Genetic and Pathogenic Analysis of a Novel Porcine Epidemic Diarrhea Virus Strain Isolated in the Republic of Korea
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Isolation of Porcine Epidemic Diarrhea Virus
2.3. Development of PEDV Strain CKK1-1
2.4. Titration of Porcine Epidemic Diarrhea Virus
2.5. Immunofluorescence Assay
2.6. Evaluation of Pathogenicity, CKK1-1
2.7. Quantitative Real-Time PCR
2.8. Histopathology and Immunochemistry
2.9. Next-Generation Sequencing
2.10. Sequence Analysis and Phylogenetic Tree
3. Results
3.1. Genetic Characterization of PEDV Strain CKK1-1
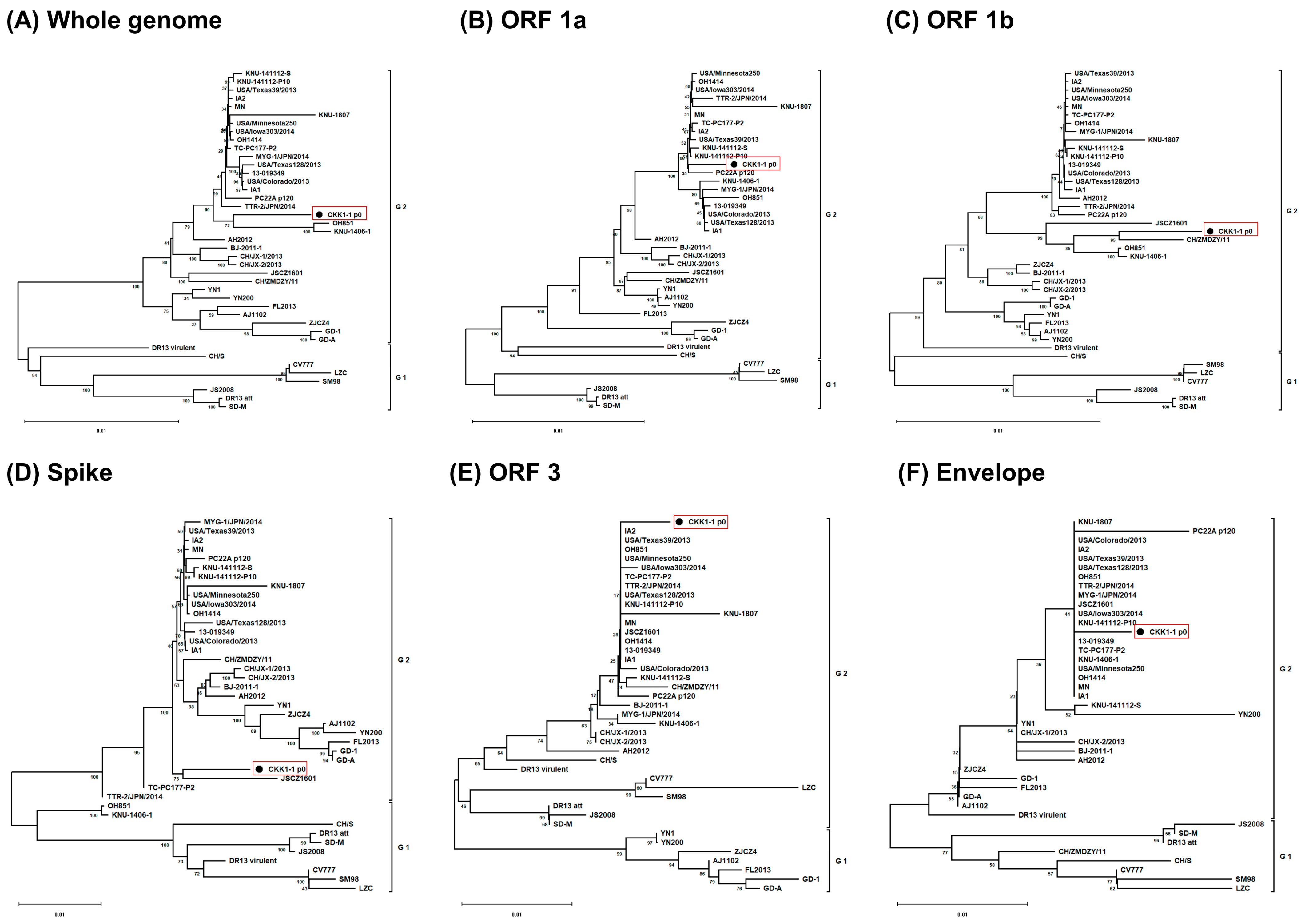
3.2. Phylogenetic Analysis
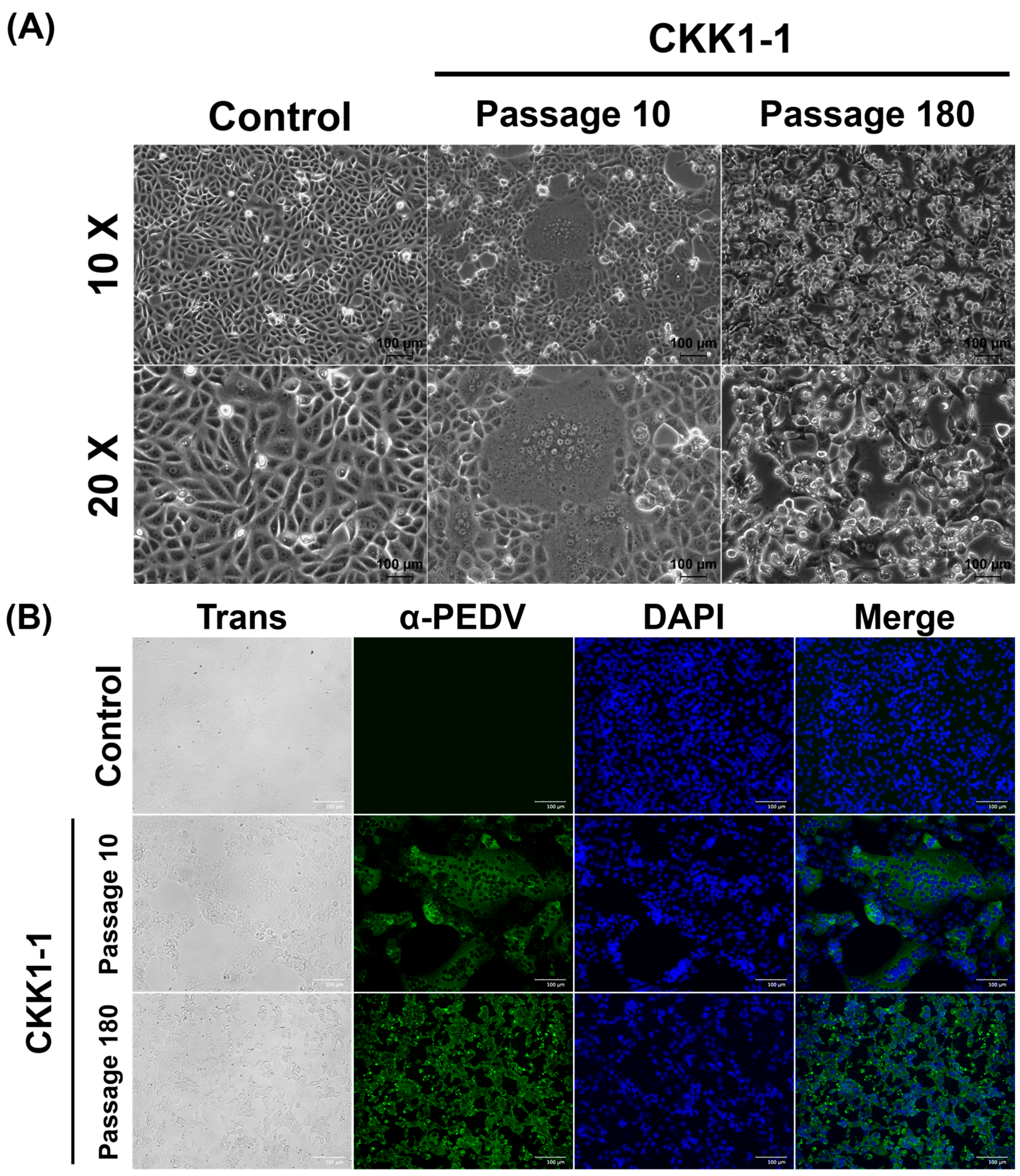
3.3. Virus Isolation and Biological Characteristics
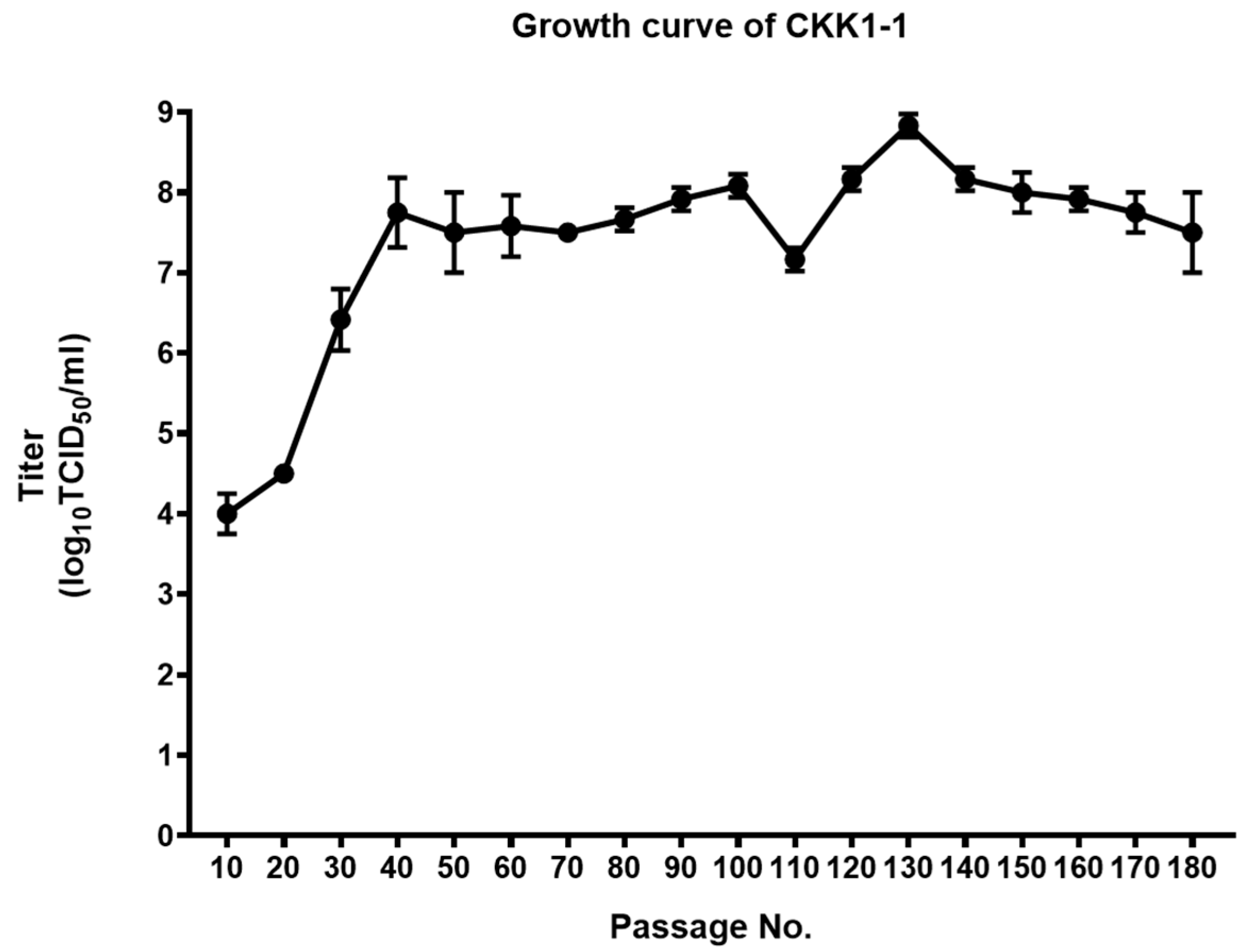
3.4. Growth Kinetics of CKK1-1
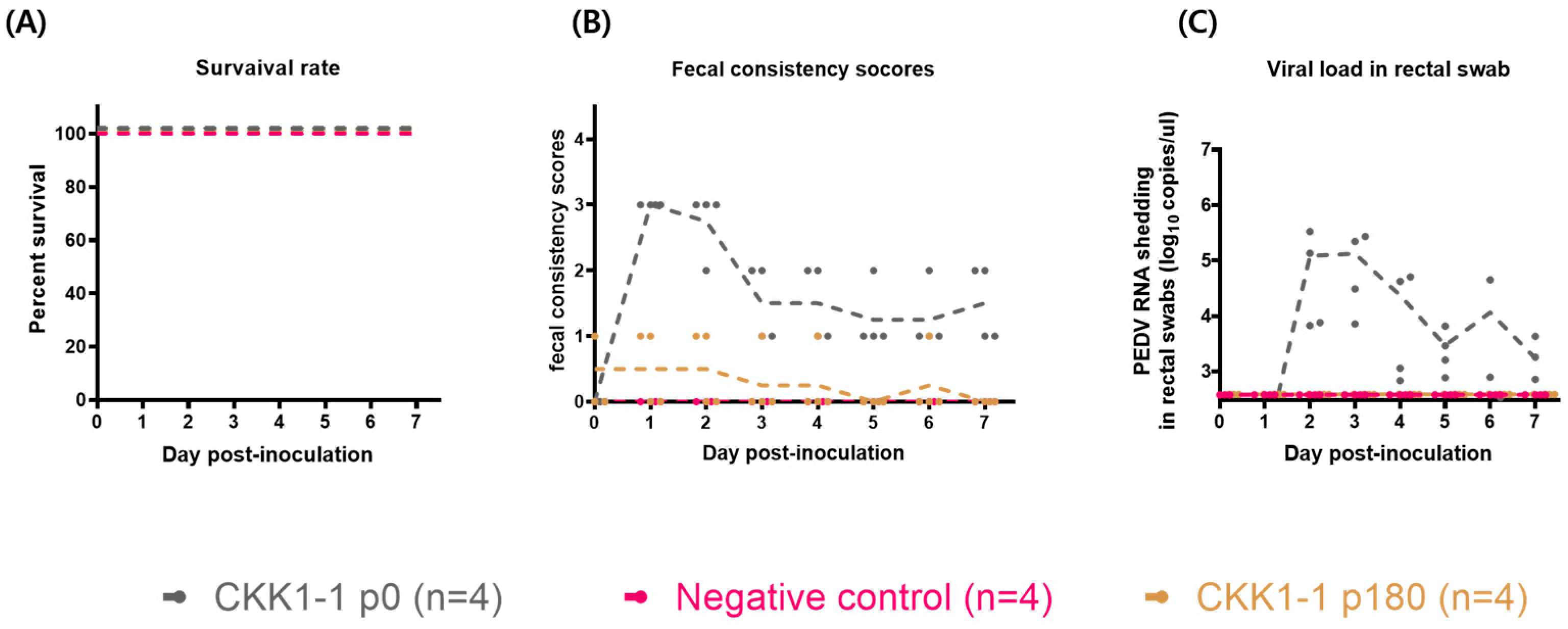
3.5. Evaluation of the Pathogenicity of CKK1-1
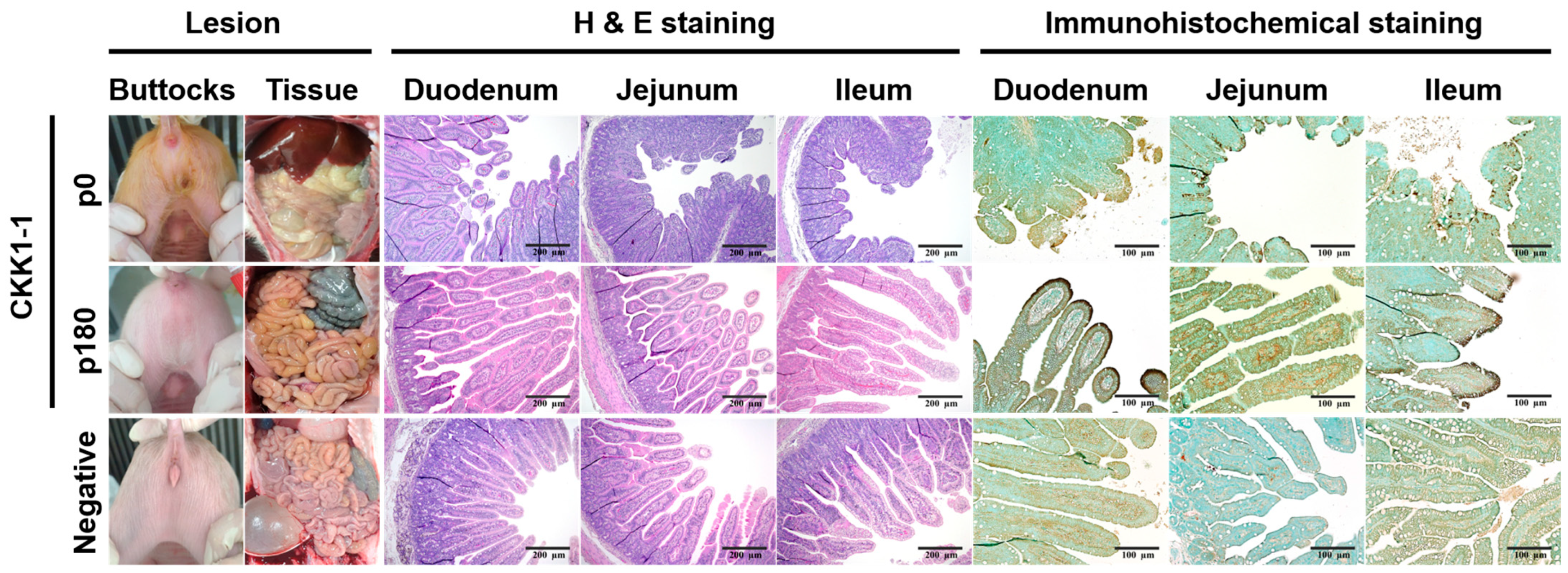
3.6. Clinical Symptoms and Histopathological and Immunohistochemical Staining
3.7. Comparison of Genetic Mutations in the CKK1-1 Strain Depending on Passage Number
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Singh, G.; Singh, P.; Pillatzki, A.; Nelson, E.; Webb, B.; Dillberger-Lawson, S.; Ramamoorthy, S. A Minimally Replicative Vaccine Protects Vaccinated Piglets Against Challenge with the Porcine Epidemic Diarrhea Virus. Front. Vet. Sci. 2019, 6, 347. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Li, G.; Stasko, J.; Thomas, J.T.; Stensland, W.R.; Pillatzki, A.E.; Gauger, P.C.; Schwartz, K.J.; Madson, D.; Yoon, K.J.; et al. Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States. J. Clin. Microbiol. 2014, 52, 234–243. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Qiao, S.; Yang, Y.; Guo, J.; Xie, S.; Zhou, E.; Zhang, G. Genome sequencing and analysis of a novel recombinant porcine epidemic diarrhea virus strain from Henan, China. Virus Genes 2016, 52, 91–98. [Google Scholar] [CrossRef]
