Exploration of the Role of Cyclophilins in Established Hepatitis B and C Infections
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Viral Production and Infection
2.3. ELISA
2.4. Transfection
2.5. Western Blot Analysis and Immunofluorescence
2.6. RT/qPCR for HCV and HBV Quantification
2.7. Statistics
3. Results
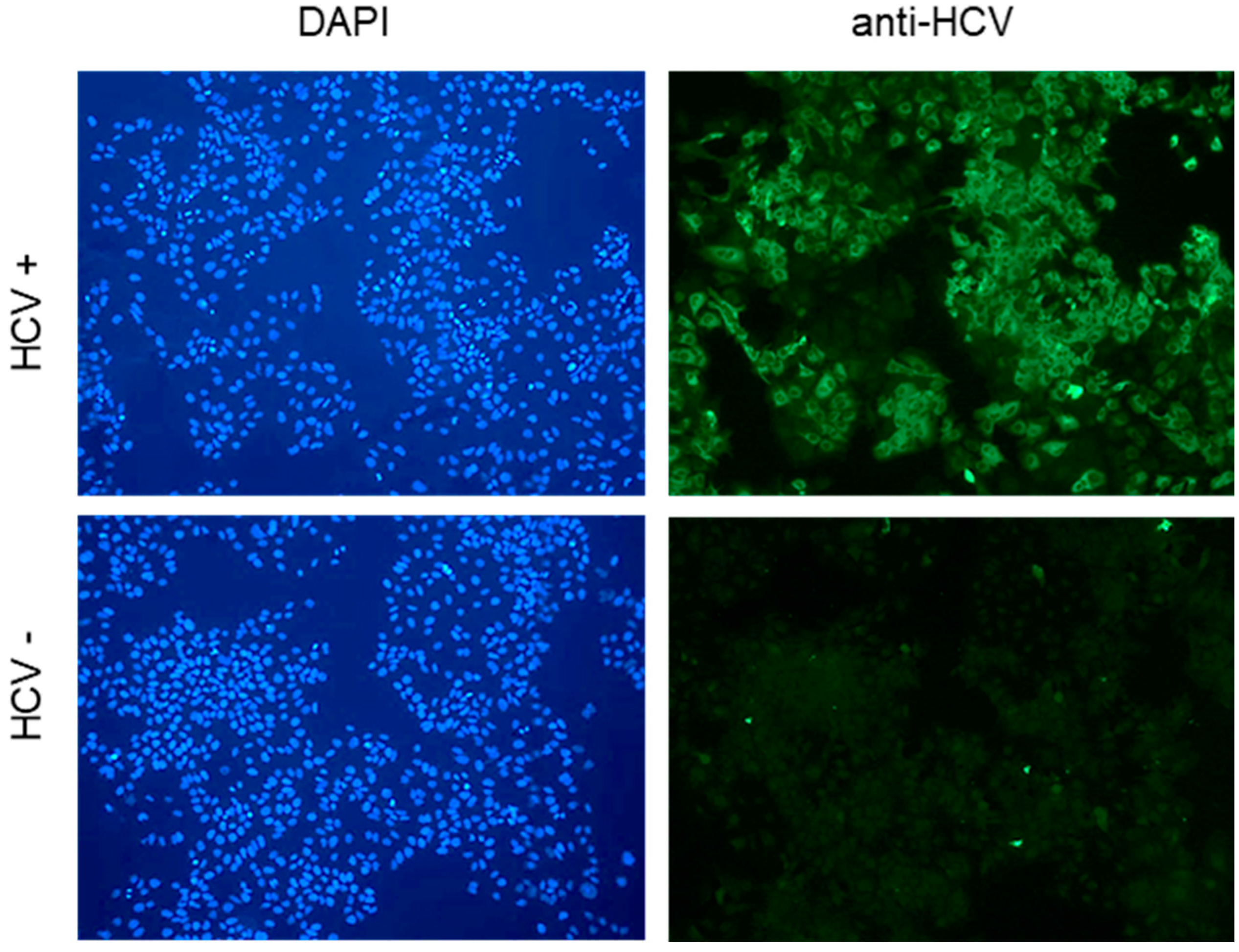
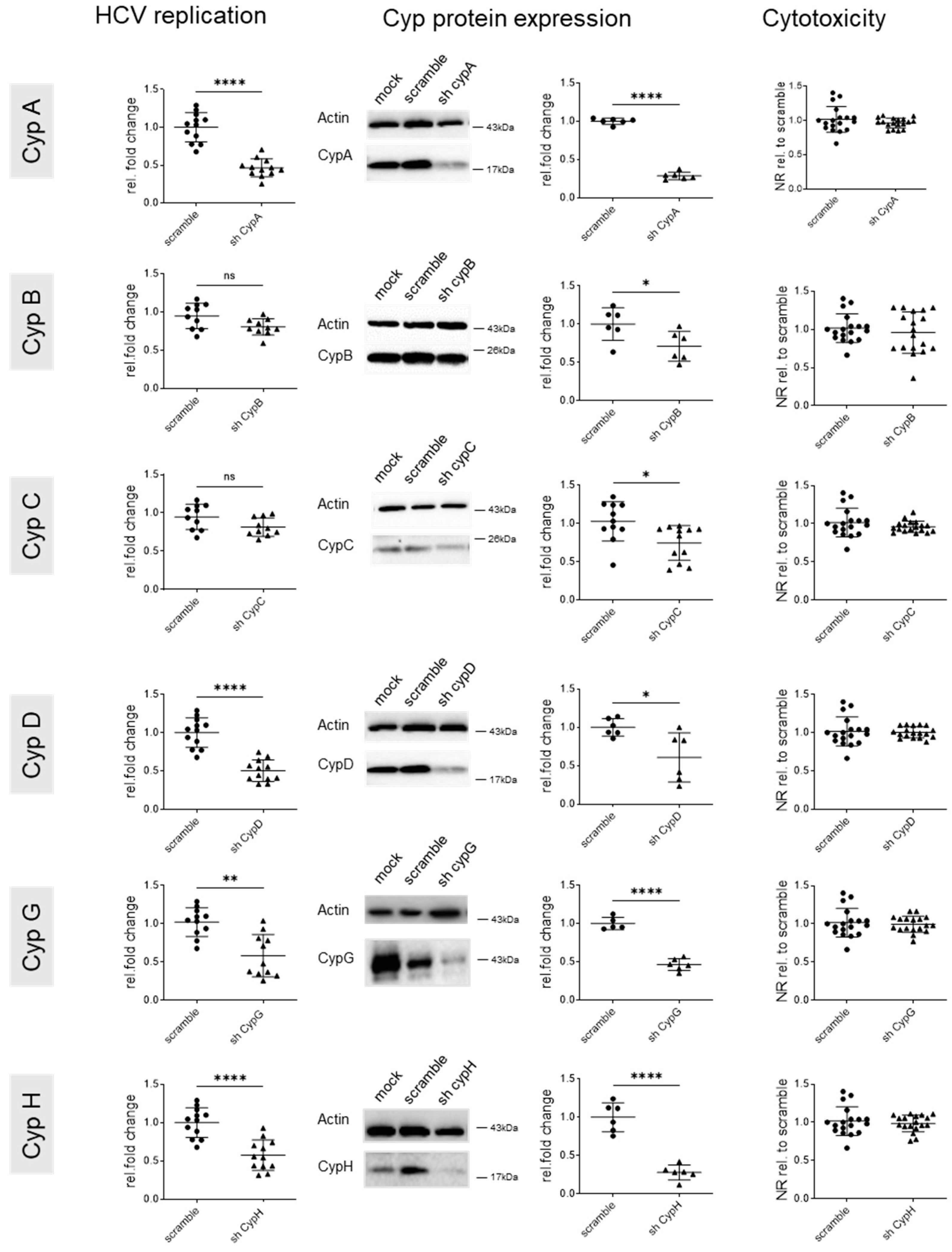
3.1. Cyclophilin G and H Are Required for Efficient HCV Replication
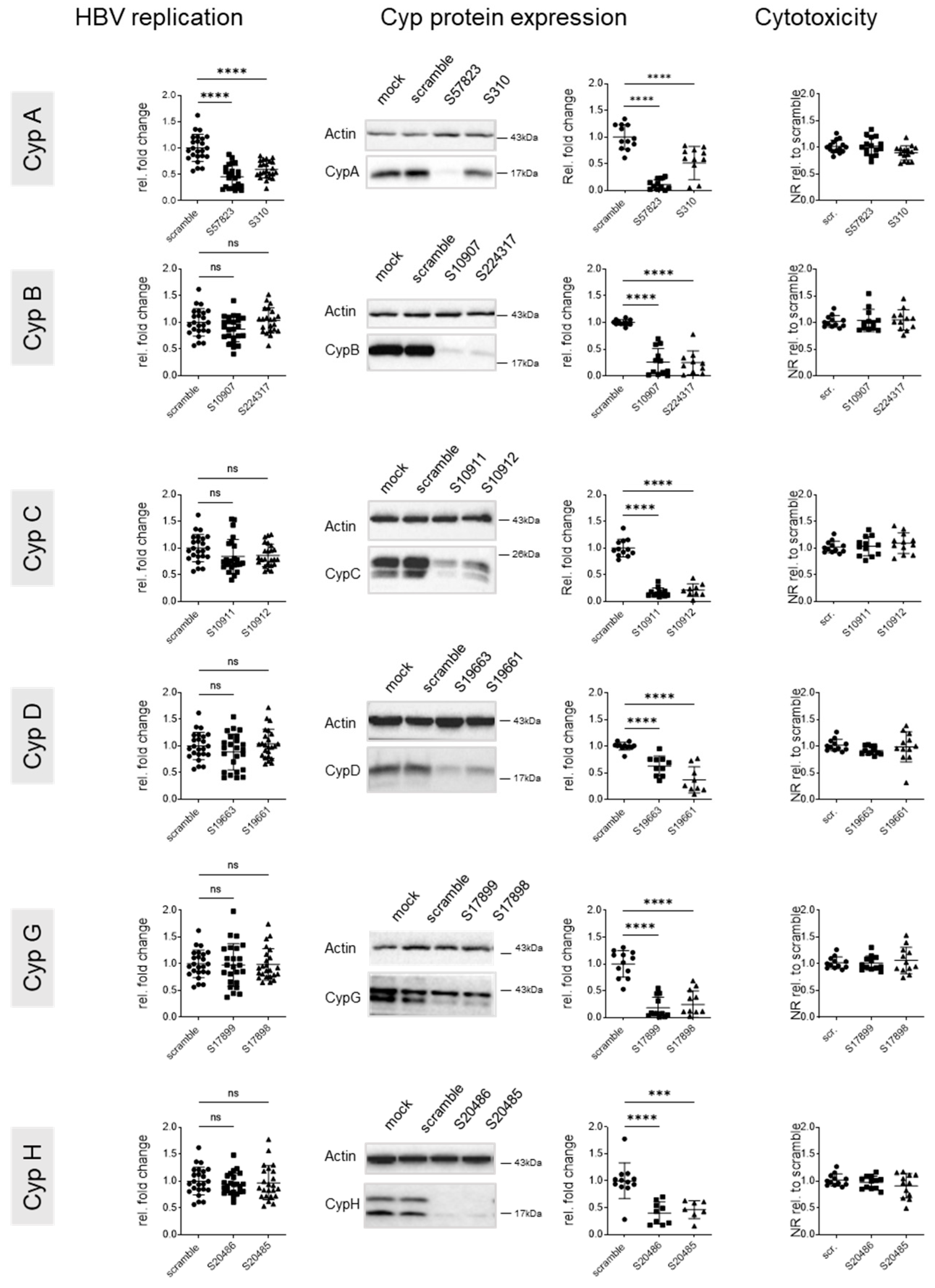
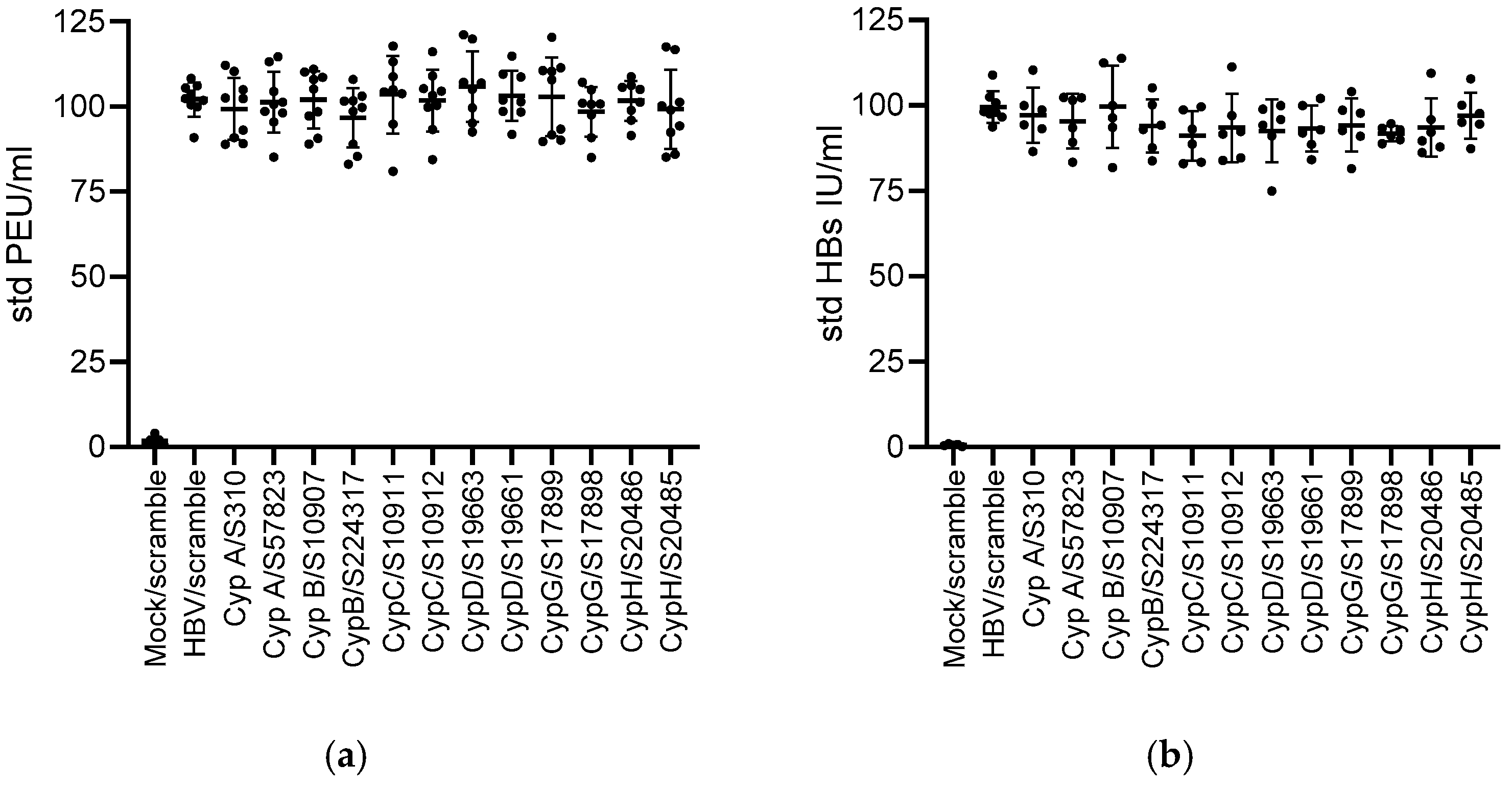
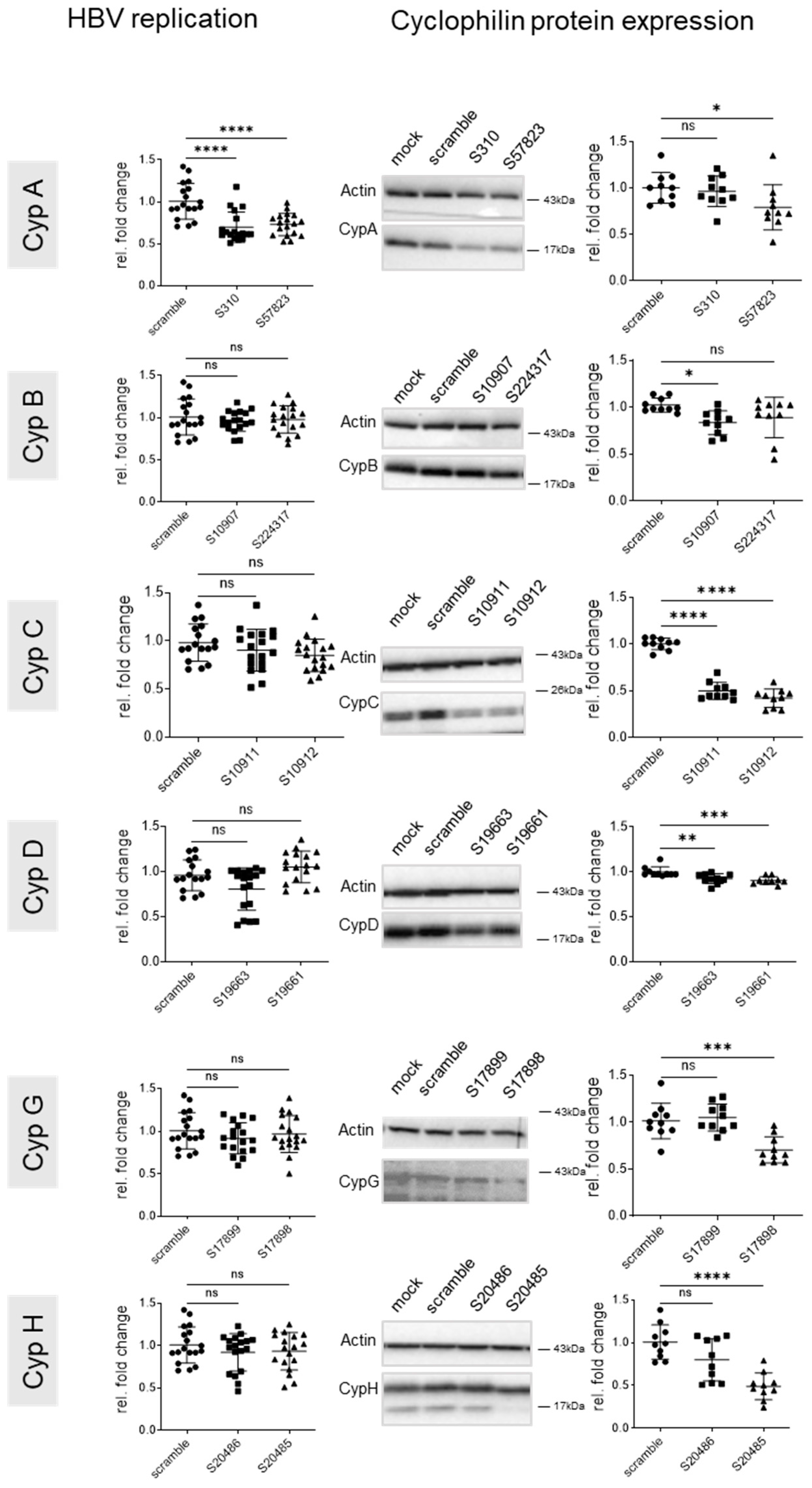
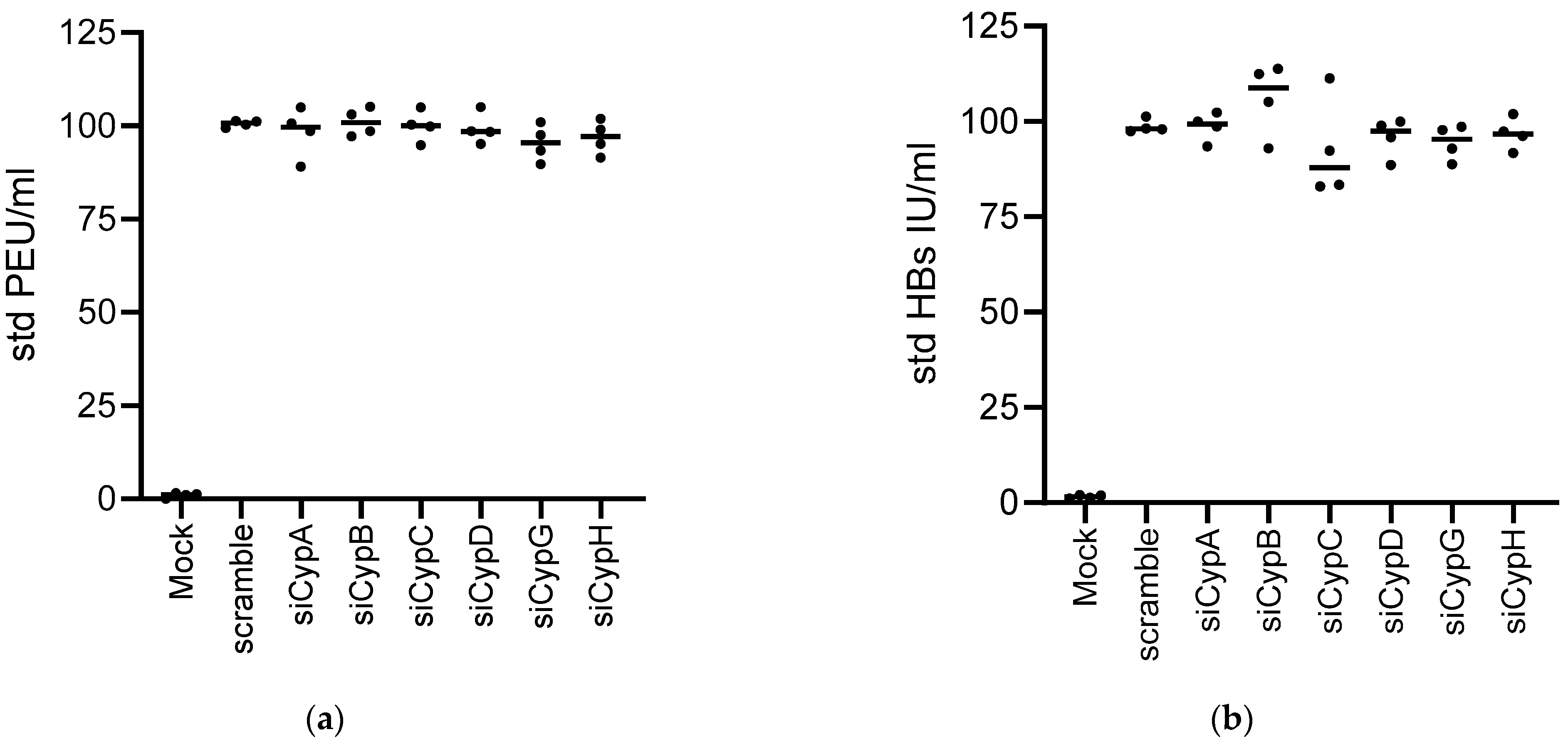
3.2. Knock Down of CypA Impacts Established HBV Infection
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stamnes, M.A.; Rutherford, S.L.; Zuker, C.S. Cyclophilins: A new family of proteins involved in intracellular folding. Trends Cell Biol. 1992, 2, 272–276. [Google Scholar] [CrossRef] [PubMed]
- Nigro, P.; Pompilio, G.; Capogrossi, M.C. Cyclophilin a: A key player for human disease. Cell Death Dis. 2013, 4, e888. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Choi, T.G.; Ding, Y.; Kim, Y.; Ha, K.S.; Lee, K.H.; Kang, I.; Ha, J.; Kaufman, R.J.; Lee, J.; et al. Overexpressed cyclophilin b suppresses apoptosis associated with ros and Ca2+ homeostasis after er stress. J. Cell Sci. 2008, 121, 3636–3648. [Google Scholar] [CrossRef] [PubMed]
- Elrod, J.W.; Molkentin, J.D. Physiologic functions of cyclophilin d and the mitochondrial permeability transition pore. Circ. J. 2013, 77, 1111–1122. [Google Scholar] [CrossRef] [PubMed]
- Rajiv, C.; Davis, T.L. Structural and functional insights into human nuclear cyclophilins. Biomolecules 2018, 8, 161. [Google Scholar] [CrossRef] [PubMed]
- Thali, M.; Bukovsky, A.; Kondo, E.; Rosenwirth, B.; Walsh, C.T.; Sodroski, J.; Gottlinger, H.G. Functional association of cyclophilin a with hiv-1 virions. Nature 1994, 372, 363–365. [Google Scholar] [CrossRef]
- Franke, E.K.; Yuan, H.E.; Luban, J. Specific incorporation of cyclophilin a into hiv-1 virions. Nature 1994, 372, 359–362. [Google Scholar] [CrossRef]
- Chatterji, U.; Bobardt, M.; Selvarajah, S.; Yang, F.; Tang, H.; Sakamoto, N.; Vuagniaux, G.; Parkinson, T.; Gallay, P. The isomerase active site of cyclophilin a is critical for hepatitis c virus replication. J. Biol. Chem. 2009, 284, 16998–17005. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Robotham, J.M.; Nelson, H.B.; Irsigler, A.; Kenworthy, R.; Tang, H. Cyclophilin a is an essential cofactor for hepatitis c virus infection and the principal mediator of cyclosporine resistance in vitro. J. Virol. 2008, 82, 5269–5278. [Google Scholar] [CrossRef] [PubMed]
