Inhibition of SARS-CoV-2 Replication by Self-Assembled siRNA Nanoparticles Targeting Multiple Highly Conserved Viral Sequences
Abstract
1. Introduction
2. Materials and Methods
2.1. Design of siRNA Sequences
2.2. Assembly of siRNA Nanoparticles
2.3. Transmission Electron Microscopy (TEM) and Dynamic Light Scattering (DLS)
2.4. Cell Cultures
2.5. SARS-CoV-2 GFP/ΔN trVLP Production
2.6. SARS-CoV-2 GFP/ΔN trVLP Infection
2.7. Confocal Microscopy
2.8. RNA Extraction and RT-qPCR
2.9. Statistical Analysis
3. Results
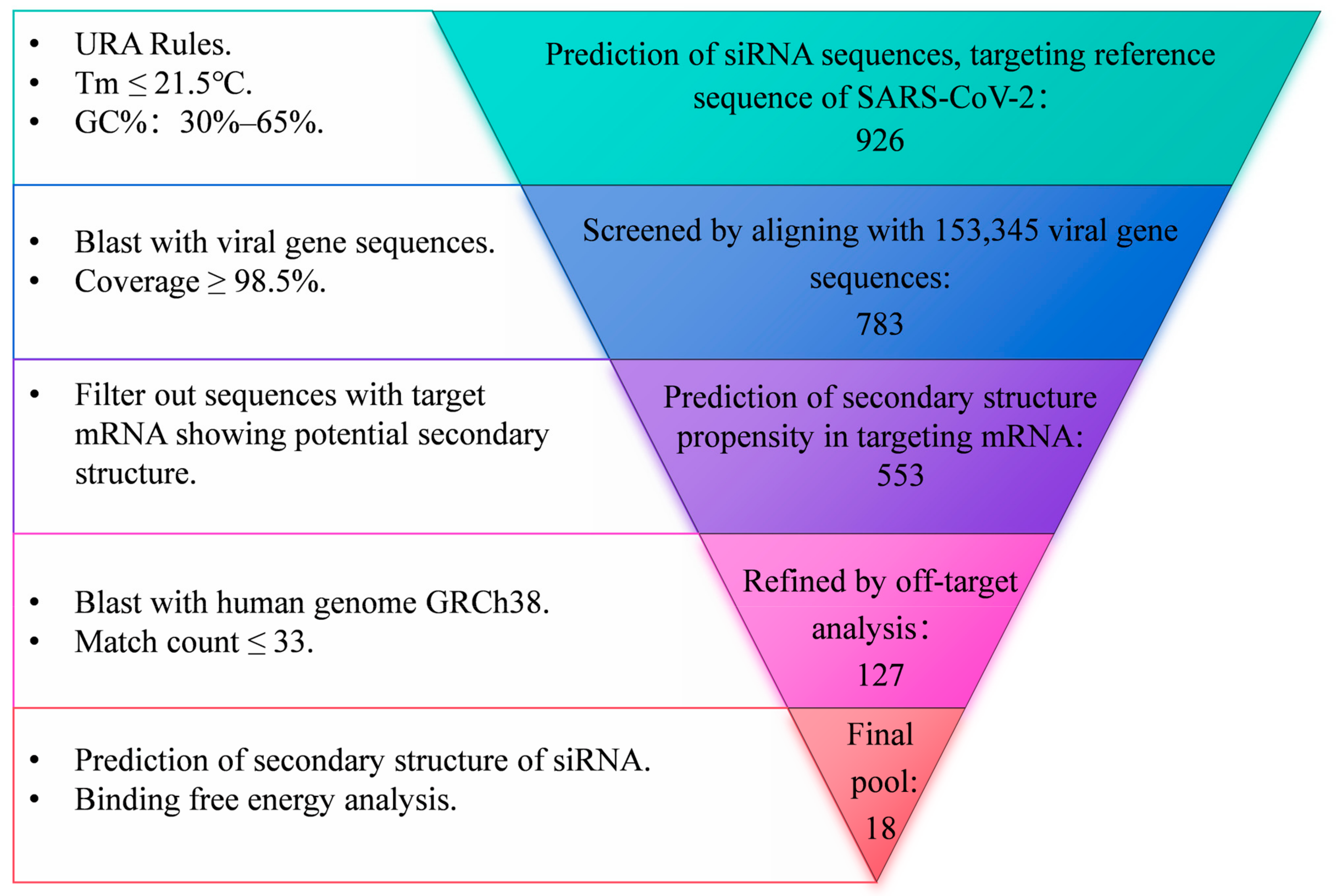
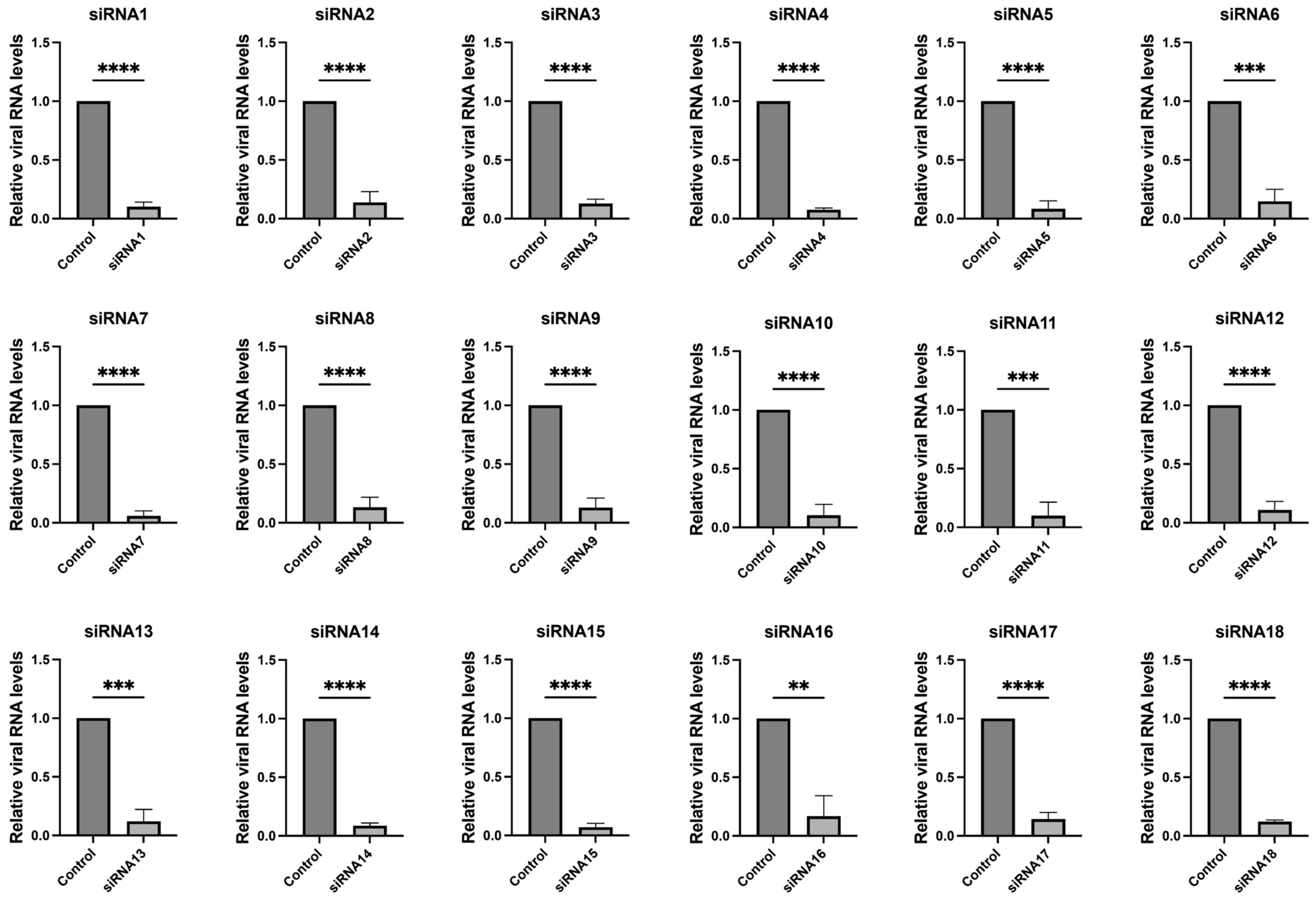
3.1. Design and Verification of siRNAs Targeting Highly Conserved Regions of SARS-CoV-2