- Dervas, E.; Hepojoki, J.; Laimbacher, A.; Romero-Palomo, F.; Jelinek, C.; Keller, S.; Smura, T.; Hepojoki, S.; Kipar, A.; Hetzel, U. Nidovirus-Associated Proliferative Pneumonia in the Green Tree Python (Morelia viridis). J. Virol. 2017, 91, e00718-17. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Xie, X.; Yang, L.; Wang, A. A Comprehensive View on the Host Factors and Viral Proteins Associated with Porcine Epidemic Diarrhea Virus Infection. Front. Microbiol 2021, 12, 762358. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Yang, D.K.; Kim, H.H.; Cho, I.S. Efficacy of inactivated variant porcine epidemic diarrhea virus vaccines in growing pigs. Clin. Exp. Vaccine Res. 2018, 7, 61–69. [Google Scholar] [CrossRef]
- Baek, P.S.; Choi, H.W.; Lee, S.; Yoon, I.J.; Lee, Y.J.; Lee, D.S.; Lee, S.; Lee, C. Efficacy of an inactivated genotype 2b porcine epidemic diarrhea virus vaccine in neonatal piglets. Vet. Immunol. Immunopathol. 2016, 174, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Park, J.E.; Kang, K.J.; Ryu, J.H.; Park, J.Y.; Jang, H.; Sung, D.J.; Kang, J.G.; Shin, H.J. Porcine epidemic diarrhea vaccine evaluation using a newly isolated strain from Korea. Vet. Microbiol. 2018, 221, 19–26. [Google Scholar] [CrossRef]
- Hou, Y.; Wang, Q. Emerging Highly Virulent Porcine Epidemic Diarrhea Virus: Molecular Mechanisms of Attenuation and Rational Design of Live Attenuated Vaccines. Int. J. Mol. Sci. 2019, 20, 5478. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A SIMPLE METHOD OF ESTIMATING FIFTY PER CENT ENDPOINTS. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Wu, Y.; Li, W.; Zhou, Q.; Li, Q.; Xu, Z.; Shen, H.; Chen, F. Characterization and pathogenicity of Vero cell-attenuated porcine epidemic diarrhea virus CT strain. Virol. J. 2019, 16, 121. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zhang, T.; Song, D.; Huang, T.; Peng, Q.; Chen, Y.; Li, A.; Zhang, F.; Wu, Q.; Ye, Y.; et al. Comparison and evaluation of conventional RT-PCR, SYBR green I and TaqMan real-time RT-PCR assays for the detection of porcine epidemic diarrhea virus. Mol. Cell. Probes 2017, 33, 36–41. [Google Scholar] [CrossRef]
- Kim, D.M.; Moon, S.H.; Kim, S.C.; Lee, T.G.; Cho, H.S.; Tark, D. Genome characterization of a Korean isolate of porcine epidemic diarrhea virus. Microbiol. Resour. Announc. 2024, 13, e0011823. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988, 16, 10881–10890. [Google Scholar] [CrossRef] [PubMed]
- Lee, C. Porcine epidemic diarrhea virus: An emerging and re-emerging epizootic swine virus. Virol. J. 2015, 12, 193. [Google Scholar] [CrossRef]
