Molecular Epidemiology and Genetic Evolution of Porcine Reproductive and Respiratory Syndrome Virus in Northern China During 2021–2023
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Sample Processing and Analysis
2.3. ORF5 Sequencing
2.4. Analysis of ORF5 Gene Phylogeny and Amino Acid Mutation
2.5. Virus Isolation and Whole-Genome Sequencing
2.6. Recombination Analysis
3. Results
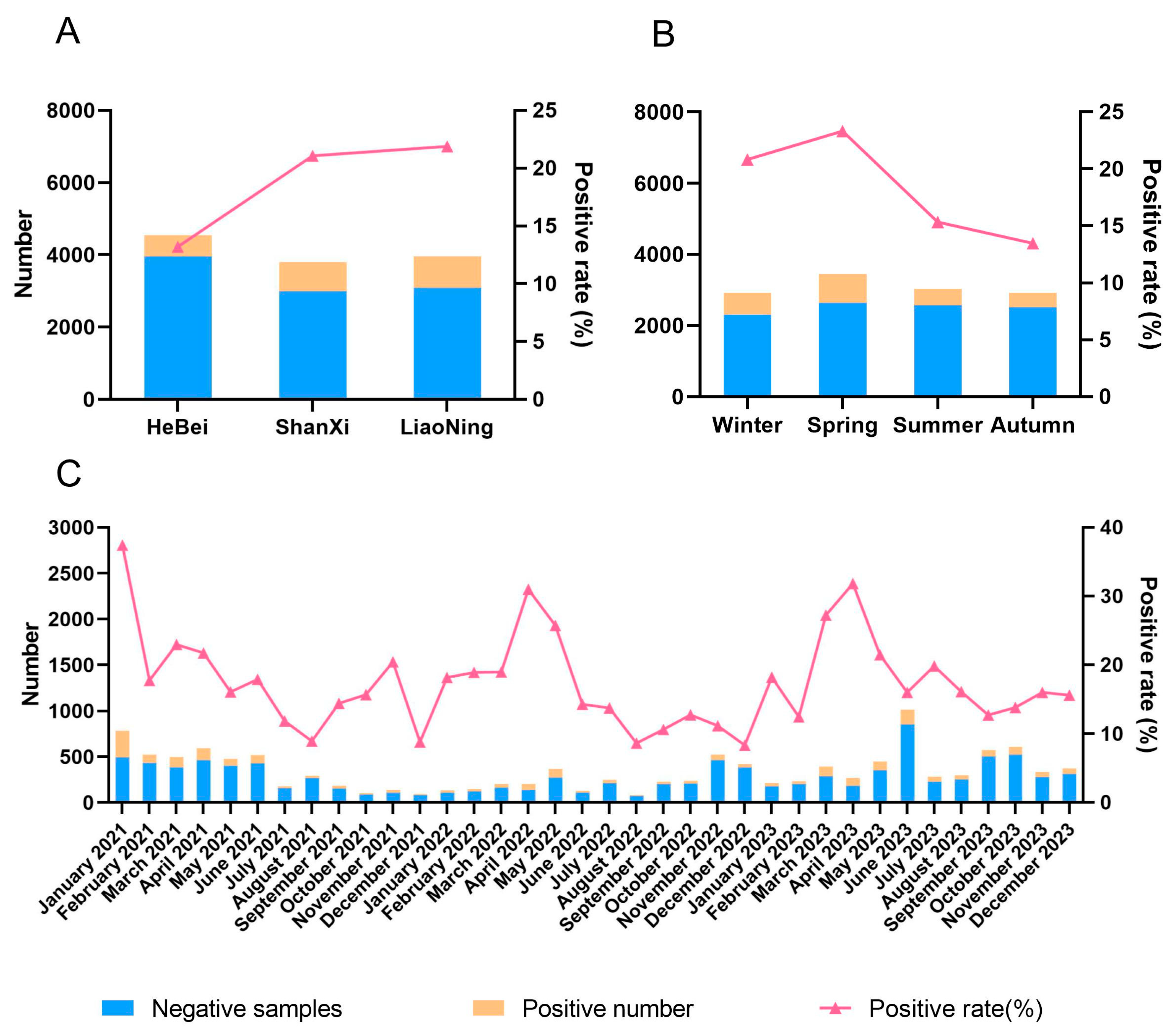
3.1. Clinical Sample Detection and Analysis
3.2. Analysis of ORF5 Gene Phylogeny
3.3. Mutational Analysis of ORF5 Amino Acid Sequences
3.4. Isolation of PRRSV Strains
3.5. Whole-Genome Recombination Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Keffaber, K. Reproduction Failure of Unknown Etiology. Am. Assoc. Swine Prac. News. 1989, 1, 1–9. [Google Scholar]
- Li, Y.H.; Tas, A.; Sun, Z.; Snijder, E.J.; Fang, Y. Proteolytic processing of the porcine reproductive and respiratory syndrome virus replicase. Virus Res. 2015, 202, 48–59. [Google Scholar] [CrossRef] [PubMed]
- Brinton, M.A.; Gulyeava, A.A.; Balasuriya, U.B.R.; Dunowska, M.; Faaberg, K.S.; Goldberg, T.; Leung, F.C.C.; Nauwynck, H.J.; Snijder, E.J.; Stadejek, T.; et al. ICTV virus taxonomy profile: Arteriviridae 2021. J. Gen. Virol. 2021, 102, 001632. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Lam, T.T.; Hon, C.C.; Murtaugh, M.P.; Davies, P.R.; Hui, R.K.; Li, J.; Wong, L.T.; Yip, C.W.; Jiang, J.W.; et al. Phylogeny-based evolutionary, demographical, and geographical dissection of North American type 2 porcine reproductive and respiratory syndrome viruses. J. Virol. 2010, 84, 8700–8711. [Google Scholar] [CrossRef] [PubMed]
- Bao, H.Y.; Li, X.D. Emergence and spread of NADC34-like PRRSV in China. Transbound. Emerg. Dis. 2021, 68, 3005–3008. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.Q.; Chen, Z.S.; Liu, W.X.; Cui, Y.Z. Isolation and identification of porcine reproductive and respiratory syndrome (PRRS) virus from aborted fetuses suspected of PRRS. Chin. J. Prevent. Vet. Med. 1996, 87, 3–7. (In Chinese) [Google Scholar]
- Yang, H.C.; Guan, S.H.; Yin, X.M.; Gan, M.H. Isolation and preliminary identification of porcine reproductive and respiratory syndrome virus. Chin. J. Vet. Med. 1997, 20, 9–10. (In Chinese) [Google Scholar]
- Tian, K.G.; Yu, X.L.; Zhao, T.Z.; Feng, Y.J.; Cao, Z.; Wang, C.B.; Hu, Y.; Chen, X.Z.; Hu, D.M.; Tian, X.S.; et al. Emergence of fatal PRRSV variants: Unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS ONE 2007, 2, e526. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.H.; Tun, H.M.; Sun, B.L.; Mo, J.; Zhou, Q.F.; Deng, Y.X.; Xie, Q.M.; Bi, Y.Z.; Leung, F.C.; Ma, J.Y. Re-emerging of porcine respiratory and reproductive syndrome virus (lineage 3) and increased pathogenicity after genomic recombination with vaccine variant. Vet. Microbiol. 