Comparative Transcriptomics Reveals Novel and Differential Circular RNA Responses Underlying Interferon-Mediated Antiviral Regulation in Porcine Alveolar Macrophages
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement and Animal Cell Sources
2.2. Virus Infection and IFN Treatment
2.3. RNA Transcriptomic Analysis
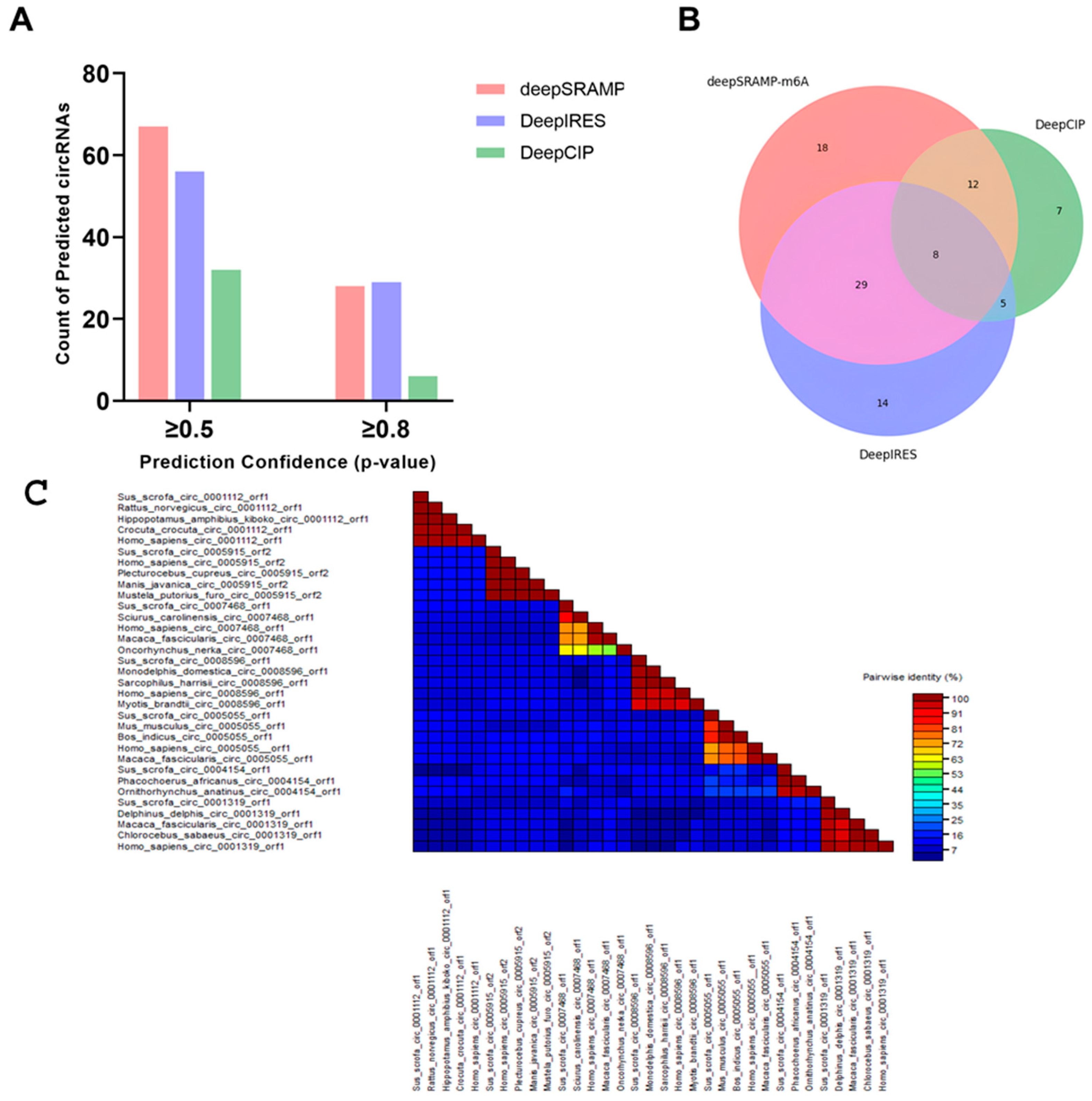
2.4. MicroRNA Target Site Prediction and Coding Potential Analysis
2.5. Statistical Analysis
3. Results
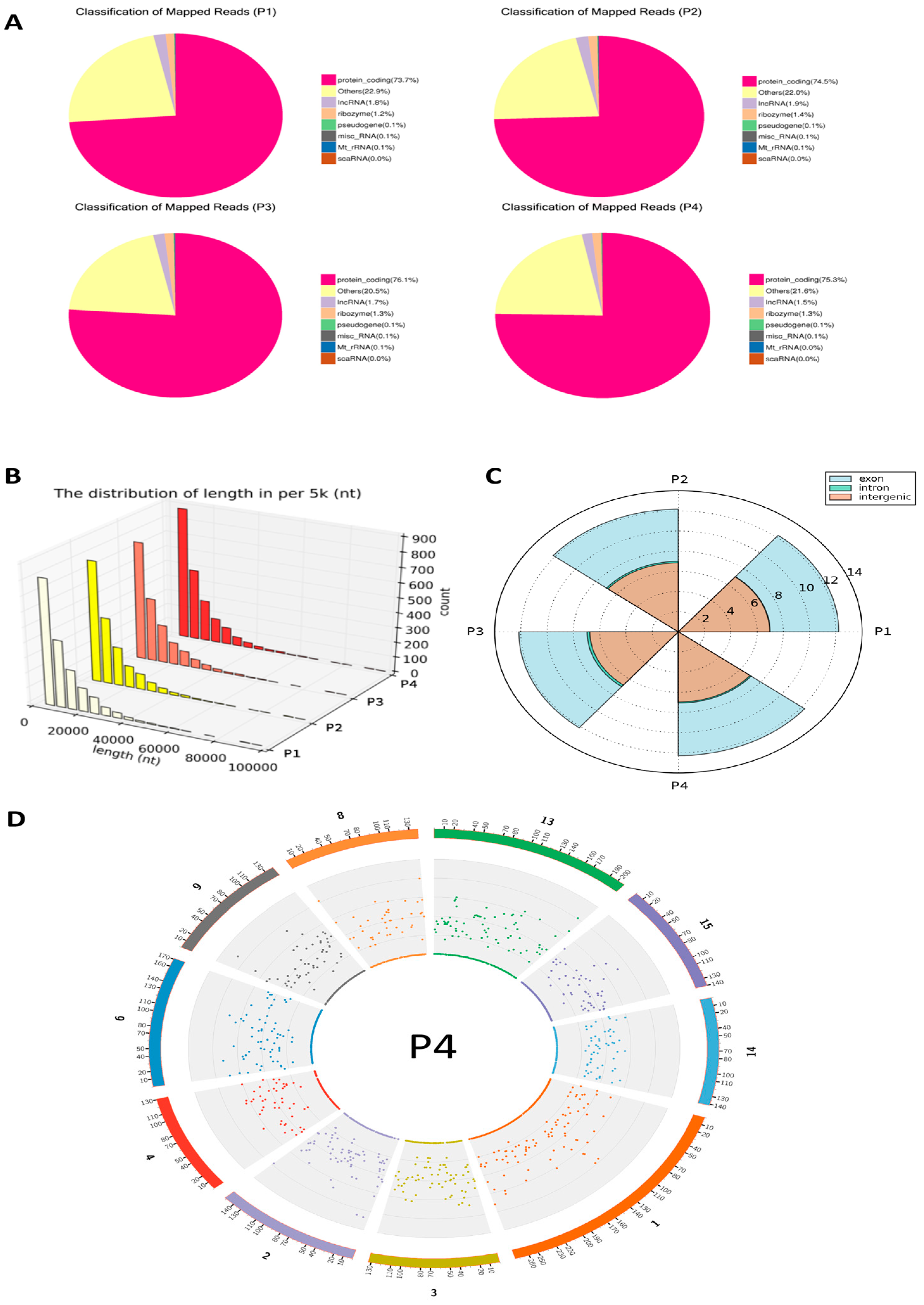
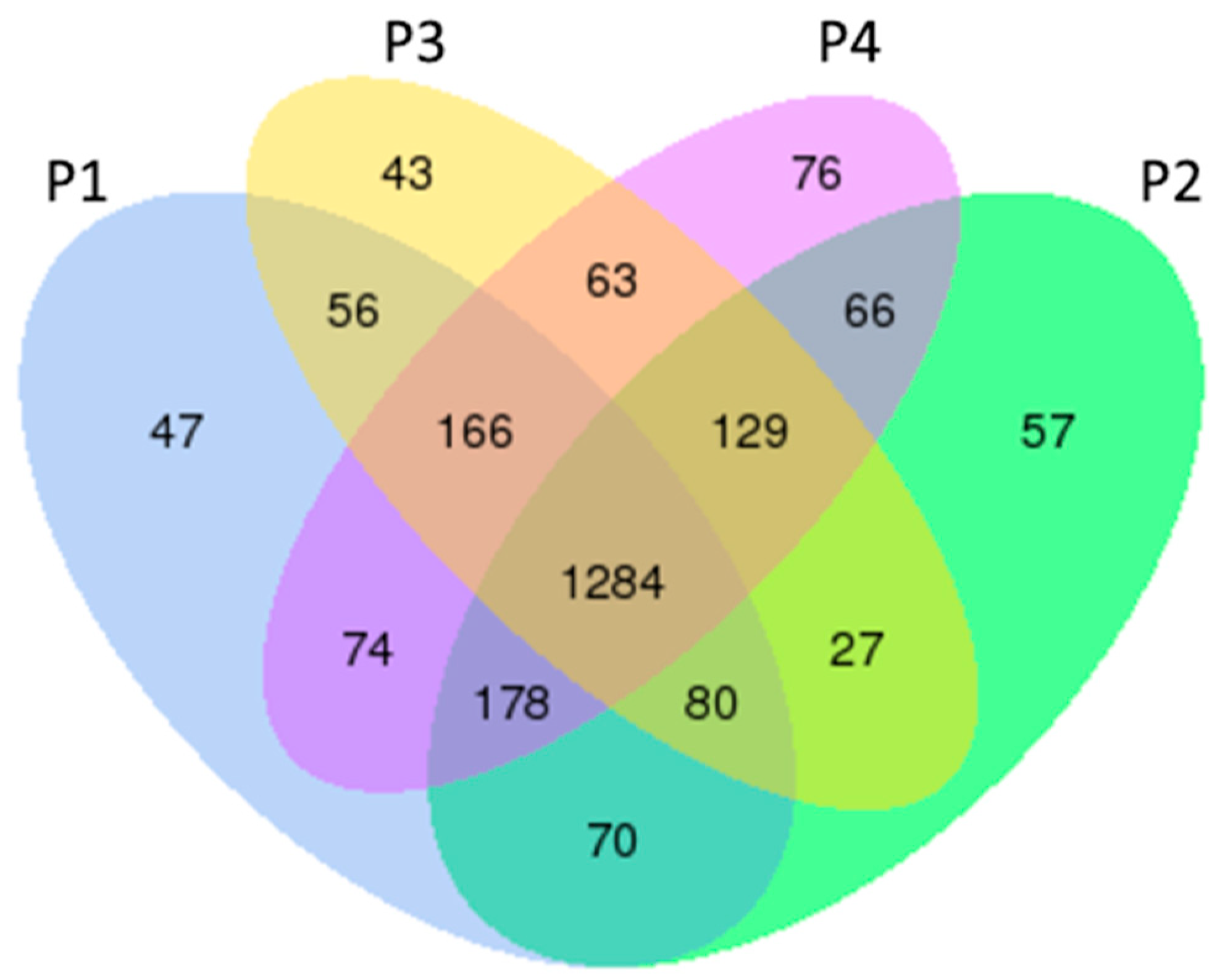
3.1. CircRNA Compositions and Identification Across the PAM Samples
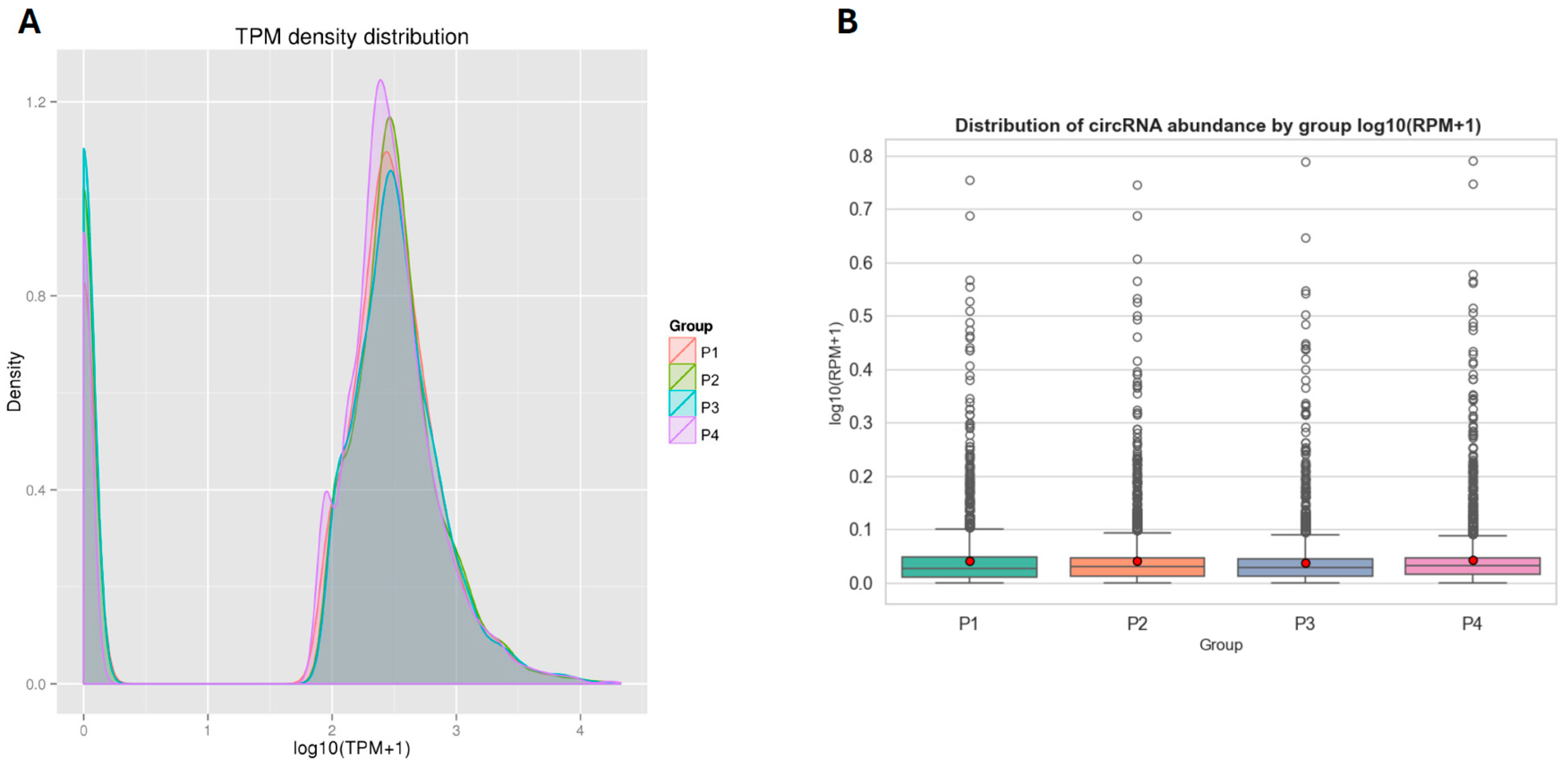
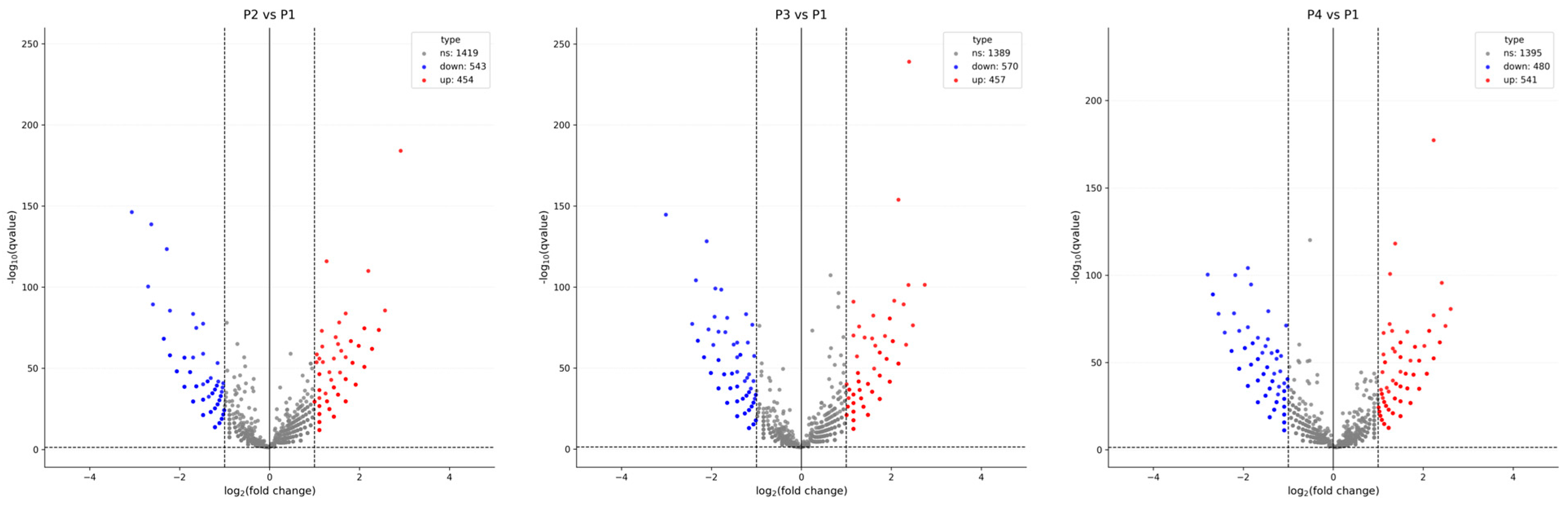
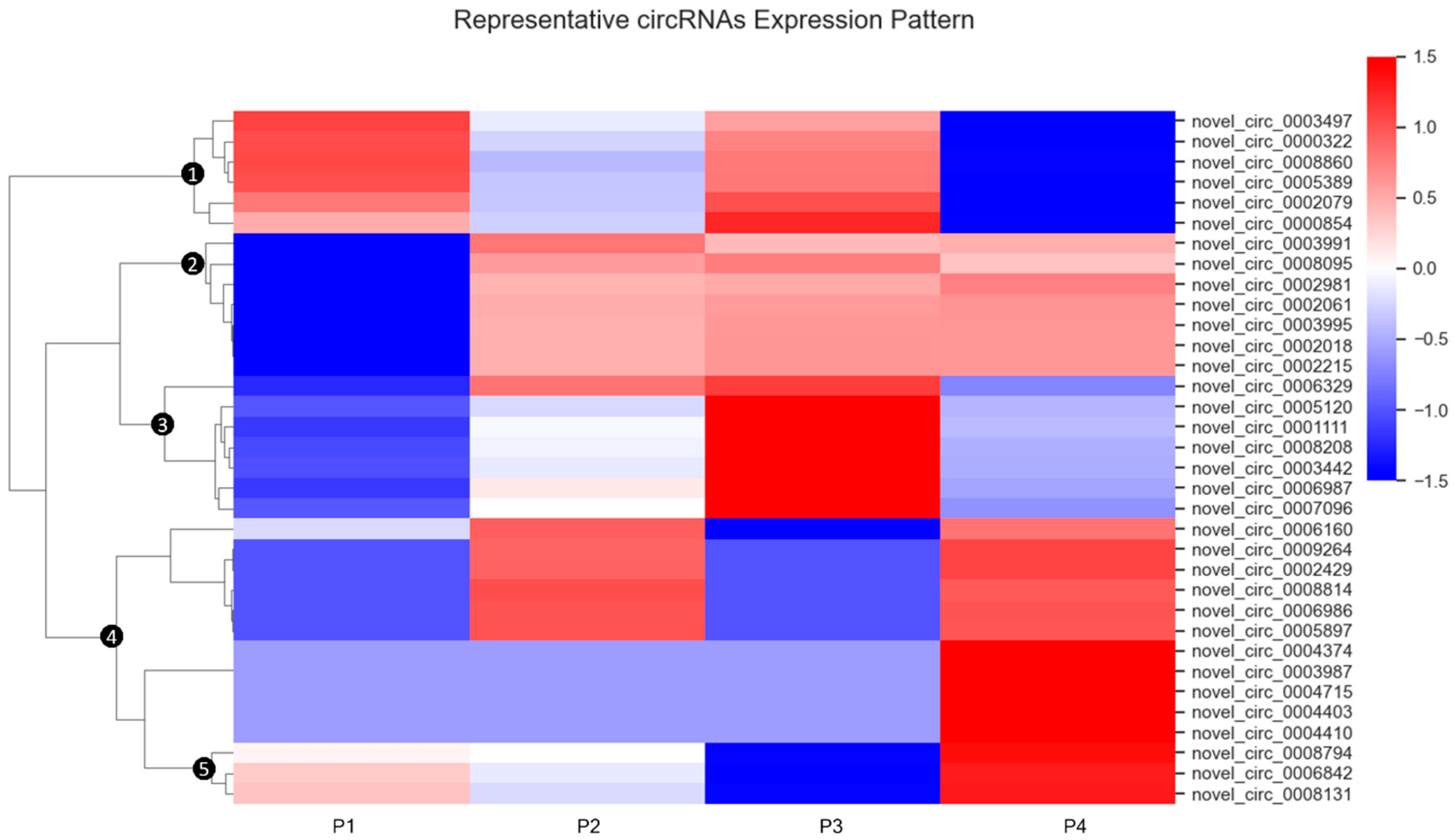
3.2. General Expression Pattern and Differential Expression of circRNAs
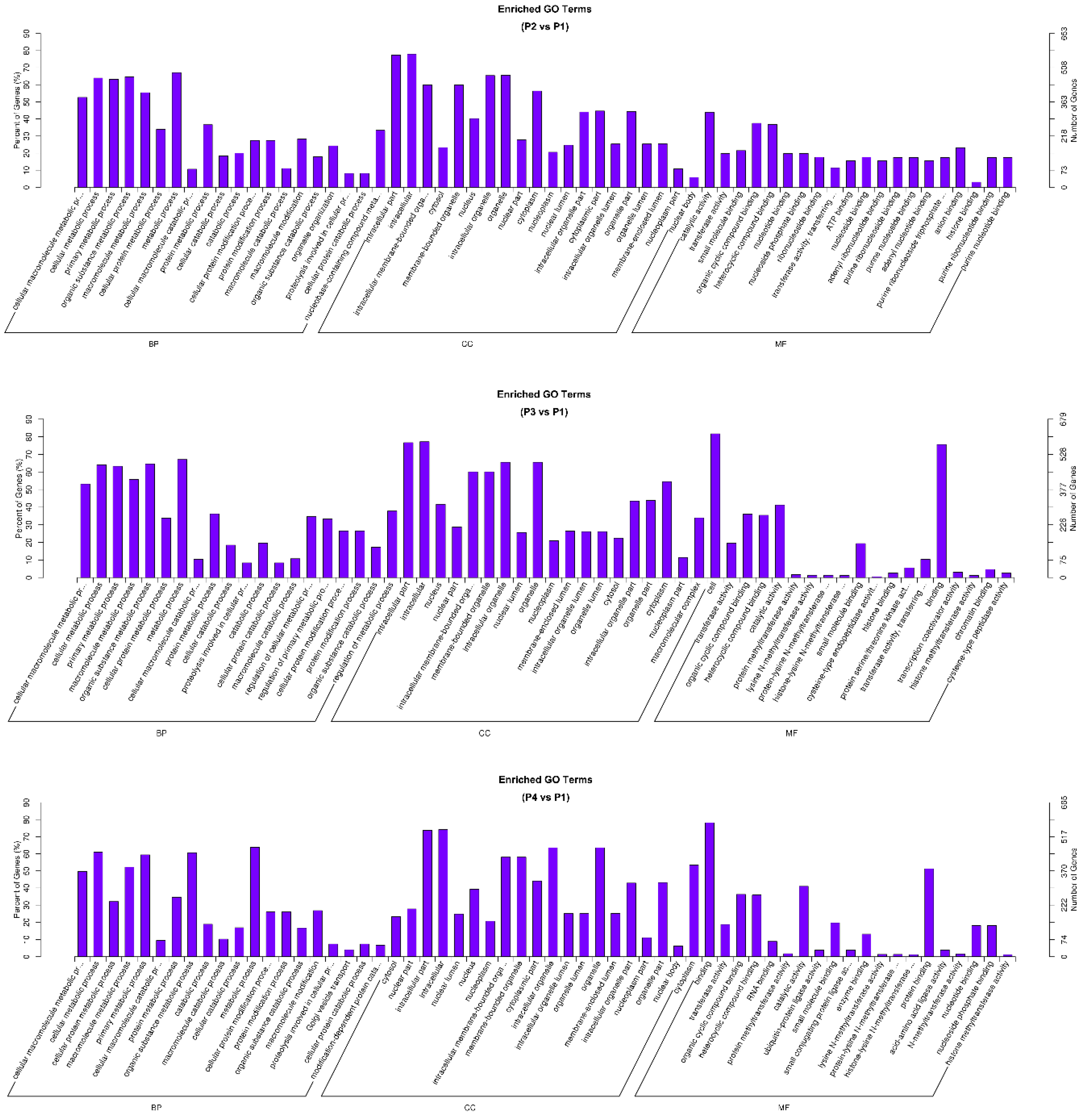
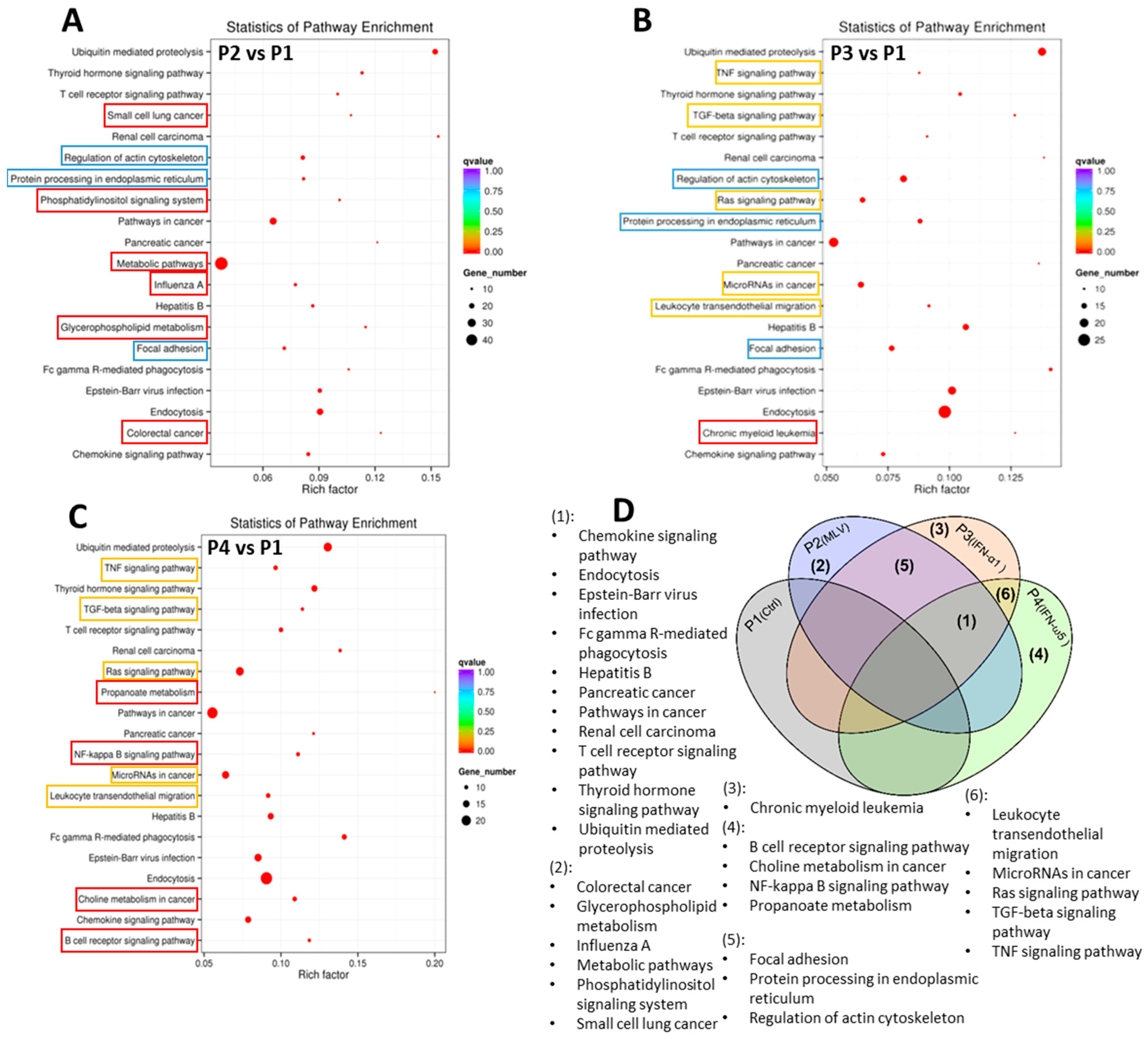
3.3. Gene Ontology and KEGG Analysis of circRNA-Targeted Genes
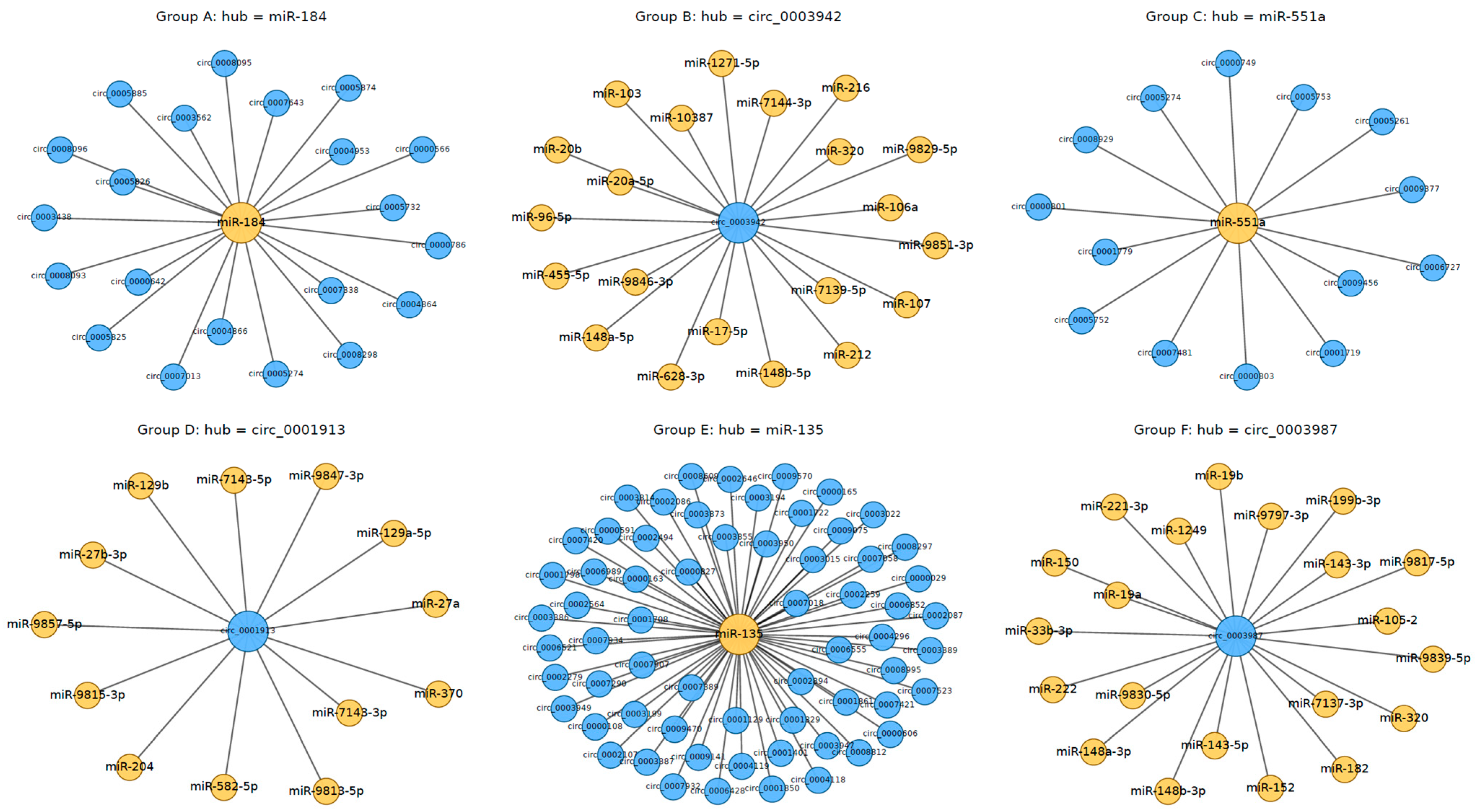
3.4. CircRNA-miRNA Interactions and Coding Potential
4. Discussion
4.1. Interpretation of circRNA Composition and Identification in PAMs
4.2. Biological Implications of circRNA Expression Patterns and Differential Profiles
4.3. Functional Significance of circRNA-Targeted Genes in GO and KEGG Pathways
4.4. Regulatory Roles and Translational Potential of circRNAs via miRNA Interactions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Meulenberg, J.J. PRRSV, the virus. Vet. Res. 2000, 31, 11–21. [Google Scholar] [CrossRef]
- Holtkamp, D.; Kliebenstein, J.; Neumann, E.; Zimmerman, J.; Rotto, H.; Yoder, T.; Wang, C.; Yeske, P.; Mowrer, C.; Haley, C. Assessment of the economic impact of porcine reproductive and respiratory syndrome virus on United States pork producers. J. Swine Health Prod. 2013, 21, 72–84. [Google Scholar] [CrossRef]
- Gorbalenya, A.E.; Enjuanes, L.; Ziebuhr, J.; Snijder, E.J. Nidovirales: Evolving the largest RNA virus genome. Virus Res. 2006, 117, 17–37. [Google Scholar] [CrossRef] [PubMed]
