Bat Rhinacoviruses Related to Swine Acute Diarrhoea Syndrome Coronavirus Evolve under Strong Host and Geographic Constraints in China and Vietnam
Abstract
1. Introduction
2. Materials and Methods
2.1. RNA Extraction, Sequencing, and Genome Assembly
2.2. Whole-Genome Alignment of Rhinacoviruses
2.3. Analysis of Synonymous Nucleotide Composition (SNC)
2.4. Phylogenetic Analyses
2.5. Construction of Genomic Bootstrap (GB) Barcodes
2.6. Construction of Coloured Genomic Bootstrap (CGB) Barcodes
2.7. Recombination and SimPlot Analyses
3. Results
3.1. Rhinacoviruses Detected in Vietnam
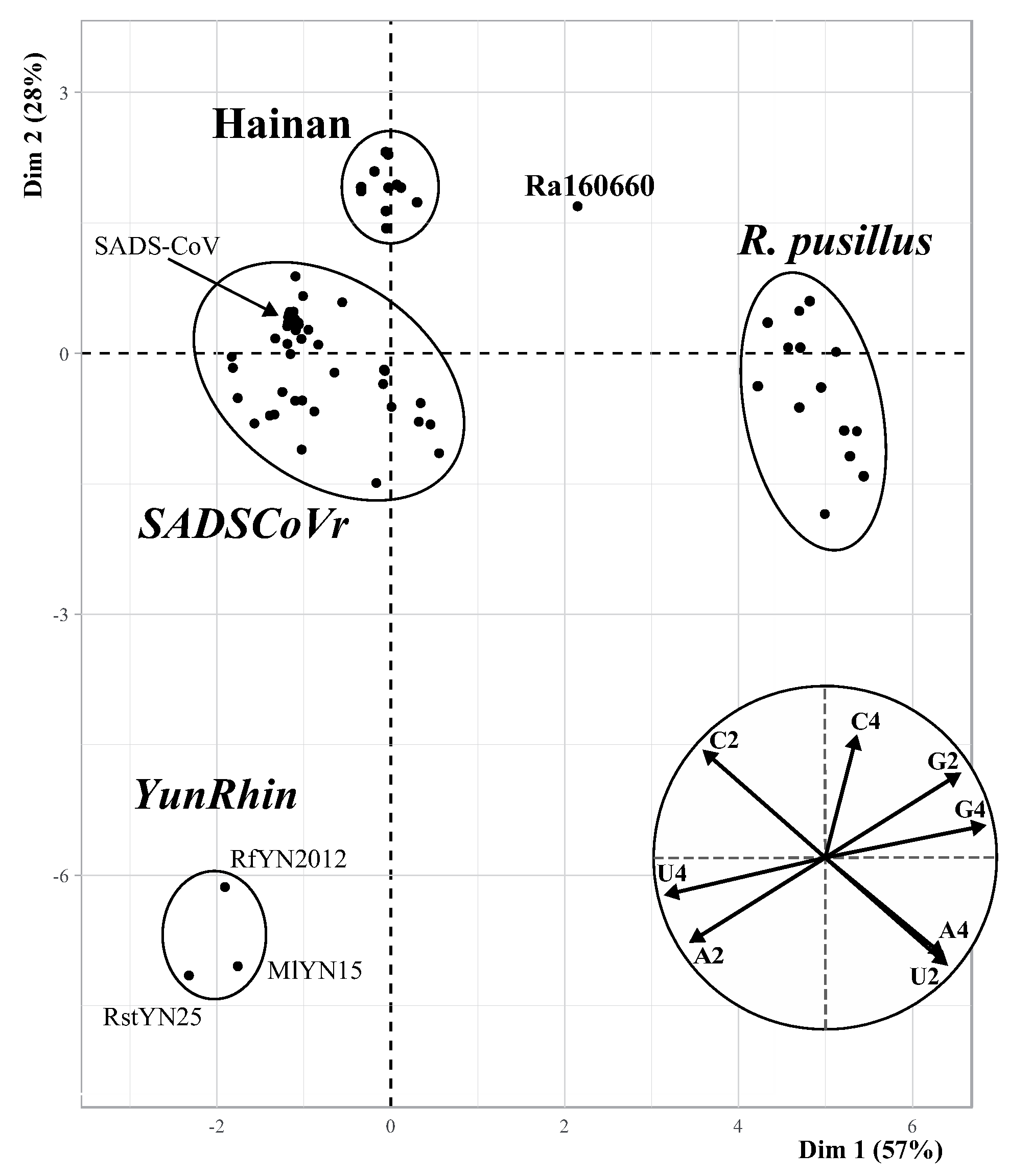
3.2. Synonymous Nucleotide Composition
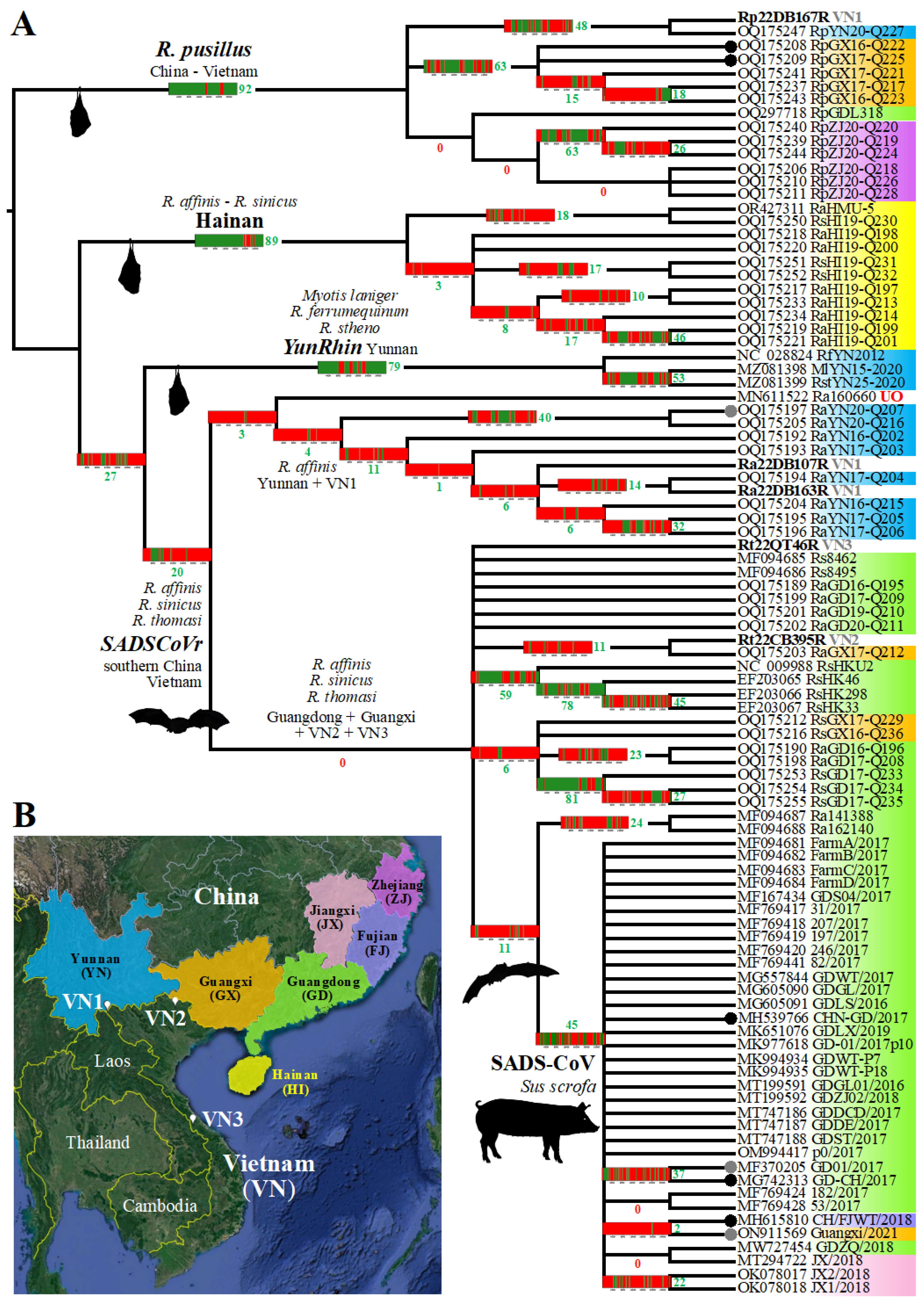
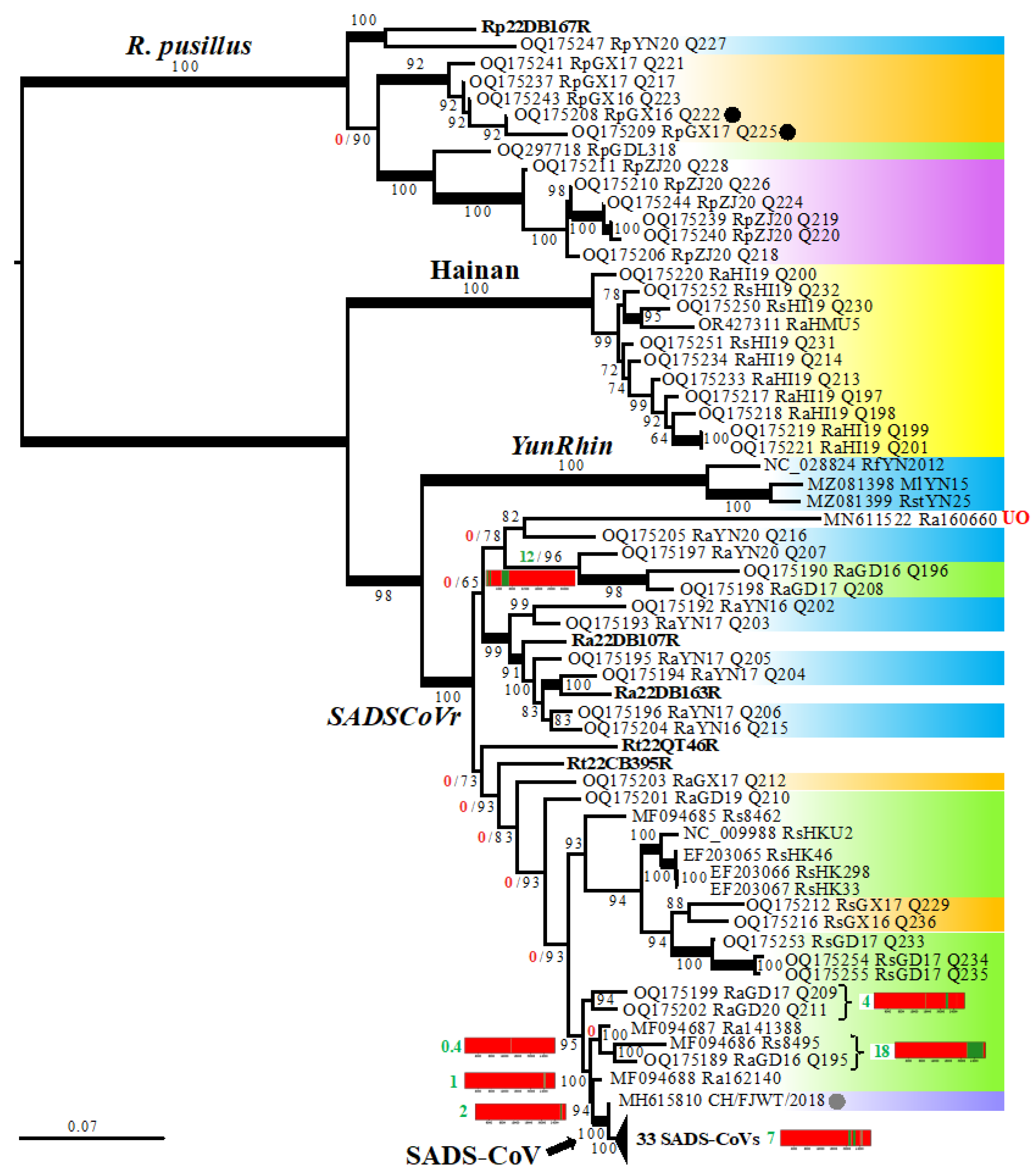
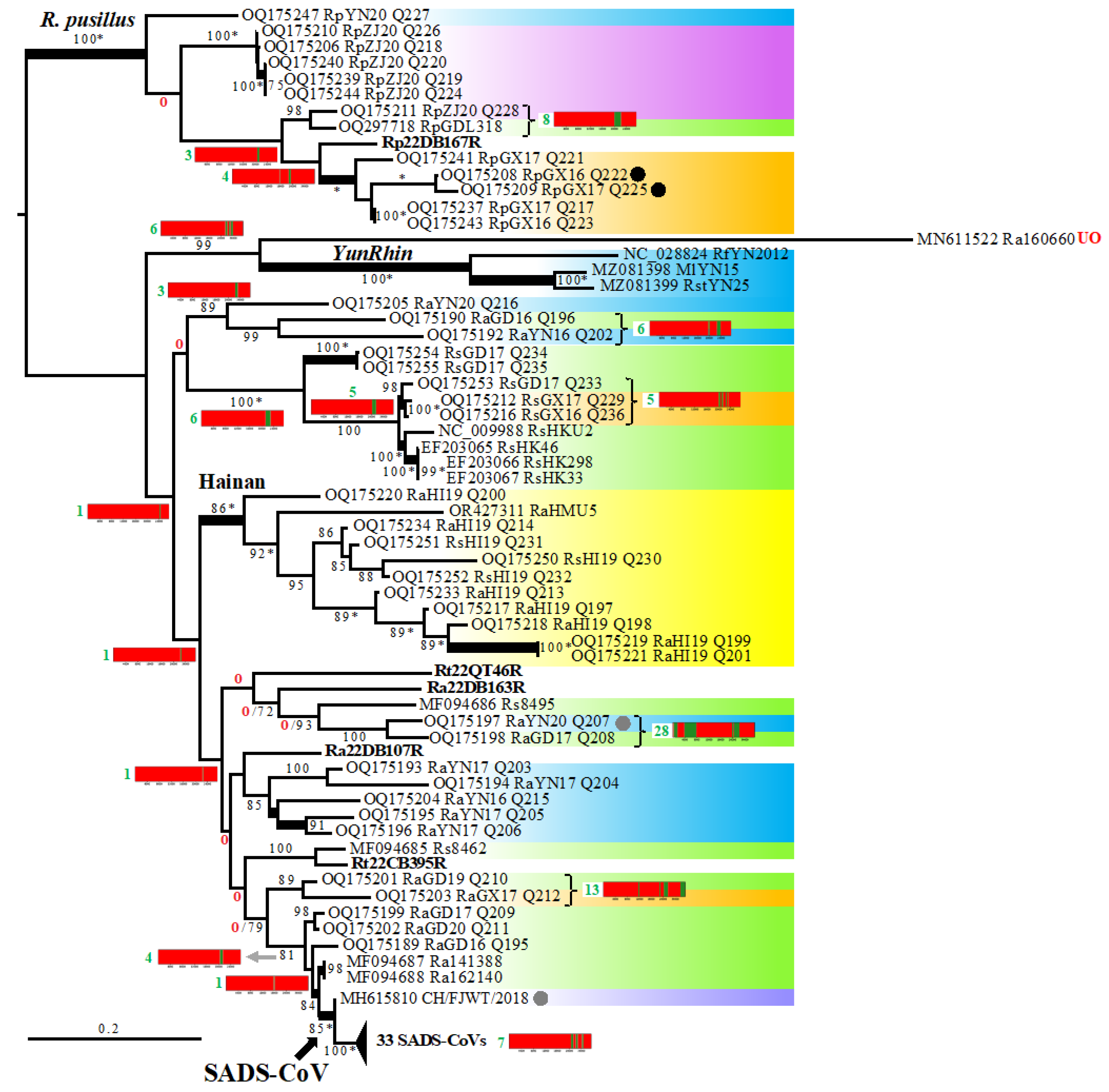
3.3. Phylogenetic Analyses Based on 95 Rhinacovirus Genomes
3.4. Detection of Several Problematic Genomes
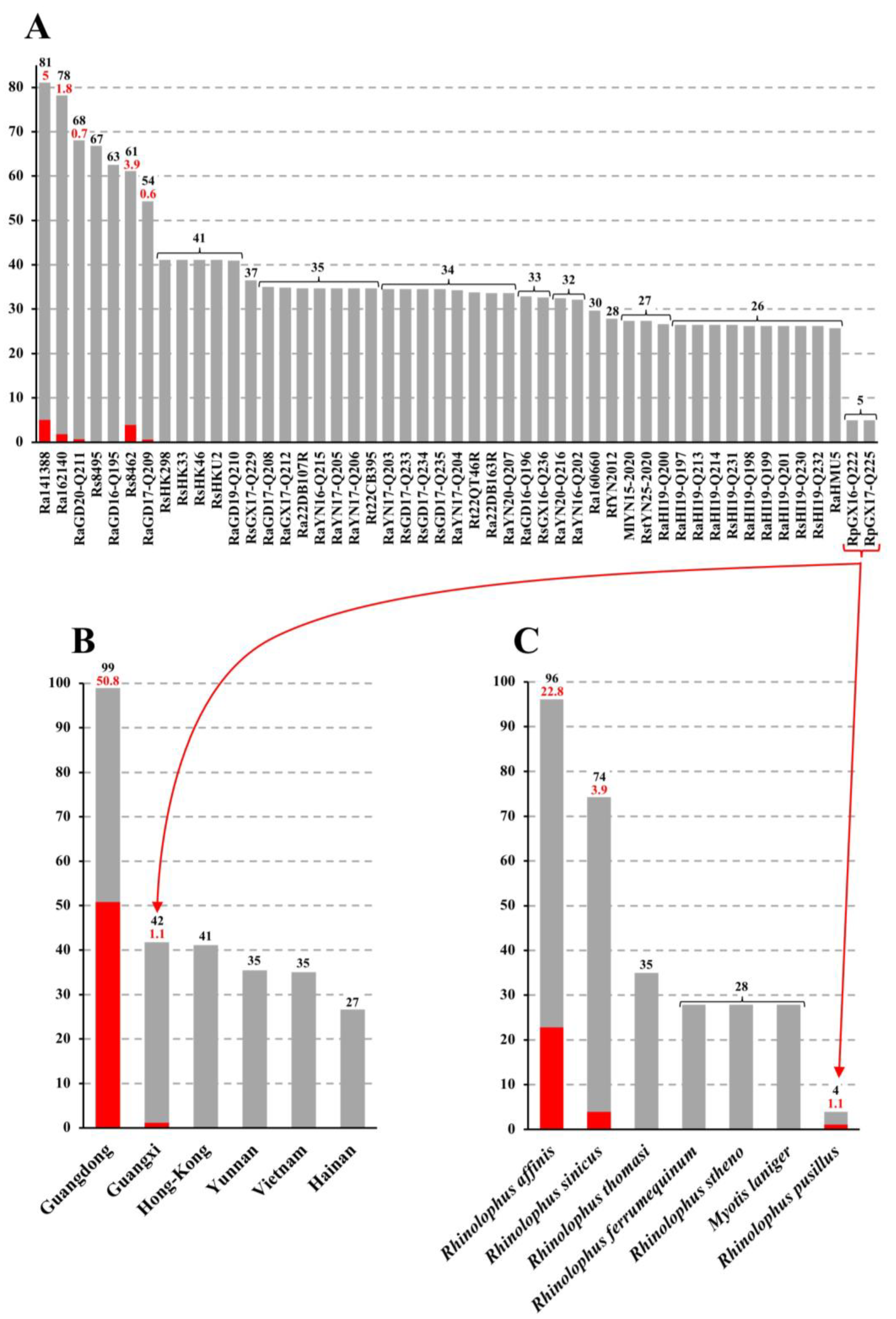
3.5. Virus, Geographic, and Host Contributions Calculated for the Ancestor of SADS-CoVs
3.6. On the Geographic Sampling of Ra160660
3.7. Phylogeography of Bat Rhinacoviruses from Vietnam
4. Discussion
4.1. Host and Geographic Evolution of the Four Divergent Rhinacovirus Groups
4.2. SADS-CoV Evolved from Rhinolophus affinis Circulating in Guangdong
4.3. On the Re-Emergences of SADS-CoV in Fujian in 2018 and Guangxi in 2021