- Li, Z.; Ma, Z.; Li, Y.; Gao, S.; Xiao, S. Porcine epidemic diarrhea virus: Molecular mechanisms of attenuation and vaccines. Microb. Pathog. 2020, 149, 104553. [Google Scholar] [CrossRef]
- Lin, C.M.; Hou, Y.; Marthaler, D.G.; Gao, X.; Liu, X.; Zheng, L.; Saif, L.J.; Wang, Q. Attenuation of an original US porcine epidemic diarrhea virus strain PC22A via serial cell culture passage. Vet. Microbiol. 2017, 201, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wu, Y.; Yan, Z.; Li, G.; Luo, J.; Huang, S.; Guo, X. A Comprehensive View on the Protein Functions of Porcine Epidemic Diarrhea Virus. Genes 2024, 15, 165. [Google Scholar] [CrossRef]
- Jang, G.; Lee, D.; Shin, S.; Lim, J.; Won, H.; Eo, Y.; Kim, C.H.; Lee, C. Porcine epidemic diarrhea virus: An update overview of virus epidemiology, vaccines, and control strategies in South Korea. J. Vet. Sci. 2023, 24, e58. [Google Scholar] [CrossRef]
- Wei, M.Z.; Chen, L.; Zhang, R.; Chen, Z.; Shen, Y.J.; Zhou, B.J.; Wang, K.G.; Shan, C.L.; Zhu, E.P.; Cheng, Z.T. Overview of the recent advances in porcine epidemic diarrhea vaccines. Vet. J. 2024, 304, 106097. [Google Scholar] [CrossRef]
- Oh, C.; Kim, Y.; Chang, K.O. Proteases facilitate the endosomal escape of porcine epidemic diarrhea virus during entry into host cells. Virus Res. 2019, 272, 197730. [Google Scholar] [CrossRef]
- Shirato, K.; Kawase, M.; Matsuyama, S. Middle East respiratory syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2. J. Virol. 2013, 87, 12552–12561. [Google Scholar] [CrossRef] [PubMed]
- Chandran, K.; Sullivan, N.J.; Felbor, U.; Whelan, S.P.; Cunningham, J.M. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 2005, 308, 1643–1645. [Google Scholar] [CrossRef] [PubMed]
- Zamolodchikova, T.S. Serine proteases of small intestine mucosa—Localization, functional properties, and physiological role. Biochem. Biokhimiia 2012, 77, 820–829. [Google Scholar] [CrossRef] [PubMed]
- Wicht, O.; Li, W.; Willems, L.; Meuleman, T.J.; Wubbolts, R.W.; van Kuppeveld, F.J.; Rottier, P.J.; Bosch, B.J. Proteolytic activation of the porcine epidemic diarrhea coronavirus spike fusion protein by trypsin in cell culture. J. Virol. 2014, 88, 7952–7961. [Google Scholar] [CrossRef]