- Balka, G.; Podgorska, K.; Brar, M.S.; Balint, A.; Cadar, D.; Celer, V.; Denes, L.; Dirbakova, Z.; Jedryczko, A.; Marton, L.; et al. Genetic diversity of PRRSV 1 in Central Eastern Europe in 1994-2014: Origin and evolution of the virus in the region. Sci. Rep. 2018, 8, 7811. [Google Scholar] [CrossRef] [PubMed]
- Dokland, T. The structural biology of PRRSV. Virus Res. 2010, 154, 86–97. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Miller, L.C.; Sang, Y. Current Status of Vaccines for Porcine Reproductive and Respiratory Syndrome: Interferon Response, Immunological Overview, and Future Prospects. Vaccines 2024, 12, 606. [Google Scholar] [CrossRef]
- Popperl, P.; Stoff, M.; Beineke, A. Alveolar Macrophages in Viral Respiratory Infections: Sentinels and Saboteurs of Lung Defense. Int. J. Mol. Sci. 2025, 26, 407. [Google Scholar] [CrossRef]
- Sanin, D.E.; Ge, Y.; Marinkovic, E.; Kabat, A.M.; Castoldi, A.; Caputa, G.; Grzes, K.M.; Curtis, J.D.; Thompson, E.A.; Willenborg, S.; et al. A common framework of monocyte-derived macrophage activation. Sci. Immunol. 2022, 7, eabl7482. [Google Scholar] [CrossRef]
- Netea, M.G.; Joosten, L.A.; Latz, E.; Mills, K.H.; Natoli, G.; Stunnenberg, H.G.; O’Neill, L.A.; Xavier, R.J. Trained immunity: A program of innate immune memory in health and disease. Science 2016, 352, aaf1098. [Google Scholar] [CrossRef]
- van der Heijden, C.; Noz, M.P.; Joosten, L.A.B.; Netea, M.G.; Riksen, N.P.; Keating, S.T. Epigenetics and Trained Immunity. Antioxid. Redox Signal 2018, 29, 1023–1040. [Google Scholar] [CrossRef]
- Flores-Concha, M.; Onate, A.A. Long Non-coding RNAs in the Regulation of the Immune Response and Trained Immunity. Front. Genet. 2020, 11, 718. [Google Scholar] [CrossRef]
- Nakayama, Y.; Fujiu, K. Innate immune memory in macrophage differentiation and cardiovascular diseases. Inflamm. Regen. 2025, 45, 17. [Google Scholar] [CrossRef] [PubMed]