- Kaul, A.; Stauffer, S.; Berger, C.; Pertel, T.; Schmitt, J.; Kallis, S.; Zayas, M.; Lohmann, V.; Luban, J.; Bartenschlager, R. Essential role of cyclophilin a for hepatitis c virus replication and virus production and possible link to polyprotein cleavage kinetics. PLoS Pathog. 2009, 5, e1000546. [Google Scholar] [CrossRef]
- Damaso, C.R.; Moussatche, N. Inhibition of vaccinia virus replication by cyclosporin a analogues correlates with their affinity for cellular cyclophilins. J. Gen. Virol. 1998, 79 Pt 2, 339–346. [Google Scholar] [CrossRef]
- Qing, M.; Yang, F.; Zhang, B.; Zou, G.; Robida, J.M.; Yuan, Z.; Tang, H.; Shi, P.Y. Cyclosporine inhibits flavivirus replication through blocking the interaction between host cyclophilins and viral ns5 protein. Antimicrob. Agents Chemother. 2009, 53, 3226–3235. [Google Scholar] [CrossRef] [PubMed]
- Nkongolo, S.; Ni, Y.; Lempp, F.A.; Kaufman, C.; Lindner, T.; Esser-Nobis, K.; Lohmann, V.; Mier, W.; Mehrle, S.; Urban, S. Cyclosporin a inhibits hepatitis b and hepatitis d virus entry by cyclophilin-independent interference with the ntcp receptor. J. Hepatol. 2014, 60, 723–731. [Google Scholar] [CrossRef]
- Bienkowska-Haba, M.; Patel, H.D.; Sapp, M. Target cell cyclophilins facilitate human papillomavirus type 16 infection. PLoS Pathog. 2009, 5, e1000524. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, H.; Mocarski, E.S.; Kosugi, I.; Tsutsui, Y. Cyclosporine inhibits mouse cytomegalovirus infection via a cyclophilin-dependent pathway specifically in neural stem/progenitor cells. J. Virol. 2007, 81, 9013–9023. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pfefferle, S.; Schopf, J.; Kogl, M.; Friedel, C.C.; Muller, M.A.; Carbajo-Lozoya, J.; Stellberger, T.; von Dall’Armi, E.; Herzog, P.; Kallies, S.; et al. The sars-coronavirus-host interactome: Identification of cyclophilins as target for pan-coronavirus inhibitors. PLoS Pathog. 2011, 7, e1002331. [Google Scholar] [CrossRef] [PubMed]
- Ma-Lauer, Y.; Zheng, Y.; Malesevic, M.; von Brunn, B.; Fischer, G.; von Brunn, A. Influences of cyclosporin a and non-immunosuppressive derivatives on cellular cyclophilins and viral nucleocapsid protein during human coronavirus 229e replication. Antiviral Res. 2020, 173, 104620. [Google Scholar] [CrossRef]