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gong, L.; Li, J.; Zhou, Q.; Xu, Z.; Chen, L.; Zhang, Y.; Xue, C.; Wen, Z.; Cao, Y. A New Bat-HKU2-like Coronavirus in Swine, China, 2017. Emerg. Infect. Dis. 2017, 23, 1607–1609. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Tian, X.; Qin, P.; Wang, B.; Zhao, P.; Yang, Y.L.; Wang, L.; Wang, D.; Song, Y.; Zhang, X.; et al. Discovery of a novel swine enteric alphacoronavirus (SeACoV) in southern China. Vet. Microbiol. 2017, 211, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Fan, H.; Lan, T.; Yang, X.L.; Shi, W.F.; Zhang, W.; Zhu, Y.; Zhang, Y.W.; Xie, Q.M.; Mani, S.; et al. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature 2018, 556, 255–258. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Sun, Y.; Lan, T.; Wu, R.; Chen, J.; Wu, Z.; Xie, Q.; Zhang, X.; Ma, J. Retrospective detection and phylogenetic analysis of swine acute diarrhoea syndrome coronavirus in pigs in southern China. Transbound. Emerg. Dis. 2019, 66, 687–695. [Google Scholar] [CrossRef]
- Li, K.; Li, H.; Bi, Z.; Gu, J.; Gong, W.; Luo, S.; Zhang, F.; Song, D.; Ye, Y.; Tang, Y. Complete genome sequence of a novel swine acute diarrhea syndrome coronavirus, CH/FJWT/2018, isolated in Fujian, China, in 2018. Microbiol. Resour. Announc. 2018, 7, e10-1128. [Google Scholar] [CrossRef]
- Zhou, L.; Li, Q.N.; Su, J.N.; Chen, G.H.; Wu, Z.X.; Luo, Y.; Wu, R.T.; Sun, Y.; Lan, T.; Ma, J.Y. The re-emerging of SADS-CoV infection in pig herds in Southern China. Transbound. Emerg. Dis. 2019, 66, 2180–2183. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Xing, J.; Xu, Z.Y.; Gao, H.; Xu, S.J.; Liu, J.; Zhu, D.H.; Guo, Y.F.; Yang, B.S.; Chen, X.N.; et al. Re-emergence of Severe Acute Diarrhea Syndrome Coronavirus (SADS-CoV) in Guangxi, China, 2021. J. Infect. 