- Jarroux, J.; Morillon, A.; Pinskaya, M. History, Discovery, and Classification of lncRNAs. Adv. Exp. Med. Biol. 2017, 1008, 1–46. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Zhang, Z.; Krause, H.M. Long Noncoding RNAs and Repetitive Elements: Junk or Intimate Evolutionary Partners? Trends Genet. 2019, 35, 892–902. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef]
- Denzler, R.; Agarwal, V.; Stefano, J.; Bartel, D.P.; Stoffel, M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol. Cell 2014, 54, 766–776. [Google Scholar] [CrossRef]
- Zhang, M.; Huang, N.; Yang, X.; Luo, J.; Yan, S.; Xiao, F.; Chen, W.; Gao, X.; Zhao, K.; Zhou, H.; et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene 2018, 37, 1805–1814. [Google Scholar] [CrossRef]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef]
- Chen, Q.; Mang, G.; Wu, J.; Sun, P.; Li, T.; Zhang, H.; Wang, N.; Tong, Z.; Wang, W.; Zheng, Y.; et al. Circular RNA circSnx5 Controls Immunogenicity of Dendritic Cells through the miR-544/SOCS1 Axis and PU.1 Activity Regulation. Mol. Ther. 2020, 28, 2503–2518. [Google Scholar] [CrossRef]
- Niu, D.; Wu, Y.; Lian, J. Circular RNA vaccine in disease prevention and treatment. Signal Transduct. Target. Ther. 2023, 8, 341. [Google Scholar] [CrossRef]
- Qin, Z.; Liu, W.; Qin, Z.; Zhang, H.; Huang, X. Host combats porcine reproductive and respiratory syndrome virus infection at non-coding RNAs level. Virulence 2024, 15, 2416551. [Google Scholar] [CrossRef]
- Shields, L.E.; Jennings, J.; Liu, Q.; Lee, J.; Ma, W.; Blecha, F.; Miller, L.C.; Sang, Y. Cross-Species Genome-Wide Analysis Reveals Molecular and Functional Diversity of the Unconventional Interferon-omega Subtype. Front. Immunol. 2019, 10, 1431. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Sang, E.R.; Adeyemi, O.; Miller, L.C.; Sang, Y. Comparative transcriptomics reveals small RNA composition and differential microRNA responses underlying interferon-mediated antiviral regulation in porcine alveolar macrophages. Front. Immunol. 2022, 13, 1016268. [Google Scholar] [CrossRef]