- Kambara, H.; Tani, H.; Mori, Y.; Abe, T.; Katoh, H.; Fukuhara, T.; Taguwa, S.; Moriishi, K.; Matsuura, Y. Involvement of cyclophilin b in the replication of japanese encephalitis virus. Virology 2011, 412, 211–219. [Google Scholar] [CrossRef]
- Liu, X.; Zhao, Z.; Li, Z.; Xu, C.; Sun, L.; Chen, J.; Liu, W. Cyclosporin a inhibits the influenza virus replication through cyclophilin a-dependent and -independent pathways. PLoS ONE 2012, 7, e37277. [Google Scholar] [CrossRef]
- Bose, S.; Mathur, M.; Bates, P.; Joshi, N.; Banerjee, A.K. Requirement for cyclophilin a for the replication of vesicular stomatitis virus new jersey serotype. J. Gen. Virol. 2003, 84, 1687–1699. [Google Scholar] [CrossRef] [PubMed]
- Nevers, Q.; Ruiz, I.; Ahnou, N.; Donati, F.; Brillet, R.; Softic, L.; Chazal, M.; Jouvenet, N.; Fourati, S.; Baudesson, C.; et al. Characterization of the anti-hepatitis c virus activity of new nonpeptidic small-molecule cyclophilin inhibitors with the potential for broad anti-flaviviridae activity. Antimicrob. Agents Chemother. 2018, 62, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Peterson, A.A.; Rangwala, A.M.; Thakur, M.K.; Ward, P.S.; Hung, C.; Outhwaite, I.R.; Chan, A.I.; Usanov, D.L.; Mootha, V.K.; Seeliger, M.A.; et al. Discovery and molecular basis of subtype-selective cyclophilin inhibitors. Nat. Chem. Biol. 2022, 18, 1184–1195. [Google Scholar] [CrossRef] [PubMed]
- Chatterji, U.; Garcia-Rivera, J.A.; Baugh, J.; Gawlik, K.; Wong, K.A.; Zhong, W.; Brass, C.A.; Naoumov, N.V.; Gallay, P.A. The combination of alisporivir plus an ns5a inhibitor provides additive to synergistic anti-hepatitis c virus activity without detectable cross-resistance. Antimicrob. Agents Chemother. 2014, 58, 3327–3334. [Google Scholar] [CrossRef]
- Pawlotsky, J.M.; Flisiak, R.; Sarin, S.K.; Rasenack, J.; Piratvisuth, T.; Chuang, W.L.; Peng, C.Y.; Foster, G.R.; Shah, S.; Wedemeyer, H.; et al. Alisporivir plus ribavirin, interferon free or in combination with pegylated interferon, for hepatitis c virus genotype 2 or 3 infection. Hepatology 2015, 62, 1013–1023. [Google Scholar] [CrossRef]
- Zeuzem, S.; Flisiak, R.; Vierling, J.M.; Mazur, W.; Mazzella, G.; Thongsawat, S.; Abdurakhmanov, D.; Van Kinh, N.; Calistru, P.; Heo, J.; et al. Randomised clinical trial: Alisporivir combined with peginterferon and ribavirin in treatment-naive patients with chronic hcv genotype 1 infection (essential ii). Aliment. Pharmacol. Ther. 2015, 42, 829–844. [Google Scholar] [CrossRef] [PubMed]