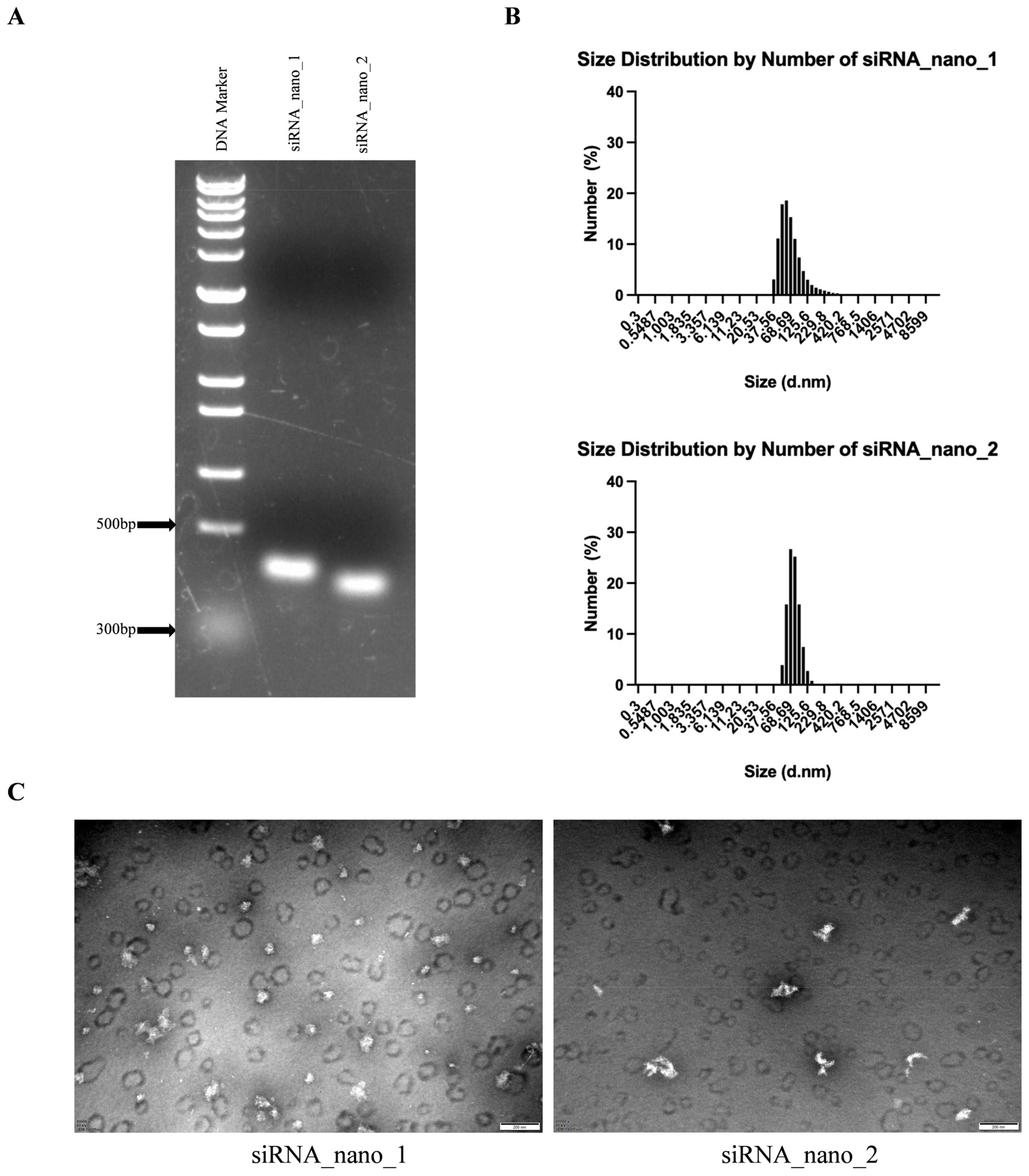
3.2. Construction and Characterization of siRNA Nanoparticles
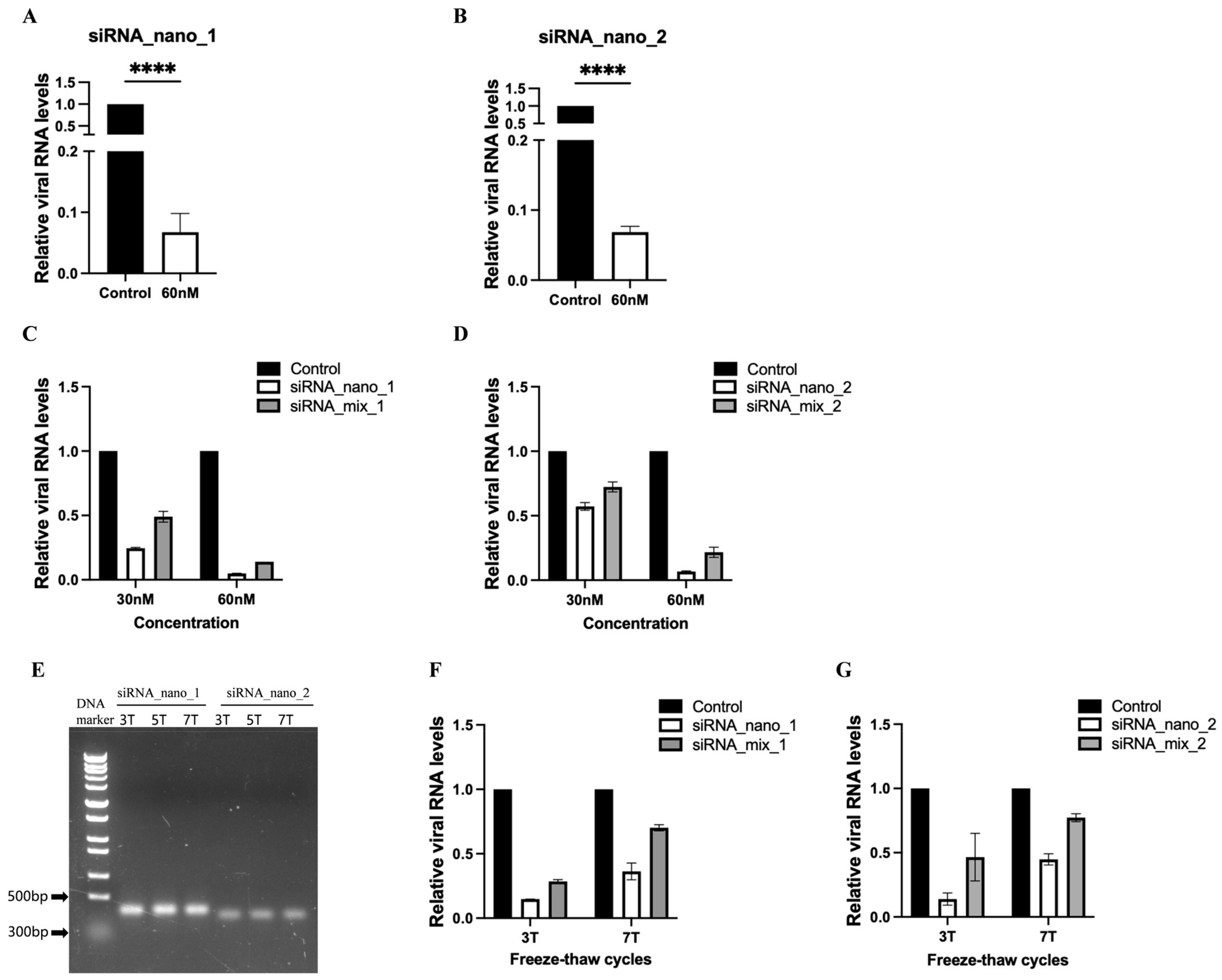
3.3. siRNA Nanoparticles Efficiently Inhibited SARS-CoV-2 Replication
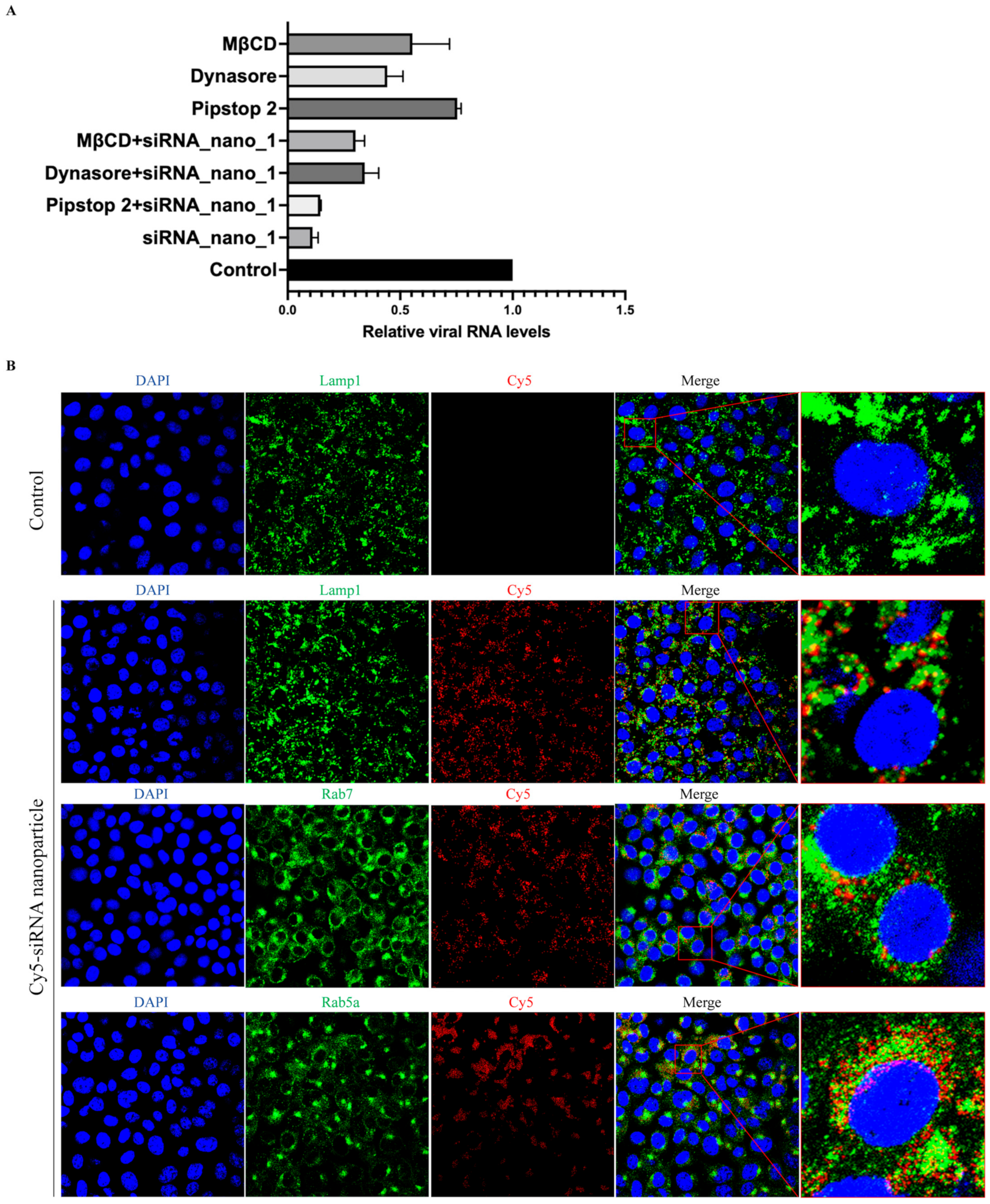
3.4. siRNA Nanoparticles Enter Cells Directly through Cellular Endocytic Pathways
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chan, J.F.; Yuan, S.; Chu, H.; Sridhar, S.; Yuen, K.Y. COVID-19 drug discovery and treatment options. Nat. Rev. Microbiol. 2024, 22, 391–407. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.M.; Chung, Y.S.; Jo, H.J.; Lee, N.J.; Kim, M.S.; Woo, S.H.; Park, S.; Kim, J.W.; Kim, H.M.; Han, M.G. Identification