- Sang, Y.; Brichalli, W.; Rowland, R.R.; Blecha, F. Genome-wide analysis of antiviral signature genes in porcine macrophages at different activation statuses. PLoS ONE 2014, 9, e87613. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zhang, J.; Zhao, F. Circular RNA identification based on multiple seed matching. Brief. Bioinform. 2018, 19, 803–810. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Zhao, J.; Chen, Z.; Zhang, M.; Zou, L.; He, S.; Liu, J.; Wang, Q.; Song, X.; Wu, J. DeepIRES: A hybrid deep learning model for accurate identification of internal ribosome entry sites in cellular and viral mRNAs. Brief. Bioinform. 2024, 25, bbae439. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wu, J.; Yao, S.; Xu, Y.; Zhao, W.; Tong, Y.; Zhou, Z. DeepCIP: A multimodal deep learning method for the prediction of internal ribosome entry sites of circRNAs. Comput. Biol. Med. 2023, 164, 107288. [Google Scholar] [CrossRef]
- Fan, R.; Cui, C.; Kang, B.; Chang, Z.; Wang, G.; Cui, Q. A combined deep learning framework for mammalian m6A site prediction. Cell Genom. 2024, 4, 100697. [Google Scholar] [CrossRef]
- Fleming, D.S.; Miller, L.C.; Li, J.; Van Geelen, A.; Sang, Y. Transcriptomic Analysis of Liver Indicates Novel Vaccine to Porcine Reproductive and Respiratory Virus Promotes Homeostasis in T-Cell and Inflammatory Immune Responses Compared to a Commercial Vaccine in Pigs. Front. Vet. Sci. 2022, 9, 791034. [Google Scholar] [CrossRef]
- Zhou, Y.H.; Xia, K.; Wright, F.A. A powerful and flexible approach to the analysis of RNA sequence count data. Bioinformatics 2011, 27, 2672–2678. [Google Scholar] [CrossRef]
- Wu, J.; Qi, X.; Liu, L.; Hu, X.; Liu, J.; Yang, J.; Yang, J.; Lu, L.; Zhang, Z.; Ma, S.; et al. Emerging Epigenetic Regulation of Circular RNAs in Human Cancer. Mol. Ther. Nucleic Acids 2019, 16, 589–596. [Google Scholar] [CrossRef]
- Verduci, L.; Strano, S.; Yarden, Y.; Blandino, G. The circRNA-microRNA code: Emerging implications for cancer diagnosis and treatment. Mol. Oncol. 2019, 13, 669–680. [Google Scholar] [CrossRef] [PubMed]