- Kim, D.M.; Moon, S.H.; Kim, S.C.; Cho, H.S.; Tark, D. Development of Effective PEDV Vaccine Candidates Based on Viral Culture and Protease Activity. Vaccines 2023, 11, 923. [Google Scholar] [CrossRef]
- Kim, Y.; Oh, C.; Shivanna, V.; Hesse, R.A.; Chang, K.O. Trypsin-independent porcine epidemic diarrhea virus US strain with altered virus entry mechanism. BMC Vet. Res. 2017, 13, 356. [Google Scholar] [CrossRef]
- Kong, F.; Saif, L.J.; Wang, Q. Roles of bile acids in enteric virus replication. Anim. Dis. 2021, 1, 2. [Google Scholar] [CrossRef]
- Chang, K.O.; Sosnovtsev, S.V.; Belliot, G.; Kim, Y.; Saif, L.J.; Green, K.Y. Bile acids are essential for porcine enteric calicivirus replication in association with down-regulation of signal transducer and activator of transcription 1. Proc. Natl. Acad. Sci. USA 2004, 101, 8733–8738. [Google Scholar] [CrossRef]
- Takagi, H.; Oka, T.; Shimoike, T.; Saito, H.; Kobayashi, T.; Takahashi, T.; Tatsumi, C.; Kataoka, M.; Wang, Q.; Saif, L.J.; et al. Human sapovirus propagation in human cell lines supplemented with bile acids. Proc. Natl. Acad. Sci. USA 2020, 117, 32078–32085. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Meulia, T.; Gao, X.; Saif, L.J.; Wang, Q. Deletion of both the Tyrosine-Based Endocytosis Signal and the Endoplasmic Reticulum Retrieval Signal in the Cytoplasmic Tail of Spike Protein Attenuates Porcine Epidemic Diarrhea Virus in Pigs. J. Virol. 2019, 93, e01758-18. [Google Scholar] [CrossRef] [PubMed]

| Passage Numbers | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Origin | 1 | 2 | 3 | 4 | … *** | 28 | 29 | 30 | 31 | … | 66 | 67 | 68 | 69 | 70 | 71 | … | 76 | 77 | 78 | 79 | 80 | 81 | 82 | 83 | 84 | 85 | … | 177 | 178 | 179 | 180 | |
| T15N * | 1 | 2 | 3 | 4 | … | ||||||||||||||||||||||||||||
| T10N ** | 1 | 2 | 3 | 4 | … | 39 | 40 | ||||||||||||||||||||||||||
| T5N | 1 | 2 | 3 | 4 | … | 9 | 10 | ||||||||||||||||||||||||||
| T2N | 1 | 2 | 3 | 4 | |||||||||||||||||||||||||||||
| N | 1 | 2 | 3 | 4 | … | 96 | 97 | 98 | 99 | ||||||||||||||||||||||||
| Identity to CKK1-1/Nucleotide (Amino Acid) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Strains | Whole Genome | 5′ UTR | ORF1a | ORF1b | Spike | ORF3 | Envelope | Membrane | Nucleocapsid | 3′ UTR |