- Buti, M.; Flisiak, R.; Kao, J.H.; Chuang, W.L.; Streinu-Cercel, A.; Tabak, F.; Calistru, P.; Goeser, T.; Rasenack, J.; Horban, A.; et al. Alisporivir with peginterferon/ribavirin in patients with chronic hepatitis c genotype 1 infection who failed to respond to or relapsed after prior interferon-based therapy: Fundamental, a phase ii trial. J. Viral Hepat. 2015, 22, 596–606. [Google Scholar] [CrossRef]
- Shimura, S.; Watashi, K.; Fukano, K.; Peel, M.; Sluder, A.; Kawai, F.; Iwamoto, M.; Tsukuda, S.; Takeuchi, J.S.; Miyake, T.; et al. Cyclosporin derivatives inhibit hepatitis b virus entry without interfering with ntcp transporter activity. J. Hepatol. 2017, 66, 685–692. [Google Scholar] [CrossRef]
- Watashi, K.; Sluder, A.; Daito, T.; Matsunaga, S.; Ryo, A.; Nagamori, S.; Iwamoto, M.; Nakajima, S.; Tsukuda, S.; Borroto-Esoda, K.; et al. Cyclosporin a and its analogs inhibit hepatitis b virus entry into cultured hepatocytes through targeting a membrane transporter, sodium taurocholate cotransporting polypeptide (ntcp). Hepatology 2014, 59, 1726–1737. [Google Scholar] [CrossRef]
- Phillips, S.; Chokshi, S.; Chatterji, U.; Riva, A.; Bobardt, M.; Williams, R.; Gallay, P.; Naoumov, N.V. Alisporivir inhibition of hepatocyte cyclophilins reduces hbv replication and hepatitis b surface antigen production. Gastroenterology 2015, 148, 403–414.e7. [Google Scholar] [CrossRef]
- Hansson, M.J.; Moss, S.J.; Bobardt, M.; Chatterji, U.; Coates, N.; Garcia-Rivera, J.A.; Elmer, E.; Kendrew, S.; Leyssen, P.; Neyts, J.; et al. Bioengineering and semisynthesis of an optimized cyclophilin inhibitor for treatment of chronic viral infection. Chem. Biol. 2015, 22, 285–292. [Google Scholar] [CrossRef][Green Version]
- Gallay, P.; Ure, D.; Bobardt, M.; Chatterji, U.; Ou, J.; Trepanier, D.; Foster, R. The cyclophilin inhibitor crv431 inhibits liver hbv DNA and hbsag in transgenic mice. PLoS ONE 2019, 14, e0217433. [Google Scholar] [CrossRef] [PubMed]
- Coelmont, L.; Hanoulle, X.; Chatterji, U.; Berger, C.; Snoeck, J.; Bobardt, M.; Lim, P.; Vliegen, I.; Paeshuyse, J.; Vuagniaux, G.; et al. Deb025 (alisporivir) inhibits hepatitis c virus replication by preventing a cyclophilin a induced cis-trans isomerisation in domain ii of ns5a. PLoS ONE 2010, 5, e13687. [Google Scholar] [CrossRef] [PubMed]
- Hanoulle, X.; Badillo, A.; Wieruszeski, J.M.; Verdegem, D.; Landrieu, I.; Bartenschlager, R.; Penin, F.; Lippens, G. Hepatitis c virus ns5a protein is a substrate for the peptidyl-prolyl cis/trans isomerase activity of cyclophilins a and b. J. Biol. Chem. 2009, 284, 13589–13601. [Google Scholar] [CrossRef]