- Thomson, D.W.; Dinger, M.E. Endogenous microRNA sponges: Evidence and controversy. Nat. Rev. Genet. 2016, 17, 272–283. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Hou, C.; Chen, C.; Guo, Y.; Yuan, W.; Yin, D.; Liu, J.; Sun, Z. The role of N(6)-methyladenosine (m(6)A) modification in the regulation of circRNAs. Mol. Cancer 2020, 19, 105. [Google Scholar] [CrossRef]
- Chen, J.; Wang, H.; Jin, L.; Wang, L.; Huang, X.; Chen, W.; Yan, M.; Liu, G. Profile analysis of circRNAs induced by porcine endemic diarrhea virus infection in porcine intestinal epithelial cells. Virology 2019, 527, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.; Chen, C.Y.; Chuang, T.J. Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip. Rev. RNA 2015, 6, 563–579. [Google Scholar] [CrossRef]
- Min, J.; Liu, W.; Li, J. Emerging Role of Interferon-Induced Noncoding RNA in Innate Antiviral Immunity. Viruses 2022, 14, 2607. [Google Scholar] [CrossRef]
- Sun, Y.; Han, M.; Kim, C.; Calvert, J.G.; Yoo, D. Interplay between interferon-mediated innate immunity and porcine reproductive and respiratory syndrome virus. Viruses 2012, 4, 424–446. [Google Scholar] [CrossRef]
- Miller, L.C.; Anderson, S.J.; Buckley, A.C.; Schirtzinger, E.E.; Hasan, M.; Sarlo Davila, K.M.; Fleming, D.S.; Lager, K.M.; Li, J.; Sang, Y. Replication-competent recombinant porcine reproductive and respiratory syndrome (PRRS) virus expressing antiviral cytokine interferon-omega5 as a modified live virus vaccine. Vet. Microbiol. 2025, 301, 110366. [Google Scholar] [CrossRef]
- Miller, L.C.; Anderson, S.J.; Buckley, A.C.; Schirtzinger, E.E.; Hasan, M.; Sarlo Davila, K.M.; Fleming, D.S.; Lager, K.M.; Li, J.; Sang, Y. Vaccine Efficacy of a Replication-Competent Interferon-Expressing Porcine Reproductive and Respiratory Syndrome (PRRS) Virus Against NADC-34 Challenge. Vaccines 2025, 13, 413. [Google Scholar] [CrossRef]
- Qiu, H.; Yang, B.; Chen, Y.; Zhu, Q.; Wen, F.; Peng, M.; Wang, G.; Guo, G.; Chen, B.; Maarouf, M.; et al. Influenza A Virus-Induced circRNA circMerTK Negatively Regulates Innate Antiviral Responses. Microbiol. Spectr. 2023, 11, e0363722. [Google Scholar] [CrossRef] [PubMed]