2022, 85, e130–e133. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Si, H.R.; Zhu, Y.; Yang, X.L.; Anderson, D.E.; Shi, Z.L.; Wang, L.F.; Zhou, P. Discovery of bat coronaviruses through surveillance and probe capture-based next-generation sequencing. mSphere 2020, 5, e00807-19. [Google Scholar] [CrossRef]
- Han, Y.; Xu, P.; Wang, Y.; Zhao, W.; Zhang, J.; Zhang, S.; Wang, J.; Jin, Q.; Wu, Z. Panoramic analysis of coronaviruses carried by representative bat species in Southern China to better understand the coronavirus sphere. Nat. Commun. 2023, 14, 5537. [Google Scholar] [CrossRef]
- Yang, Y.L.; Yu, J.Q.; Huang, Y.W. Swine enteric alphacoronavirus (swine acute diarrhea syndrome coronavirus): An update three years after its discovery. Virus Res. 2020, 285, 198024. [Google Scholar] [CrossRef]
- Hassanin, A.; Tu, V.T.; Görföl, T.; Ngon, L.Q.; Pham, P.V.; Hang, C.T.; Tuan, T.A.; Prot, M.; Simon-Lorière, E.; Kemenesi, G.; et al. Phylogeographic evolution of horseshoe bat sarbecoviruses in Vietnam and implications for the origins of SARS-CoV and SARS-CoV-2. Res. Sq. Preprint 2023. [Google Scholar] [CrossRef]
- Lau, S.K.; Woo, P.C.; Li, K.S.; Huang, Y.; Wang, M.; Lam, C.S.; Xu, H.; Guo, R.; Chan, K.H.; Zheng, B.J.; et al. Complete genome sequence of bat coronavirus HKU2 from Chinese horseshoe bats revealed a much smaller spike gene with a different evolutionary lineage from the rest of the genome. J. Virol. 2007, 367, 428–439. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Yang, L.; Ren, X.; He, G.; Zhang, J.; Yang, J.; Qian, Z.; Dong, J.; Sun, L.; Zhu, Y.; et al. Deciphering the bat virome catalog to better understand the ecological diversity of bat viruses and the bat origin of emerging infectious diseases. ISME J. 2016, 10, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Cheng, J.; Luo, Y.; Yan, X.L.; Wu, Z.X.; He, L.L.; Tan, Y.R.; Zhou, Z.H.; Li, Q.N.; Zhou, L.; et al. Attenuation of a virulent swine acute diarrhea syndrome coronavirus strain via cell culture passage. Virol. J. 2019, 538, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usa-bility. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Larsson, A. AliView: A fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef] [PubMed]
- Hassanin, A. Variation in synonymous nucleotide composition among genomes of sarbecoviruses and consequences for the origin of COVID-19. Gene 2022, 835, 146641. [Google Scholar] [CrossRef]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R package for multivariate analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Hassanin, A.; Rambaud, O.; Klein, D. Genomic bootstrap barcodes and their application to study the evolution of sarbecoviruses. Viruses 2022, 14, 440. [Google Scholar] [CrossRef]
- Hassanin, A.; Rambaud, O. Retracing phylogenetic, host and geographic origins of coronaviruses with coloured genomic bootstrap barcodes: SARS-CoV and SARS-CoV-2 as case studies. Viruses 2023, 15, 406. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Ropiquet, A.; Li, B.; Hassanin, A. SuperTRI: A new approach based on branch support analyses of multiple independent data sets for assessing reliability of phylogenetic inferences. C. R. Biol. 2009, 332, 832–847. [Google Scholar] [CrossRef] [PubMed]