2015, 175, 332–340. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Y.; Ji, G.B.; Wang, J.; Tan, F.F.; Zhuang, J.S.; Li, X.D.; Tian, K.G. Complete genome sequence of an NADC30-like porcine reproductive and respiratory syndrome virus characterized by recombination with other strains. Genome Announc. 2016, 4, e00330-16. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Yan, Y.; Shi, M.; Liu, H.Z.; Zhang, H.L.; Yang, Y.B.; Huang, X.Y.; Gauger, P.C.; Zhang, J.Q.; Zhang, Y.H.; et al. Phylogenetics, genomic recombination, and NSP2 polymorphic patterns of porcine reproductive and respiratory syndrome virus in China and the United States in 2014–2018. J. Virol. 2020, 94, e01813-19. [Google Scholar] [CrossRef]
- van Geelen, A.G.M.; Anderson, T.K.; Lager, K.M.; Das, P.B.; Otis, N.J.; Montiel, N.A.; Miller, L.C.; Kulshreshtha, V.; Buckley, A.C.; Brockmeier, S.L.; et al. Porcine reproductive and respiratory disease virus: Evolution and recombination yields distinct ORF5 RFLP 1-7-4 viruses with individual pathogenicity. Virology 2018, 513, 168–179. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.Z.; Ha, Z.; Nan, F.L.; Zhang, Y.; Zhang, H.; Li, J.F.; Zhang, P.; Han, J.C.; Zhang, H.; Zhuang, X.Y.; et al. Characterization of porcine reproductive and respiratory syndrome virus (ORF5 RFLP 1-7-4 viruses) in northern China. Microb. Pathog. 2020, 140, 103941. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Kang, R.M.; Zhang, Y.; Ding, M.D.; Xie, B.; Tian, Y.M.; Wu, X.; Zuo, L.; Yang, X.; Wang, H.N. Whole genome analysis of two novel type 2 porcine reproductive and respiratory syndrome viruses with complex genome recombination between lineage 8, 3, and 1 strains identified in southwestern China. Viruses 2018, 10, 328. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Kang, R.M.; Yu, J.F.; Xie, B.; Chen, C.Y.; Li, X.Y.; Xie, J.; Ye, Y.G.; Xiao, L.; Zhang, J.L.; et al. Genetic characterization and pathogenicity of a novel recombined porcine reproductive and respiratory syndrome virus 2 among Nadc30-like, Jxa1-like, and Mlv-like strains. Viruses 2018, 10, 551. [Google Scholar] [CrossRef]
- Wu, Y.; Peng, O.Y.; Xu, Q.P.; Li, Q.H.; Li, W.; Lin, L.M.; Zhou, Q.F.; Cai, X.B.; Hu, G.L.; He, Z.Y.; et al. Characterization and pathogenicity of two novel PRRSVs recombined by NADC30-like and NADC34-like strains in China. Viruses 2022, 14, 2174. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.D.; Qu, X.Y.; Zhang, H.L.; Tang, X.D.; Bian, T.; Sun, Y.J.; Zhou, M.M.; Ren, F.B.; Wu, P. Evolutionary and recombination analysis of porcine reproductive and respiratory syndrome isolates in China. Virus Genes 2020, 56, 354–360. [Google Scholar] [CrossRef] [PubMed]
