in pigs with cardiac and multi-systemic inflammation. Virol. J. 2016, 13, 184. [Google Scholar] [CrossRef]
- Niu, G.; Chen, S.; Li, X.; Zhang, L.; Ren, L. Advances in Crosstalk between Porcine Circoviruses and Host. Viruses 2022, 14, 1419. [Google Scholar] [CrossRef]
- Chang, C.C.; Wu, C.Y.; Wu, C.M.; Wu, C.W.; Wang, Y.C.; Lin, G.J.; Chien, M.S.; Huang, C. Cytotoxicity effect and transcriptome analysis of PCV3-infected cells revealed potential viral pathogenic mechanisms. Microb. Pathog. 2024, 192, 106715. [Google Scholar] [CrossRef]
- Molossi, F.A.; de Almeida, B.A.; de Cecco, B.S.; da Silva, M.S.; Mosena, A.C.S.; Brandalise, L.; Simão, G.M.R.; Canal, C.W.; Vanucci, F.; Pavarini, S.P.; et al. A putative PCV3-associated disease in piglets from Southern Brazil. Braz. J. Microbiol. 2022, 53, 491–498. [Google Scholar] [CrossRef]
- Palinski, R.; Pineyro, P.; Shang, P.; Yuan, F.; Guo, R.; Fang, Y.; Byers, E.; Hause, B.M. A Novel Porcine Circovirus Distantly Related to Known Circoviruses Is Associated with Porcine Dermatitis and Nephropathy Syndrome and Reproductive Failure. J. Virol. 2017, 91, e01879-16. [Google Scholar] [CrossRef]
- Mora-Diaz, J.; Pineyro, P.; Shen, H.; Schwartz, K.; Vannucci, F.; Li, G.; Arruda, B.; Giménez-Lirola, L. Isolation of PCV3 from Perinatal and Reproductive Cases of PCV3-Associated Disease and In Vivo Characterization of PCV3 Replication in CD/CD Growing Pigs. Viruses 2020, 12, 219. [Google Scholar] [CrossRef]
- Saporiti, V.; Franzo, G.; Sibila, M.; Segales, J. Porcine circovirus 3 (PCV-3) as a causal agent of disease in swine and a proposal of PCV-3 associated disease case definition. Transbound. Emerg. Dis. 2021, 68, 2936–2948. [Google Scholar] [CrossRef]
- Temeeyasen, G.; Lierman, S.; Arruda, B.L.; Main, R.; Vannucci, F.; Gimenez-Lirola, L.G.; Piñeyro, P.E. Pathogenicity and immune response against porcine circovirus type 3 infection in caesarean-derived, colostrum-deprived pigs. J. Gen. Virol. 2021, 102, jgv001502. [Google Scholar] [CrossRef]
- Kroeger, M.; Temeeyasen, G.; Pineyro, P.E. Five years of porcine circovirus 3: What have we learned about the clinical disease, immune pathogenesis, and diagnosis. Virus Res. 2022, 314, 198764. [Google Scholar] [CrossRef]
- Zhang, B.; Cai, J.; Zhu, C.; Zhang, Y.; Wu, J.; Li, Y. Rescue of naive porcine circovirus type 3 and its pathogenesis in CD pigs. J. Virol. 2025, 99, e0034125. [Google Scholar] [CrossRef]
- Tan, C.Y.; Lin, C.N.; Ooi, P.T. What do we know about porcine circovirus 3 (PCV3) diagnosis so far?: A review. Transbound. Emerg. Dis. 2021, 68, 2915–2935. [Google Scholar] [CrossRef]
- Qi, S.; Su, M.; Guo, D.; Li, C.; Wei, S.; Feng, L.; Sun, D. Molecular detection and phylogenetic analysis of porcine circovirus type 3 in 21 Provinces of China during 2015–2017. Transbound. Emerg. Dis. 2019, 66, 1004–1015. [Google Scholar] [CrossRef]
- Ge, M.; Ren, J.; Xie, Y.L.; Zhao, D.; Fan, F.C.; Song, X.Q.; Li, M.X.; Xiao, C.T. Prevalence and Genetic Analysis of Porcine Circovirus 3 in China From 2019 to 2020. Front. Vet. Sci. 2021, 8, 773912. [Google Scholar] [CrossRef]