- Fernandes, F.; Ansari, I.U.; Striker, R. Cyclosporine inhibits a direct interaction between cyclophilins and hepatitis c ns5a. PLoS ONE 2010, 5, e9815. [Google Scholar] [CrossRef]
- Hopkins, S.; Bobardt, M.; Chatterji, U.; Garcia-Rivera, J.A.; Lim, P.; Gallay, P.A. The cyclophilin inhibitor scy-635 disrupts hepatitis c virus ns5a-cyclophilin a complexes. Antimicrob. Agents Chemother. 2012, 56, 3888–3897. [Google Scholar] [CrossRef]
- Fernandes, F.; Poole, D.S.; Hoover, S.; Middleton, R.; Andrei, A.C.; Gerstner, J.; Striker, R. Sensitivity of hepatitis c virus to cyclosporine a depends on nonstructural proteins ns5a and ns5b. Hepatology 2007, 46, 1026–1033. [Google Scholar] [CrossRef] [PubMed]
- Chatterji, U.; Bobardt, M.; Tai, A.; Wood, M.; Gallay, P.A. Cyclophilin and ns5a inhibitors, but not other anti-hepatitis c virus (hcv) agents, preclude hcv-mediated formation of double-membrane-vesicle viral factories. Antimicrob. Agents Chemother. 2015, 59, 2496–2507. [Google Scholar] [CrossRef]
- Chatterji, U.; Bobardt, M.D.; Lim, P.; Gallay, P.A. Cyclophilin a-independent recruitment of ns5a and ns5b into hepatitis c virus replication complexes. J. Gen. Virol. 2010, 91, 1189–1193. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Rivera, J.A.; Bobardt, M.; Chatterji, U.; Hopkins, S.; Gregory, M.A.; Wilkinson, B.; Lin, K.; Gallay, P.A. Multiple mutations in hepatitis c virus ns5a domain ii are required to confer a significant level of resistance to alisporivir. Antimicrob. Agents Chemother. 2012, 56, 5113–5121. [Google Scholar] [CrossRef] [PubMed]
- Ciesek, S.; Steinmann, E.; Wedemeyer, H.; Manns, M.P.; Neyts, J.; Tautz, N.; Madan, V.; Bartenschlager, R.; von Hahn, T.; Pietschmann, T. Cyclosporine a inhibits hepatitis c virus nonstructural protein 2 through cyclophilin a. Hepatology 2009, 50, 1638–1645. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, M.; Sakamoto, N.; Tanabe, Y.; Koyama, T.; Itsui, Y.; Takeda, Y.; Chen, C.H.; Kakinuma, S.; Oooka, S.; Maekawa, S.; et al. Suppression of hepatitis c virus replication by cyclosporin a is mediated by blockade of cyclophilins. Gastroenterology 2005, 129, 1031–1041. [Google Scholar] [CrossRef] [PubMed]
- Watashi, K.; Hijikata, M.; Hosaka, M.; Yamaji, M.; Shimotohno, K. Cyclosporin a suppresses replication of hepatitis c virus genome in cultured hepatocytes. Hepatology 2003, 38, 1282–1288. [Google Scholar] [CrossRef] [PubMed]
- Goto, K.; Watashi, K.; Inoue, D.; Hijikata, M.; Shimotohno, K. Identification of cellular and viral factors related to anti-hepatitis c virus activity of cyclophilin inhibitor. Cancer Sci. 2009, 100, 1943–1950. [Google Scholar] [CrossRef]