- Paploski, I.A.D.; Pamornchainavakul, N.; Makau, D.N.; Rovira, A.; Corzo, C.A.; Schroeder, D.C.; Cheeran, M.C.J.; Doeschl-Wilson, A.; Kao, R.R.; Lycett, S.; et al. Phylogenetic structure and sequential dominance of sub-lineages of PRRSV type-2 lineage 1 in the United States. Vaccines 2021, 9, 608. [Google Scholar] [CrossRef]
- Jiao, D. Isolation and Identification of a Multilineage Recombinant PRRSV and Analysis of Its Genomic Characteristics and Pathogenicity. Master’s Thesis, Northwest A&F University, Yangling, China, 2022. (In Chinese). [Google Scholar]
- Wang, P.P.; Dong, J.G.; Zhang, L.Y.; Liang, P.S.; Liu, Y.L.; Wang, L.; Fan, F.H.; Song, C.X. Sequence and phylogenetic analyses of the Nsp2 and ORF5 genes of porcine reproductive and respiratory syndrome virus in boars from south China in 2015. Transbound. Emerg. Dis. 2017, 64, 1953–1964. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.S.; Liang, W.; Wang, X.Y.; Chen, H.J.; Fan, J.; Song, W.B.; Hua, L.; Tang, X.B.; Chen, H.C.; Peng, Z.; et al. Epidemiological and genetic characteristics of porcine reproduction and respiratory syndrome virus 2 in mainland China. Arch. Virol. 2020, 165, 1621–1632. [Google Scholar] [CrossRef]
- Sun, Y.F.; Yu, H.; Jiang, X.; Ma, J.F.; Xu, C.Q.; Yu, X.X.; Li, L.A. Novel ORF5 deletion of NADC30-like porcine reproductive and respiratory syndrome viruses circulating in northern China from 2016 to 2018. J. Vet. Diagn. Investig. 2020, 32, 928–932. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.Y.; Chang, T.; Wang, D.C.; Zhang, H.L.; Liu, H.Z.; Huang, X.Y.; Tian, Z.J.; Tian, X.X.; Liu, D.; An, T.Q.; et al. Genomic surveillance and evolutionary dynamics of type 2 porcine reproductive and respiratory syndrome virus in China spanning the African swine fever outbreak. Virus Evol. 2024, 10, veae016. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Li, C.; Li, W.S.; Zhao, J.; Gong, B.J.; Sun, Q.; Tang, Y.D.; Xiang, L.R.; Leng, C.L.; Peng, J.M.; et al. Novel characteristics of Chinese NADC34-like PRRSV during 2020–2021. Transbound. Emerg. Dis. 2022, 69, e3215–e3224. [Google Scholar] [CrossRef]
- Kang, P.; Zhao, M.P.; Huang, Y.H.; Lv, Z.J.; Wang, X.H. Progress in structure and function of gp5 protein of porcine reproductive and respiratory syndrome virus. Guangdong Agric. Sci. 2023, 50, 104–114. (In Chinese) [Google Scholar]