- Mai, J.; Wang, D.; Zou, Y.; Zhang, S.; Meng, C.; Wang, A.; Wang, N. High Co-infection Status of Novel Porcine Parvovirus 7 With Porcine Circovirus 3 in Sows That Experienced Reproductive Failure. Front. Vet. Sci. 2021, 8, 695553. [Google Scholar] [CrossRef]
- Donneschi, A.; Recchia, M.; Romeo, C.; Pozzi, P.; Salogni, C.; Maisano, A.M.; Santucci, G.; Scali, F.; Faccini, S.; Boniotti, M.B.; et al. Infectious Agents Associated with Abortion Outbreaks in Italian Pig Farms from 2011 to 2021. Vet. Sci. 2024, 11, 496. [Google Scholar] [CrossRef]
- Zheng, S.; Shi, J.; Wu, X.; Peng, Z.; Xin, C.; Zhang, L.; Liu, Y.; Gao, M.; Xu, S.; Han, H.; et al. Presence of Torque teno sus virus 1 and 2 in porcine circovirus 3-positive pigs. Transbound. Emerg. Dis. 2018, 65, 327–330. [Google Scholar] [CrossRef]
- Ngo, T.N.T.; Nguyen, N.M.; Thanawongnuwech, R.; Thong, L.M.; Nguyen, T.P.T.; Nguyen, T.T.; Do, D.T. Coinfection of Mycoplasma suis and porcine circovirus type 3 is linked to reproductive failure in pig farms. Vet. World 2024, 17, 2477–2487. [Google Scholar] [CrossRef]
- Zhang, P.; Shen, H.; Liu, X.; Wang, S.; Liu, Y.; Xu, Z.; Song, C. Porcine Circovirus Type 3 Cap Inhibits Type I Interferon Induction Through Interaction with G3BP1. Front. Vet. Sci. 2020, 7, 594438. [Google Scholar] [CrossRef]
- Yan, Y.R.; Sun, Y.H. Genotypic diversity and immunological implications of porcine circovirus: Inspiration from PCV1 to PCV4. Microb. Pathog. 2024, 196, 106997. [Google Scholar] [CrossRef]
- Gao, Y.Y.; Wang, Q.; Li, H.W.; Zhang, S.; Zhao, J.; Bao, D.; Zhao, H.; Wang, K.; Hu, G.-X.; Gao, F.-S. Genomic composition and pathomechanisms of porcine circoviruses: A review. Virulence 2024, 15, 2439524. [Google Scholar] [CrossRef] [PubMed]
- Nath, B.K.; Das, S.; Roby, J.A.; Sarker, S.; Luque, D.; Raidal, S.R.; Forwood, J.K. Structural Perspectives of Beak and Feather Disease Virus and Porcine Circovirus Proteins. Viral Immunol. 2021, 34, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Qiu, H.; Sun, M.; Wang, N.; Zhang, S.; Deng, Z.; Xu, H.; Yang, H.; Gu, H.; Fang, W.; He, F. Efficacy comparison in cap VLPs of PCV2 and PCV3 as swine vaccine vehicle. Int. J. Biol. Macromol. 2024, 278, 134955. [Google Scholar] [CrossRef] [PubMed]
- Maity, H.K.; Samanta, K.; Deb, R.; Gupta, V.K. Revisiting Porcine Circovirus Infection: Recent Insights and Its Significance in the Piggery Sector. Vaccines 2023, 11, 1308. [Google Scholar] [CrossRef] [PubMed]
- Franzo, G.; He, W.; Correa-Fiz, F.; Li, G.; Legnardi, M.; Su, S.; Segalés, J. A Shift in Porcine Circovirus 3 (PCV-3) History Paradigm: Phylodynamic Analyses Reveal an Ancient Origin and Prolonged Undetected Circulation in the Worldwide Swine Population. Adv. Sci. 2019, 6, 1901004. [Google Scholar] [CrossRef]
- Cui, Y.; Hou, L.; Pan, Y.; Feng, X.; Zhou, J.; Wang, D.; Guo, J.; Liu, C.; Shi, Y.; Sun, T.; et al. Reconstruction of the Evolutionary Origin, Phylodynamics, and Phylogeography of the Porcine Circovirus Type 3. Front. Microbiol. 2022, 13, 898212. [Google Scholar] [CrossRef]
- Wozniak, A.; Milek, D.; Baska, P.; Stadejek, T. Does porcine circovirus type 3 (PCV3) interfere with porcine circovirus type 2 (PCV2) vaccine efficacy? Transbound. Emerg. Dis. 2019, 66, 1454–1461. [Google Scholar] [CrossRef]