- Swofford, D.L. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4; Sinauer Associates: Sunderland, MA, USA, 2021. [Google Scholar]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015, 1, vev003. [Google Scholar] [CrossRef] [PubMed]
- Samson, S.; Lord, É.; Makarenkov, V. SimPlot++: A Python application for representing sequence similarity and detecting recombination. Bioinformatics 2022, 38, 3118–3120. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Fang, L.; Liu, S.; Ke, W.; Wang, D.; Peng, G.; Xiao, S. Susceptibility of porcine IPI-2I intestinal epithelial cells to infection with swine enteric coronaviruses. Vet. Microbiol. 2019, 233, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Zhao, Z.Y.; Yang, Y.L.; Qin, Y.; Pan, D.; Yuan, L.X.; Huang, Y.W.; Wang, B. The origin and evolution of emerged swine acute diarrhea syndrome coronavirus with zoonotic potential. J. Med. Virol. 2023, 95, e28672. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Fang, B.; Liu, Y.; Cai, M.; Jun, J.; Ma, J.; Bu, D.; Wang, L.; Zhou, P.; Wang, H.; et al. Newly emerged porcine enteric alphacoronavirus in southern China: Identification, origin and evolutionary history analysis. Infect. Genet. Evol. 2018, 62, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Dinerstein, E.; Olson, D.; Joshi, A.; Vynne, C.; Burgess, N.D.; Wikramanayake, E.; Hahn, N.; Palminteri, S.; Hedao, P.; Noss, R.; et al. An Ecoregion-Based Approach to Protecting Half the Terrestrial Realm. Bioscience 2017, 67, 534–545. [Google Scholar] [CrossRef]