of Coronavirus Isolated from a Patient in Korea with COVID-19. Osong Public Health Res. Perspect. 2020, 11, 3–7. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef] [PubMed]
- Steiner, S.; Kratzel, A.; Barut, G.T.; Lang, R.M.; Aguiar Moreira, E.; Thomann, L.; Kelly, J.N.; Thiel, V. SARS-CoV-2 biology and host interactions. Nat. Rev. Microbiol. 2024, 22, 206–225. [Google Scholar] [CrossRef]
- Brant, A.C.; Tian, W.; Majerciak, V.; Yang, W.; Zheng, Z.M. SARS-CoV-2: From its discovery to genome structure, transcription, and replication. Cell Biosci. 2021, 11, 136. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, M.; Aigner, A. Therapeutic siRNA: State-of-the-Art and Future Perspectives. Biodrugs 2022, 36, 549–571. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.J.; Huang, H.W.; Liu, C.Y.; Hong, C.F.; Chan, Y.L. Inhibition of SARS-CoV replication by siRNA. Antivir. Res. 2005, 65, 45–48. [Google Scholar] [CrossRef] [PubMed]
- Idris, A.; Davis, A.; Supramaniam, A.; Acharya, D.; Kelly, G.; Tayyar, Y.; West, N.; Zhang, P.; McMillan, C.L.D.; Soemardy, C.; et al. A SARS-CoV-2 targeted siRNA-nanoparticle therapy for COVID-19. Mol. Ther. 2021, 29, 2219–2226. [Google Scholar] [CrossRef] [PubMed]
- Tolksdorf, B.; Heinze, J.; Niemeyer, D.; Röhrs, V.; Berg, J.; Drosten, C.; Kurreck, J. Development of a highly stable, active small