| CV777 | 96.50 (-) | 97.14 (-) | 96.89 (93.36) | 97.48 (98.99) | 93.27 (93.09) | 96.31 (96.36) | 96.38 (98.68) | 97.91 (98.21) | 95.59 (97.01) | 97.21 (-) |
| CH/S | 97.06 (-) | 98.95 (-) | 97.78 (94.77) | 97.92 (98.88) | 93.00 (92.93) | 97.72 (100) | 95.93 (97.33) | 97.75 (98.21) | 96.27 (97.47) | 97.84 (-) |
| SM98 | 96.31 (-) | 97.15 (-) | 96.68 (93.02) | 97.38 (98.80) | 92.93 (92.34) | 96.15 (95.70) | 95.47 (95.97) | 97.75 (97.30) | 95.43 (96.77) | 96.88 (-) |
| DR13_virulent | 97.37 (-) | 98.95 (-) | 97.76 (94.66) | 98.21 (99.73) | 94.31 (94.17) | 98.03 (99.55) | 97.76 (100) | 98.06 (98.21) | 96.90 (97.71) | 97.86 (-) |
| DR13_att | 96.97 (-) | 98.94 (-) | 97.73 (95.14) | 97.58 (99.23) | 93.19 (92.92) | 97.22 (99.03) | 96.03 (95.49) | 97.61 (97.75) | 96.34 (97.71) | 98.78 (-) |
| LZC | 96.23 (-) | 94.94 (-) | 96.75 (93.09) | 97.41 (98.84) | 92.70 (92.07) | 94.90 (94.49) | 95.47 (95.97) | 96.85 (96.38) | 95.35 (96.54) | 96.59 (-) |
| JS2008 | 97.01 (-) | 98.22 (-) | 97.77 (95.29) | 97.65 (99.15) | 93.41 (93.00) | 96.88 (98.54) | 95.44 (94.59) | 97.45 (97.30) | 96.34 (97.71) | 98.79 (-) |
| AJ1102 | 98.38 (-) | 99.30 (-) | 99.08 (97.97) | 98.46 (99.69) | 97.18 (97.43) | 95.65 (97.28) | 98.68 (100) | 98.37 (98.21) | 96.02 (97.24) | 98.79 (-) |
| GD-1 | 97.89 (-) | 98.95 (-) | 98.05 (95.60) | 98.45 (99.54) | 97.13 (97.21) | 94.84 (96.36) | 98.23 (98.68) | 98.37 (98.21) | 96.09 (97.47) | 98.48 (-) |
| ZJCZ4 | 97.91 (-) | 99.30 (-) | 97.93 (95.39) | 98.33 (99.65) | 97.71 (97.73) | 95.50 (96.83) | 98.68 (100) | 98.06 (98.21) | 96.26 (97.71) | 98.79 (-) |
| BJ-2011-1 | 98.74 (-) | 98.95 (-) | 99.03 (97.88) | 98.33 (99.65) | 98.41 (98.32) | 99.10 (99.55) | 98.67 (100) | 99.41 (98.66) | 99.08 (99.77) | 98.48 (-) |
| SD-M | 96.94 (-) | 98.94 (-) | 97.72 (95.17) | 97.58 (99.19) | 93.12 (92.53) | 97.22 (99.03) | 95.93 (95.97) | 97.76 (97.75) | 96.18 (97.47) | 98.48 (-) |
| AH2012 | 98.91 (-) | 99.30 (-) | 99.39 (98.77) | 98.54 (99.57) | 98.12 (97.81) | 98.19 (99.55) | 98.67 (100) | 99.41 (99.55) | 99.08 (99.55) | 98.79 (-) |
| 13-019349 | 99.17 (-) | 98.95 (-) | 99.56 (98.99) | 98.57 (99.65) | 98.86 (98.10) | 99.55 (100) | 99.56 (100) | 99.71 (99.55) | 99.62 (100) | 99.10 (-) |