- Pranoto, S.; Wu, H.C.; Chu, C.Y. Porcine Circovirus Type 3: Diagnostics, Genotyping, and Challenges in Vaccine Development. Transbound. Emerg. Dis. 2023, 2023, 8858447. [Google Scholar] [CrossRef]
- Pan, J.; Zeng, M.; Zhao, M.; Huang, L. Research Progress on the detection methods of porcine reproductive and respiratory syndrome virus. Front. Microbiol. 2023, 14, 1097905. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, G.; Duan, W.T.; Sun, M.X.; Wang, M.H.; Wang, S.H.; Cai, X.H.; Tu, Y.B. Self-assembly into virus-like particles of the recombinant capsid protein of porcine circovirus type 3 and its application on antibodies detection. AMB Express 2020, 10, 3. [Google Scholar] [CrossRef]
- Wang, J.; Lei, B.; Zhang, W.; Li, L.; Ji, J.; Liu, M.; Zhao, K.; Yuan, W. Preparation of Monoclonal Antibodies against the Capsid Protein and Development of an Epitope-Blocking Enzyme-Linked Immunosorbent Assay for Detection of the Antibody against Porcine Circovirus 3. Animals 2024, 14, 235. [Google Scholar] [CrossRef] [PubMed]
- Hayrapetyan, H.; Tran, T.; Tellez-Corrales, E.; Madiraju, C. Enzyme-Linked Immunosorbent Assay: Types and Applications. Methods Mol. Biol. 2023, 2612, 1–17. [Google Scholar] [PubMed]
- Sun, M.; Wang, S.; Fang, Z.; Zhao, M.; Gao, Y.; An, T.; Tu, Y.; Wang, H.; Cai, X. A Sandwich ELISA for Quality Control of PCV2 Virus-like Particles Vaccine. Vaccines 2022, 10, 2175. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.C.; Nguyen, V.G.; Park, Y.H.; Park, B.K. Genotyping of PCV3 based on reassembled viral gene sequences. Vet. Med. Sci. 2021, 7, 474–482. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, L.; Li, X.; Niu, G.; Ren, L. Recent Progress on Epidemiology and Pathobiology of Porcine Circovirus 3. Viruses 2021, 13, 1944. [Google Scholar] [CrossRef]
- Hayashi, S.; Katakura, F.; Moritomo, T.; Tsutsumi, N.; Sugiura, K.; Sato, T. Isolation of porcine circovirus 3 using primary porcine bone marrow-derived cells. Virol. J. 2024, 21, 184. [Google Scholar] [CrossRef]
- Oh, T.; Chae, C. First isolation and genetic characterization of porcine circovirus type 3 using primary porcine kidney cells. Vet. Microbiol. 2020, 241, 108576. [Google Scholar] [CrossRef]
- Moraes, J.Z.; Hamaguchi, B.; Braggion, C.; Speciale, E.R.; Cesar, F.B.V.; Soares, G.; Osaki, J.H.; Pereira, T.M.; Aguiar, R.B. Hybridoma technology: Is it still useful? Curr. Res. Immunol. 2021, 2, 32–40. [Google Scholar] [CrossRef]
- Li, X.; Bai, Y.; Zhang, H.; Zheng, D.; Wang, T.; Wang, Y.; Deng, J.; Sun, Z.; Tian, K. Production of a monoclonal antibody against Porcine circovirus type 3 cap protein. J. Virol. Methods. 2018, 261, 10–13. [Google Scholar] [CrossRef]
- Lanzavecchia, A.; Fruhwirth, A.; Perez, L.; Corti, D. Antibody-guided vaccine design: Identification of protective epitopes. Curr. Opin. Immunol. 2016, 41, 62–67. [Google Scholar] [CrossRef]
- Jiang, M.; Guo, J.; Zhang, G.; Jin, Q.; Liu, Y.; Jia, R.; Wang, A. Fine mapping of linear B cell epitopes on capsid protein of porcine circovirus 3. Appl. Microbiol. Biotechnol. 2020, 104, 6223–6234. [Google Scholar] [CrossRef]
- Chang, C.C.; Wu, C.Y.; Ciou, J.G.; Wu, C.W.; Wang, Y.C.; Chang, H.W.; Chien, M.S.; Huang, C. Exploring the surface epitope and nuclear localization analysis of porcine circovirus type 3 capsid protein. AMB Express 2023, 13, 141. [Google Scholar] [CrossRef]