- Yang, Y.L.; Liang, Q.Z.; Xu, S.Y.; Mazing, E.; Xu, G.H.; Peng, L.; Qin, P.; Wang, B.; Huang, Y.W. Characterization of a novel bat-HKU2-like swine enteric alphacoronavirus (SeACoV) infection in cultured cells and development of a SeACoV infectious clone. Virology 2019, 536, 110–118. [Google Scholar] [CrossRef]
- Mei, X.Q.; Qin, P.; Yang, Y.L.; Liao, M.; Liang, Q.Z.; Zhao, Z.; Shi, F.S.; Wang, B.; Huang, Y.W. First evidence that an emerging mammalian alphacoronavirus is able to infect an avian species. Transbound. Emerg. Dis. 2022, 69, e2006–e2019. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Yao, J.; Yang, Z.; Wang, J.; Yang, K.; Yao, L. Re-emergence of severe acute diarrhea syndrome coronavirus (SADS-CoV) in Henan, central China, 2023. Vet. Microbiol. 2024, 292, 110049. [Google Scholar] [CrossRef] [PubMed]


| Virus Name | Date | Sample Code(s) | Host Species | S | Loc. | NovSeq Reads | Virus Reads | Length (nt) | GenBank |
|---|---|---|---|---|---|---|---|---|---|
| Ra22DB107R | 24 June 2022 | DB107 | R. affinis | ♀ | VN1 | 147,368,172 | 76,009 | 27,137 | PP746501 |
| Ra22DB163R | 26 June 2022 | DB163 | R. affinis | ♀ | VN1 | 221,895,122 | 6084 | 27,130 | PP746502 |
| Rp22DB167R | 26 June 2022 | DB167 | R. pusillus | ♀ | VN1 | 167,424,614 | 51,354 | 27,149 | PP746503 |
| Rt22CB395R | 28 February 2022 | PO395+399 | R. thomasi | ♀ | VN2 | 152,809,588 | 3,159,553 | 27,165 | PP746500 |