| USA/Colorado/2013 | 99.20 (-) | 99.30 (-) | 99.58 (99.02) | 98.57 (99.65) | 98.98 (98.47) | 99.40 (99.55) | 99.56 (100) | 99.71 (99.55) | 99.62 (100) | 99.10 (-) |
| YN1 | 98.60 (-) | 99.30 (-) | 99.08 (98.00) | 98.38 (99.61) | 97.76 (97.59) | 96.3 (98.20) | 99.12 (100) | 99.11 (99.11) | 98.7 (99.55) | 98.48 (-) |
| USA/Texas128/2013 | 99.15 (-) | 100 (-) | 99.56 (98.99) | 98.53 (99.54) | 98.71 (97.81) | 99.55 (100) | 99.56 (100) | 99.71 (99.55) | 99.55 (99.55) | 99.08 (-) |
| IA1 | 99.18 (-) | 99.65 (-) | 99.55 (98.96) | 98.53 (99.54) | 98.96 (98.47) | 99.55 (100) | 99.56 (100) | 99.56 (99.55) | 99.62 (100) | 99.10 (-) |
| IA2 | 99.24 (-) | 99.65 (-) | 99.74 (99.47) | 98.54 (99.65) | 98.83 (68.40) | 99.55 (100) | 99.56 (100) | 99.71 (99.55) | 99.54 (99.77) | 99.10 (-) |
| MN | 99.25 (-) | 99.65 (-) | 99.76 (99.55) | 98.54 (99.61) | 98.83 (98.32) | 99.55 (100) | 99.56 (100) | 99.71 (99.55) | 99.62 (100) | 98.48 (-) |
| TC_PC177-P2 | 99.27 (-) | 100 (-) | 99.73 (99.50) | 98.54 (99.61) | 98.92 (98.47) | 99.55 (100) | 99.56 (100) | 99.71 (99.55) | 99.62 (100) | 99.10 (-) |
| FL2013 | 98.19 (-) | 98.59 (-) | 98.79 (97.34) | 98.42 (99.57) | 96.92 (96.76) | 95.5 (96.36) | 98.23 (98.68) | 98.37 (98.21) | 96.02 (98.21) | 98.48 (-) |
| USA/Iowa303/2014 | 99.25 (-) | 100 (-) | 99.76 (99.52) | 98.54 (99.61) | 98.83 (98.47) | 99.40 (100) | 99.56 (100) | 99.71 (99.55) | 99.54 (100) | 99.10 (-) |
| USA/Minnesota250 | 99.24 (-) | 100 (-) | 99.73 (99.44) | 98.54 (99.65) | 98.81 (98.40) | 99.55 (100) | 99.56 (100) | 99.71 (99.55) | 99.54 (100) | 99.10 (-) |
| OH1414 | 99.25 (-) | 99.65 (-) | 99.74 (99.47) | 99.57 (99.61) | 98.81 (98.47) | 99.55 (100) | 99.56 (100) | 99.71 (99.55) | 99.54 (100) | 99.10 (-) |
| YN200 | 98.46 (-) | 99.30 (-) | 99.03 (97.83) | 98.42 (99.61) | 96.79 (96.53) | 96.30 (98.19) | 97.77 (97.33) | 99.26 (98.66) | 99.09 (99.55) | 98.48 (-) |
| KNU-141112-P10 | 99.26 (-) | 100 (-) | 99.76 (99.58) | 98.56 (99.65) | 98.81 (98.18) | 99.55 (100) | 99.56 (100) | 99.71 (99.55) | 99.62 (100) | 99.10 (-) |
| MYG-1/JPN/2014 | 99.10 (-) | 100 (-) | 99.50 (98.85) | 98.53 (99.57) | 98.69 (97.96) | 98.8 (100) | 99.56 (100) | 99.71 (99.55) | 99.62 (100) | 98.83 (-) |
| OH851 | 98.85 (-) | 99.65 (-) | 99.36 (98.43) | 99.02 (99.73) | 96.43 (95.85) | 99.55 (100) | 99.56 (100) | 99.56 (99.55) | 99.47 (99.77) | 99.10 (-) |