- Anderson, L.J.; Lin, K.; Compton, T.; Wiedmann, B. Inhibition of cyclophilins alters lipid trafficking and blocks hepatitis c virus secretion. Virol. J. 2011, 8, 329. [Google Scholar] [CrossRef] [PubMed]
- Gaither, L.A.; Borawski, J.; Anderson, L.J.; Balabanis, K.A.; Devay, P.; Joberty, G.; Rau, C.; Schirle, M.; Bouwmeester, T.; Mickanin, C.; et al. Multiple cyclophilins involved in different cellular pathways mediate hcv replication. Virology 2010, 397, 43–55. [Google Scholar] [CrossRef] [PubMed]
- Duponchel, S.; Monnier, L.; Molle, J.; Bendridi, N.; Alam, M.R.; Gaballah, A.; Grigorov, B.; Ivanov, A.; Schmiel, M.; Odenthal, M.; et al. Hepatitis c virus replication requires integrity of mitochondria-associated er membranes. JHEP Rep. 2023, 5, 100647. [Google Scholar] [CrossRef] [PubMed]
- Quarato, G.; D’Aprile, A.; Gavillet, B.; Vuagniaux, G.; Moradpour, D.; Capitanio, N.; Piccoli, C. The cyclophilin inhibitor alisporivir prevents hepatitis c virus-mediated mitochondrial dysfunction. Hepatology 2012, 55, 1333–1343. [Google Scholar] [CrossRef] [PubMed]
- Ishii, N.; Watashi, K.; Hishiki, T.; Goto, K.; Inoue, D.; Hijikata, M.; Wakita, T.; Kato, N.; Shimotohno, K. Diverse effects of cyclosporine on hepatitis c virus strain replication. J. Virol. 2006, 80, 4510–4520. [Google Scholar] [CrossRef]
- Hopkins, S.; Gallay, P. Cyclophilin inhibitors: An emerging class of therapeutics for the treatment of chronic hepatitis c infection. Viruses 2012, 4, 2558–2577. [Google Scholar] [CrossRef]
- Stanciu, C.; Trifan, A.; Muzica, C.; Sfarti, C. Efficacy and safety of alisporivir for the treatment of hepatitis c infection. Expert. Opin. Pharmacother. 2019, 20, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Inoue, K.; Yoshiba, M. Interferon combined with cyclosporine treatment as an effective countermeasure against hepatitis c virus recurrence in liver transplant patients with end-stage hepatitis c virus related disease. Transplant. Proc. 2005, 37, 1233–1234. [Google Scholar] [CrossRef] [PubMed]
- Strain, A.J.; Ismail, T.; Tsubouchi, H.; Arakaki, N.; Hishida, T.; Kitamura, N.; Daikuhara, Y.; McMaster, P. Native and recombinant human hepatocyte growth factors are highly potent promoters of DNA synthesis in both human and rat hepatocytes. J. Clin. Investig. 1991, 87, 1853–1857. [Google Scholar] [CrossRef]
- Kato, T.; Date, T.; Murayama, A.; Morikawa, K.; Akazawa, D.; Wakita, T. Cell culture and infection system for hepatitis c virus. Nat. Protoc. 2006, 1, 2334–2339. [Google Scholar] [CrossRef]