| Rt22QT46R | 2 November 2022 | QT46+47+55+57 | R. thomasi | ♂ | VN3 | 192,686,690 | 17,037,975 | 27,129 | PP746504 |
| Myotis laniger (n = 1) | R. affinis (n = 28) | R. f. nippon * (n = 1) | R. pusillus (n = 14) | R. sinicus (n = 14) | R. stheno (n = 1) | R. thomasi (n = 2) | Sus scrofa (n = 34) | |
|---|---|---|---|---|---|---|---|---|
| SADS-CoV 1 | 28/0 | 96/23 | 28/0 | 4/1 | 74/4 | 28/0 | 35/0 | NA |
| SADS-CoV 2 | 16/0 | 94/35 | 17/0 | 0/0 | 64/5 | 16/0 | 21/1 | NA |
| Ra22DB107R | 28/0 | 98/30 | 39/2 | 1/0 | 37/0 | 28/0 | 50/0 | 37/0 |
| Ra22DB163R | 22/0 | 100/57 | 30/0 | 1/0 | 26/0 | 22/0 | 36/0 | 25/0 |
| Rp22DB167R | 33/0 | 33/0 | 33/0 | 100/67 | 33/0 | 33/0 | 33/0 | 33/0 |
| Rt22CB395R | 30/0 | 90/38 | 31/0 | 1/0 | 59/7 | 30/0 | 46/2 | 49/0 |
| Rt22QT46R | 55/0 | 95/3 | 59/0 | 6/0 | 88/2 | 55/0 | 87/2 | 83/0 |
| Ra160660 | 55/0 | 91/25 | 63/0 | 2/0 | 61/0 | 55/0 | 65/1 | 58/0 |
| NEV (n = 1) | CV (n = 1) | NWV (n = 3) | VN (n = 5) | YN (n = 12) | GX (n = 8) | GD (n = 14) | HK (n = 4) | HI (n = 11) | ZJ (n = 6) | |
|---|---|---|---|---|---|---|---|---|---|---|
| SADS-CoV 1 | 35/0 | 34/0 | 34/0 | 35/0 | 35/0 | 42/1 | 99/51 | 41/0 | 27/0 | 0/0 |
| SADS-CoV 2 | 21/1 | 20/0 | 21/1 | 22/1 | 20/0 | 24/0 | 99/65 | 30/0 | 16/0 | 0/0 |
| Ra22DB107R | 49/0 | 35/0 | 74/6 | 74/6 | 94/21 | 37/0 | 37/0 | 33/0 | 25/0 | 0/0 |
| Ra22DB163R | 34/0 | 27/0 | 56/6 | 61/6 | 89/34 | 31/4 | 26/0 | 26/0 | 19/1 | 1/0 |
| Rp22DB167R | 33/0 | 33/0 | 33/0 | 33/0 | 88/48 | 50/6 | 39/0 | 33/0 | 33/0 | 8/0 |