- Sang, L.; Yang, L.; Ge, Q.; Xie, S.; Zhou, T.; Lin, A. Subcellular distribution, localization, and function of noncoding RNAs. Wiley Interdiscip. Rev. RNA 2022, 13, e1729. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yang, T.; Wang, W.; Xi, W.; Zhang, T.; Li, Q.; Yang, A.; Wang, T. Circular RNAs in immune responses and immune diseases. Theranostics 2019, 9, 588–607. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, W.; Zhou, Q.; Chen, C.; Yuan, W.; Liu, J.; Li, X.; Sun, Z. Roles of circRNAs in the tumour microenvironment. Mol. Cancer 2020, 19, 14. [Google Scholar] [CrossRef] [PubMed]
- Awan, F.M.; Yang, B.B.; Naz, A.; Hanif, A.; Ikram, A.; Obaid, A.; Malik, A.; Janjua, H.A.; Ali, A.; Sharif, S. The emerging role and significance of circular RNAs in viral infections and antiviral immune responses: Possible implication as theranostic agents. RNA Biol. 2021, 18, 1–15. [Google Scholar] [CrossRef]
- Wang, K.; Gan, T.Y.; Li, N.; Liu, C.Y.; Zhou, L.Y.; Gao, J.N.; Chen, C.; Yan, K.W.; Ponnusamy, M.; Zhang, Y.H.; et al. Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ. 2017, 24, 1111–1120. [Google Scholar] [CrossRef]
- Li, H.; Tang, W.; Jin, Y.; Dong, W.; Yan, Y.; Zhou, J. Differential CircRNA Expression Profiles in PK-15 Cells Infected with Pseudorabies Virus Type II. Virol. Sin. 2021, 36, 75–84. [Google Scholar] [CrossRef]
- Li, C.; Sun, Y.; Jiang, C.; Cao, H.; Zeng, W.; Zhang, X.; Li, Z.; He, Q. Porcine circovirus type 2 infection activates NF-κB pathway and cellular inflammatory responses through circPDCD4/miR-21/PDCD4 axis in porcine kidney 15 cell. Virus Res. 2021, 298, 198385. [Google Scholar] [CrossRef]
- Dai, C.-H.; Gao, Z.-C.; Cheng, J.-H.; Yang, L.; Wu, Z.-C.; Wu, S.-L.; Bao, W.-B. The competitive endogenous RNA (ceRNA) regulation in porcine alveolar macrophages (3D4/21) infected by swine influenza virus (H1N1 and H3N2). Int. J. Mol. Sci. 2022, 23, 1875. [Google Scholar] [CrossRef]