| TTR-2/JPN/2014 | 99.14 (-) | 99.57 (-) | 99.66 (99.33) | 98.41 (99.57) | 98.76 (98.22) | 99.55 (100) | 99.56 (100) | 99.41 (95.55) | 99.32 (100) | 98.43 (-) |
| PC22A_p120 | 99.15 (-) | 100 (-) | 99.64 (99.24) | 98.49 (99.61) | 98.68 (98.02) | 99.25 (99.10) | 98.67 (98.68) | 99.56 (99.11) | 99.62 (100) | 98.78 (-) |
| JSCZ1601 | 98.77 (-) | 98.94 (-) | 98.92 (97.65) | 98.81 (99.65) | 98.04 (97.51) | 99.55 (100) | 99.56 (100) | 99.17 (99.55) | 98.46 (99.09) | 98.48 (-) |
| Groups | Inoculum | Route | No. of Pigs | Mortality Rate [% (No/Total)] | Severe Diarrhea Rate * [% (No/Total)] | Clinical Symptoms | Virus Shedding | Peak Fecal Virus Shedding Titer (log10copies/uL); dpi |
|---|---|---|---|---|---|---|---|---|
| 1 | CKK1-1 p0 | Oral | 4 | 0 (0/4) | 100 (4/4) | Severe | Started at 2 dpi | 4.78 ± 0.75, 3 |
| 2 | Negative control | 4 | 0 (0/4) | 0 (0/4) | No diarrhea | N/D ** | NA *** | |
| 3 | CKK1-1 p180 | 4 | 0 (0/4) | 0 (0/4) | No diarrhea | N/D | NA |
| Gene | Nucleotide Position | Amino-Acid Position | CKK1-1 Strains | |
|---|---|---|---|---|
| P0 | P180 | |||
| ORF 1a/1b | 99 | 33 | GAT(D) | GAG(E) |
| 132 | 44 | TTC(F) | TTT(F) | |
| 833 | 278 | TGC(C) | TTC(F) | |
| 2098 | 700 | TCA(S) | GCA(A) | |
| 2858 | 953 | GTT(V) | GCT(A) | |
| 2941 | 981 | CCC(P) | TCC(S) | |
| 3790 | 1264 | CTG(L) | TTG(L) | |
| 3966 | 1322 | AAT(N) | AAC(N) | |
| 4106 | 1369 | GGT(G) | GAT(D) | |
| 4691 | 1564 | TCT(S) | TTT(F) | |
| 4718 | 1573 | GAC(D) | GCC(A) | |
| 8129 | 2710 | ATC(I) | ACC(T) | |
| 8712 | 2904 | GAG(E) | GAT(D) | |
| 9264 | 3088 | AAT(N) | AAG(K) | |
| 10,301 | 3434 | TCT(S) | TTT(F) | |
| 11,451 | 3817 | AAT(N) | AAC(N) | |
| 13,388 | 4463 | TCA(S) | TTA(L) | |
| 13,979 | 4660 | CTA(L) | CCA(P) | |
| 14,003 | 4668 | AGA(R) | AAA(K) | |
| 14,042 | 4681 | CAG(Q) | CGG(R) | |
| 14,693 | 4898 | CTG(L) | CCG(P) | |
| 16,607 | 5536 | ATG(M) | AGG(R) | |
| 16,609 | 5537 | CTG(L) | TTG(L) | |
| 17,870 | 5957 | ACC(T) | ATC(I) | |
| 18,265 | 6089 | GAT(D) | AAT(N) | |
| 18,311 | 6104 | ACC(T) | ATC(I) | |
| 18,677 | 6226 | TCT(S) | TTT(F) | |
| Spike | 79–80 | 27 | TCA(S) | AAA(K) |
| 397 | 131 | ACA(T) | CCA(P) | |
| 469 | 157 | CAT(H) | TAT(Y) | |
| 974 | 325 | GAC(D) | GCC(A) | |