| Rt22CB395R | NA | 46/2 | 62/0 | 65/2 | 60/3 | 68/11 | 70/14 | 47/0 | 26/0 | 0/0 |
| Rt22QT46R | 87/2 | NA | 87/0 | 90/2 | 91/0 | 89/0 | 90/2 | 84/0 | 45/0 | 2/0 |
| Ra160660 | 61/0 | 64/1 | 69/0 | 70/1 | 96/28 | 62/0 | 61/0 | 60/1 | 39/0 | 2/0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hassanin, A.; Tu, V.T.; Pham, P.V.; Ngon, L.Q.; Chabane, T.; Moulin, L.; Wurtzer, S. Bat Rhinacoviruses Related to Swine Acute Diarrhoea Syndrome Coronavirus Evolve under Strong Host and Geographic Constraints in China and Vietnam. Viruses 2024, 16, 1114. https://doi.org/10.3390/v16071114
Hassanin A, Tu VT, Pham PV, Ngon LQ, Chabane T, Moulin L, Wurtzer S. Bat Rhinacoviruses Related to Swine Acute Diarrhoea Syndrome Coronavirus Evolve under Strong Host and Geographic Constraints in China and Vietnam. Viruses. 2024; 16(7):1114. https://doi.org/10.3390/v16071114
Chicago/Turabian StyleHassanin, Alexandre, Vuong Tan Tu, Phu Van Pham, Lam Quang Ngon, Thanina Chabane, Laurent Moulin, and Sébastien Wurtzer. 2024. "Bat Rhinacoviruses Related to Swine Acute Diarrhoea Syndrome Coronavirus Evolve under Strong Host and Geographic Constraints in China and Vietnam" Viruses 16, no. 7: 1114. https://doi.org/10.3390/v16071114
APA StyleHassanin, A., Tu, V. T., Pham, P. V., Ngon, L. Q., Chabane, T., Moulin, L., & Wurtzer, S. (2024). Bat Rhinacoviruses Related to Swine Acute Diarrhoea Syndrome Coronavirus Evolve under Strong Host and Geographic Constraints in China and Vietnam. Viruses, 16(7), 1114. https://doi.org/10.3390/v16071114