- Chen, T.C.; Tallo-Parra, M.; Cao, Q.M.; Kadener, S.; Bottcher, R.; Perez-Vilaro, G.; Boonchuen, P.; Somboonwiwat, K.; Diez, J.; Sarnow, P. Host-derived circular RNAs display proviral activities in Hepatitis C virus-infected cells. PLoS Pathog. 2020, 16, e1008346. [Google Scholar] [CrossRef]
- Zhang, X.; Yan, Y.; Lei, X.; Li, A.; Zhang, H.; Dai, Z.; Li, X.; Chen, W.; Lin, W.; Chen, F.; et al. Circular RNA alterations are involved in resistance to avian leukosis virus subgroup-J-induced tumor formation in chickens. Oncotarget 2017, 8, 34961–34970. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, H.; An, M.; Zhao, B.; Ding, H.; Zhang, Z.; He, Y.; Shang, H.; Han, X. Crosstalk in competing endogenous RNA networks reveals new circular RNAs involved in the pathogenesis of early HIV infection. J. Transl. Med. 2018, 16, 332. [Google Scholar] [CrossRef] [PubMed]
- Ji, P.; Wu, W.; Chen, S.; Zheng, Y.; Zhou, L.; Zhang, J.; Cheng, H.; Yan, J.; Zhang, S.; Yang, P.; et al. Expanded Expression Landscape and Prioritization of Circular RNAs in Mammals. Cell Rep. 2019, 26, 3444–3460. [Google Scholar] [CrossRef] [PubMed]
- Yaylak, B.; Erdogan, I.; Akgul, B. Transcriptomics Analysis of Circular RNAs Differentially Expressed in Apoptotic HeLa Cells. Front. Genet. 2019, 10, 176. [Google Scholar] [CrossRef]
| Sample | Raw Reads | Clean Reads | Clean Bases | Error Rate | Q20 | Q30 | GC Content |
|---|---|---|---|---|---|---|---|
| P1 (Ctrl) | 90284642 | 88711244 | 13.3 G | 0.03 | 97.56 | 93.53 | 53.74 |
| P2 (MLV) | 83084248 | 81698196 | 12.3 G | 0.03 | 97.28 | 92.88 | 53.74 |
| P3 (IFN-α1) | 85481978 | 83905840 | 12.6 G | 0.03 | 97.67 | 93.8 | 53.34 |
| P4 (IFN-ω5) | 91860194 | 90253536 | 13.5 G | 0.03 | 97.46 | 93.27 | 52.2 |
| CircRNA_id | Chr. | Start Position | End Position | Strand | Full Length | Spliced Length | Gene Id | Gene Name |
|---|---|---|---|---|---|---|---|---|
| novel_circ_0001167 | 12 | 60348041 | 60353600 | - | 5559 | 367 | ENSSSCG00000018049 | SLC5A10 |