- Zhou, L.; Kang, R.M.; Ji, G.S.; Tian, Y.M.; Ge, M.Y.; Xie, B.; Yang, X.; Wang, H.N. Molecular characterization and recombination analysis of porcine reproductive and respiratory syndrome virus emerged in southwestern China during 2012–2016. Virus Genes 2018, 54, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.Y.; Xia, D.S.; Luo, L.Z.; An, T.Q. Recombination of Porcine Reproductive and Respiratory Syndrome Virus: Features, Possible Mechanisms, and Future Directions. Viruses 2024, 16, 929. [Google Scholar] [CrossRef]
- Ouyang, Y.; Du, Y.B.; Zhang, H.J.; Guo, J.H.; Sun, Z.; Luo, X.X.; Mei, X.W.; Xiao, S.B.; Fang, L.R.; Zhou, Y.R. Genetic characterization and pathogenicity of a recombinant porcine reproductive and respiratory syndrome virus strain in China. Viruses 2024, 16, 993. [Google Scholar] [CrossRef]
- Cui, X.Y.; Xia, D.S.; Huang, X.Y.; Sun, Y.; Shi, M.; Zhang, J.Q.; Li, G.W.; Yang, Y.B.; Wang, H.W.; Cai, X.H.; et al. Analysis of recombinant characteristics based on 949 PRRSV-2 genomic sequences obtained from 1991 to 2021 shows that viral multiplication ability contributes to dominant recombination. Microbiol. Spectr. 2022, 10, e0293422. [Google Scholar] [CrossRef]




| Age of Piglet | Type of Sample | Sampling Time | Sample Size |
|---|---|---|---|
| Boars | Semen/oropharyngeal swab | Quarterly | 20% of each batch with pools of 5 |
| Sows | Serum/tissue samples | When abortion occurs | Each pig |
| Suckling piglets | Processing fluid | Each batch of piglets | Pools of 20–30 litters |
| Weaned piglets | Serum/oropharyngeal swab | 3 days before and after weaning | Weak pigs with pools of 5 |
| Fattening pig | Serum/oral fluid | Suspected clinical symptoms | Pools of 15–20 litters |
| Dead pigs | Tissue samples | Immediately | Each pig |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, N.; Yang, Z.; Lv, F.; Dou, L.; Li, X.; Zhao, B.; Dong, S. Molecular Epidemiology and Genetic Evolution of Porcine Reproductive and Respiratory Syndrome Virus in Northern China During 2021–2023. Viruses 2025, 17, 85. https://doi.org/10.3390/v17010085
Yuan N, Yang Z, Lv F, Dou L, Li X, Zhao B, Dong S. Molecular Epidemiology and Genetic Evolution of Porcine Reproductive and Respiratory Syndrome Virus in Northern China During 2021–2023. Viruses. 2025; 17(1):85. https://doi.org/10.3390/v17010085
Chicago/Turabian StyleYuan, Na, Zuofeng Yang, Fengxia Lv, Lina Dou, Xiangqing Li, Baokai Zhao, and Shishan Dong. 2025. "Molecular Epidemiology and Genetic Evolution of Porcine Reproductive and Respiratory Syndrome Virus in Northern China During 2021–2023" Viruses 17, no. 1: 85. https://doi.org/10.3390/v17010085
APA StyleYuan, N., Yang, Z., Lv, F., Dou, L., Li, X., Zhao, B., & Dong, S. (2025). Molecular Epidemiology and Genetic Evolution of Porcine Reproductive and Respiratory Syndrome Virus in Northern China During 2021–2023. Viruses, 17(1), 85. https://doi.org/10.3390/v17010085