| 1056 | 352 | TCC(S) | TCT(S) | |
| 1148 | 383 | ACT(T) | ATT(I) | |
| 1571 | 524 | CCT(P) | CAT(H) | |
| 1903 | 635 | CTT(L) | ATT(I) | |
| 1906 | 636 | GAA(E) | CAA(Q) | |
| 1984 | 662 | ATC(I) | GTC(V) | |
| 2148 | 716 | GTT(V) | GTC(V) | |
| 2180 | 727 | AGT(S) | ATT(I) | |
| 2336 | 779 | ACT(T) | AAT(N) | |
| 2675 | 892 | AAA(K) | AGA(R) | |
| 2678 | 893 | AAA(K) | ACA(T) | |
| 2770 | 924 | CGC(R) | AGC(S) | |
| 2894 | 965 | GGT(G) | GCT(A) | |
| 2910 | 970 | GCA(A) | GCG(A) | |
| 2915 | 972 | TTG(L) | TCG(S) | |
| 3153 | 1051 | CAA(Q) | CAC(H) | |
| 3485 | 1162 | GAT(D) | GCT(A) | |
| 3500 | 1167 | ATT(I) | AGT(S) | |
| 3533 | 1178 | ATT(I) | ACT(T) | |
| 3667 | 1223 | ATT(I) | GTT(V) | |
| 3818 | 1273 | AAT(N) | ACT(T) | |
| 3823 | 1275 | ACT(T) | GCT(A) | |
| 3842 | 1281 | GGT(G) | GAT(D) | |
| 4006 | 1336 | GTT(V) | ATT(I) | |
| 4138–4140 | 1380 | ㅡㅡㅡ(ㅡ) | TGA(*) | |
| ORF3 | 571 | 191 | GAT(D) | TAT(Y) |
| Envelope | 126 | 42 | CAT(H) | CAC(H) |
| 209 | 70 | CCT(P) | CTT(L) | |
| Membrane | 34 | 12 | ATT(I) | GTT(V) |
| 70 | 24 | ATC(I) | CTC(L) | |
| Nucleocapsid | 228 | 76 | TAT(Y) | TAC(Y) |
| 497 | 166 | AGA(R) | ATA(I) | |
| 551 | 184 | CGT(R) | CCT(P) | |
| 1130 | 377 | GGG(G) | GAG(E) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, D.-M.; Moon, S.-H.; Kim, S.-C.; Cho, H.-S.; Tark, D. Genetic and Pathogenic Analysis of a Novel Porcine Epidemic Diarrhea Virus Strain Isolated in the Republic of Korea. Viruses 2024, 16, 1108. https://doi.org/10.3390/v16071108
Kim D-M, Moon S-H, Kim S-C, Cho H-S, Tark D. Genetic and Pathogenic Analysis of a Novel Porcine Epidemic Diarrhea Virus Strain Isolated in the Republic of Korea. Viruses. 2024; 16(7):1108. https://doi.org/10.3390/v16071108
Chicago/Turabian StyleKim, Dae-Min, Sung-Hyun Moon, Seung-Chai Kim, Ho-Seong Cho, and Dongseob Tark. 2024. "Genetic and Pathogenic Analysis of a Novel Porcine Epidemic Diarrhea Virus Strain Isolated in the Republic of Korea" Viruses 16, no. 7: 1108. https://doi.org/10.3390/v16071108
APA StyleKim, D.-M., Moon, S.-H., Kim, S.-C., Cho, H.-S., & Tark, D. (2024). Genetic and Pathogenic Analysis of a Novel Porcine Epidemic Diarrhea Virus Strain Isolated in the Republic of Korea. Viruses, 16(7), 1108. https://doi.org/10.3390/v16071108