| novel_circ_0003958 | 1 | 120176021 | 120185745 | + | 9724 | 480 | ENSSSCG00000032517 | DMXL2 |
| novel_circ_0005029 | 2 | 151040091 | 151052939 | + | 12,848 | 370 | ENSSSCG00000014440 | HMGXB3 |
| novel_circ_0009109 | 9 | 19852497 | 19861801 | - | 9304 | 560 | ENSSSCG00000014913 | PICALM |
| novel_circ_0000601 | 11 | 533608 | 534240 | - | 632 | 303 | ENSSSCG00000009569 | PSPC1 |
| novel_circ_0000083 | 10 | 24760508 | 24779098 | - | 18,590 | 284 | ENSSSCG00000010928 | KDM5B |
| novel_circ_0001614 | 13 | 200007333 | 200012649 | + | 5316 | 206 | ENSSSCG00000012055 | MORC3 |
| novel_circ_0004019 | 1 | 121731740 | 121738042 | + | 6302 | 452 | ENSSSCG00000004646 | ATP8B4 |
| novel_circ_0007230 | 6 | 109117106 | 109141734 | - | 24,628 | 411 | ENSSSCG00000003712 | OSBPL1A |
| novel_circ_0009337 | 9 | 92047368 | 92049234 | + | 1866 | 297 | ENSSSCG00000032241 | GPNMB |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Adeyemi, O.; Miller, L.C.; Sang, Y. Comparative Transcriptomics Reveals Novel and Differential Circular RNA Responses Underlying Interferon-Mediated Antiviral Regulation in Porcine Alveolar Macrophages. Viruses 2025, 17, 1307. https://doi.org/10.3390/v17101307
Li J, Adeyemi O, Miller LC, Sang Y. Comparative Transcriptomics Reveals Novel and Differential Circular RNA Responses Underlying Interferon-Mediated Antiviral Regulation in Porcine Alveolar Macrophages. Viruses. 2025; 17(10):1307. https://doi.org/10.3390/v17101307
Chicago/Turabian StyleLi, Jiuyi, Oluwaseun Adeyemi, Laura C. Miller, and Yongming Sang. 2025. "Comparative Transcriptomics Reveals Novel and Differential Circular RNA Responses Underlying Interferon-Mediated Antiviral Regulation in Porcine Alveolar Macrophages" Viruses 17, no. 10: 1307. https://doi.org/10.3390/v17101307
APA StyleLi, J., Adeyemi, O., Miller, L. C., & Sang, Y. (2025). Comparative Transcriptomics Reveals Novel and Differential Circular RNA Responses Underlying Interferon-Mediated Antiviral Regulation in Porcine Alveolar Macrophages. Viruses, 17(10), 1307. https://doi.org/10.3390/v17101307