- Yang, S.Q.; Yang, K.; Li, X.R.; Zheng, Y.; Cao, S.J.; Yan, Q.G.; Huang, X.B.; Wen, Y.P.; Zhao, Q.; Du, S.Y.; et al. A novel linear B cell epitope of the porcine circovirus type 3 capsid protein identified by phage display technology. J. Virol. Methods 2025, 333, 115080. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Guan, S.; Zhao, J.; Chen, Y.; Han, Y.; Tian, A.; Zhou, S.; Chen, H.; Song, Y. Multiplex quantitative PCR assay for porcine circoviruses 2 and 3 and its application for measuring infection rates at different stages of pig production. Arch. Virol. 2025, 170, 158. [Google Scholar] [CrossRef]
- Cobos, A.; Sibila, M.; Segales, J. Review of porcine circovirus 3-associated lesions in swine: Challenges and advances in diagnostics. Vet. Pathol. 2025; ahead of print. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, Z.; Imamura, M.; Osawa, M.; Teraoka, Y.; Piotrowski, J.; Ishida, Y.; Sozzi, V.; Revill, P.A.; Saito, T.; et al. Infection courses, virological features and IFN-alpha responses of HBV genotypes in cell culture and animal models. J. Hepatol. 2021, 75, 1335–1345. [Google Scholar] [CrossRef]
| Sample | OD450 | Mean Value | Standard Deviation | Coefficient of Variation | ||
|---|---|---|---|---|---|---|
| 1 | 1.354 | 1.435 | 1.377 | 1.389 | 0.0417 | 3.01% |
| 2 | 0.777 | 0.858 | 0.885 | 0.840 | 0.0562 | 6.69% |
| 3 | 0.269 | 0.265 | 0.254 | 0.263 | 0.0078 | 2.96% |
| 4 | 0.517 | 0.572 | 0.51 | 0.533 | 0.034 | 6.37% |
| Sample | OD450 | Mean Value | Standard Deviation | Coefficient of Variation | ||
|---|---|---|---|---|---|---|
| 1 | 1.393 | 1.331 | 1.317 | 1.347 | 0.0404 | 3.00% |
| 2 | 0.823 | 0.812 | 0.799 | 0.8113 | 0.012 | 1.48% |
| 3 | 0.21 | 0.206 | 0.215 | 0.2103 | 0.0045 | 2.14% |
| 4 | 0.591 | 0.58 | 0.647 | 0.606 | 0.0359 | 5.93% |
| DAS-ELISA | ||||||
|---|---|---|---|---|---|---|
| Positive | Negative | Total | Coincidence rate | Kappa value | ||
| qPCR | Positive | 51 | 3 | 54 | 93.33% | 0.837 |
| Negative | 2 | 19 | 21 | |||
| Total | 55 | 10 | 75 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Zhao, J.; Tian, A.; Wu, H.; Chen, H.; Song, Y. Development of a Highly Specific Monoclonal Antibody-Based Sandwich ELISA for Rapid Detection of Porcine Circovirus Type 3. Viruses 2025, 17, 1340. https://doi.org/10.3390/v17101340
Li Z, Zhao J, Tian A, Wu H, Chen H, Song Y. Development of a Highly Specific Monoclonal Antibody-Based Sandwich ELISA for Rapid Detection of Porcine Circovirus Type 3. Viruses. 2025; 17(10):1340. https://doi.org/10.3390/v17101340
Chicago/Turabian StyleLi, Zhen, Jiaying Zhao, Ang Tian, Hao Wu, Huanchun Chen, and Yunfeng Song. 2025. "Development of a Highly Specific Monoclonal Antibody-Based Sandwich ELISA for Rapid Detection of Porcine Circovirus Type 3" Viruses 17, no. 10: 1340. https://doi.org/10.3390/v17101340
APA StyleLi, Z., Zhao, J., Tian, A., Wu, H., Chen, H., & Song, Y. (2025). Development of a Highly Specific Monoclonal Antibody-Based Sandwich ELISA for Rapid Detection of Porcine Circovirus Type 3. Viruses, 17(10), 1340. https://doi.org/10.3390/v17101340