- Wakita, T.; Pietschmann, T.; Kato, T.; Date, T.; Miyamoto, M.; Zhao, Z.; Murthy, K.; Habermann, A.; Krausslich, H.G.; Mizokami, M.; et al. Production of infectious hepatitis c virus in tissue culture from a cloned viral genome. Nat. Med. 2005, 11, 791–796. [Google Scholar] [CrossRef]
- Hossain, T.; Romal, S.; Mahmoudi, T. Production of recombinant hepatitis b virus (hbv) and detection of hbv in infected human liver organoids. Bio Protoc. 2022, 12, e4392. [Google Scholar] [CrossRef] [PubMed]
- Gallard, C.; Lebsir, N.; Khursheed, H.; Reungoat, E.; Plissonnier, M.L.; Bre, J.; Michelet, M.; Chouik, Y.; Zoulim, F.; Pecheur, E.I.; et al. Heparanase-1 is upregulated by hepatitis c virus and favors its replication. J. Hepatol. 2022, 77, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Samanta, S.; Akhter, F.; Roy, A.; Chen, D.; Turner, B.; Wang, Y.; Clemente, N.; Wang, C.; Swerdlow, R.H.; Battaile, K.P.; et al. New cyclophilin d inhibitor rescues mitochondrial and cognitive function in alzheimer’s disease. Brain 2024, 147, 1710–1725. [Google Scholar] [CrossRef]
- Stauffer, W.T.; Goodman, A.Z.; Bobardt, M.; Ure, D.R.; Foster, R.T.; Gallay, P. Mice lacking cyclophilin b, but not cyclophilin a, are protected from the development of nash in a diet and chemical-induced model. PLoS ONE 2024, 19, e0298211. [Google Scholar] [CrossRef]
- Adams, B.M.; Coates, M.N.; Jackson, S.R.; Jurica, M.S.; Davis, T.L. Nuclear cyclophilins affect spliceosome assembly and function in vitro. Biochem. J. 2015, 469, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Karakama, Y.; Sakamoto, N.; Itsui, Y.; Nakagawa, M.; Tasaka-Fujita, M.; Nishimura-Sakurai, Y.; Kakinuma, S.; Oooka, M.; Azuma, S.; Tsuchiya, K.; et al. Inhibition of hepatitis c virus replication by a specific inhibitor of serine-arginine-rich protein kinase. Antimicrob. Agents Chemother. 2010, 54, 3179–3186. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Zhu, C.; Hong, J.; Zhao, L.; Jilg, N.; Fusco, D.N.; Schaefer, E.A.; Brisac, C.; Liu, X.; Peng, L.F.; et al. The spliceosome factor sart1 exerts its anti-hcv action through mrna splicing. J. Hepatol. 2015, 62, 1024–1032. [Google Scholar] [CrossRef]


| Nomenclature | Target | Reference |
|---|---|---|
| silencer siRNA PPIA ID:s310 | Hs peptidylprolylisomerase A (PPIA) | 4392420 |
| silencer siRNA PPIA ID:s57823 | Hs peptidylprolylisomerase A (PPIA) | 4392420 |
| silencer select validated siRNA PPIB ID:s10907 | Hs peptidylprolylisomerase B (PPIB) | 4390824 |
| silencer select validated siRNA PPIB ID:s224317 | Hs peptidylprolylisomerase B (PPIB) | 4390824 |
| silencer select validated siRNA PPIC ID:s10911 | Hs peptidylprolylisomerase C (PPIC) | 4390824 |
| silencer select validated siRNA PPIC ID:s10912 | Hs peptidylprolylisomerase C (PPIC) | 4390824 |
| silencer select validated siRNA PPIF ID:s19661 | Hs peptidylprolylisomerase F (PPIF) * | 4390824 |
| silencer select validated siRNA PPIF ID:s19663 | Hs peptidylprolylisomerase F (PPIF) * | 4390824 |
| silencer siRNA PPIG ID:s17898 | Hs peptidylprolylisomerase G (PPIG) | 4392420 |
| silencer siRNA PPIG ID:s17899 | Hs peptidylprolylisomerase G (PPIG) | 4392420 |
| silencer select validated siRNA PPIH ID:s20485 | Hs peptidylprolylisomerase H (PPIH) | 4390824 |
| silencer select validated siRNA PPIH ID:s20486 | Hs peptidylprolylisomerase H (PPIH) | 4390824 |
| Nomenclature | Target | Reference |
|---|---|---|
| NM_021130/TRCN0000049232/PLKO.1 | Hs peptidylprolylisomerase A (PPIA) | TRCN0000049232 |
| NM_000942/TRCN0000296764/PLKO.1 | Hs peptidylprolylisomerase B (PPIB) | TRCN0000296764 |
| NM_000943/TRCN0000049253/PLKO.1 | Hs peptidylprolylisomerase C (PPIC) | TRCN0000049253 |
| NM_005729/TRCN0000232682/PLKO.1 | Hs peptidylprolylisomerase F (PPIF) | TRCN0000232682 |
| NM_006347/TRCN0000289678/PLKO.1 | Hs peptidylprolylisomerase G (PPIG) | TRCN0000289678 |
| NM_004792/TRCN0000049326/PLKO.1 | Hs peptidylprolylisomerase H (PPIH) | TRCN0000049326 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Molle, J.; Duponchel, S.; Rieusset, J.; Ovize, M.; Ivanov, A.V.; Zoulim, F.; Bartosch, B. Exploration of the Role of Cyclophilins in Established Hepatitis B and C Infections. Viruses 2025, 17, 11. https://doi.org/10.3390/v17010011
Molle J, Duponchel S, Rieusset J, Ovize M, Ivanov AV, Zoulim F, Bartosch B. Exploration of the Role of Cyclophilins in Established Hepatitis B and C Infections. Viruses. 2025; 17(1):11. https://doi.org/10.3390/v17010011
Chicago/Turabian StyleMolle, Jennifer, Sarah Duponchel, Jennifer Rieusset, Michel Ovize, Alexander V. Ivanov, Fabien Zoulim, and Birke Bartosch. 2025. "Exploration of the Role of Cyclophilins in Established Hepatitis B and C Infections" Viruses 17, no. 1: 11. https://doi.org/10.3390/v17010011
APA StyleMolle, J., Duponchel, S., Rieusset, J., Ovize, M., Ivanov, A. V., Zoulim, F., & Bartosch, B. (2025). Exploration of the Role of Cyclophilins in Established Hepatitis B and C Infections. Viruses, 17(1), 11. https://doi.org/10.3390/v17010011