interfering RNA with broad activity against SARS-CoV viruses. Antivir. Res. 2024, 226, 105879. [Google Scholar] [CrossRef] [PubMed]
- Medeiros, I.G.; Khayat, A.S.; Stransky, B.; Santos, S.; Assumpção, P.; de Souza, J.E.S. A small interfering RNA (siRNA) database for SARS-CoV-2. Sci. Rep. 2021, 11, 8849. [Google Scholar] [CrossRef] [PubMed]
- Ambike, S.; Cheng, C.C.; Feuerherd, M.; Velkov, S.; Baldassi, D.; Afridi, S.Q.; Porras-Gonzalez, D.; Wei, X.; Hagen, P.; Kneidinger, N.; et al. Targeting genomic SARS-CoV-2 RNA with siRNAs allows efficient inhibition of viral replication and spread. Nucleic Acids Res. 2021, 50, 333–349. [Google Scholar] [CrossRef] [PubMed]
- Idris, A.; Supramaniam, A.; Tayyar, Y.; Kelly, G.; McMillan, N.A.J.; Morris, K.V. An intranasally delivered ultra-conserved siRNA prophylactically represses SARS-CoV-2 infection in the lung and nasal cavity. Antivir. Res. 2024, 222, 105815. [Google Scholar] [CrossRef] [PubMed]
- Brady, D.K.; Gurijala, A.R.; Huang, L.; Hussain, A.A.; Lingan, A.L.; Pembridge, O.G.; Ratangee, B.A.; Sealy, T.T.; Vallone, K.T.; Clements, T.P. A guide to COVID-19 antiviral therapeutics: A summary and perspective of the antiviral weapons against SARS-CoV-2 infection. FEBS J. 2024, 291, 1632–1662. [Google Scholar] [CrossRef] [PubMed]
- Bowden-Reid, E.; Ledger, S.; Zhang, Y.; Di Giallonardo, F.; Aggarwal, A.; Stella, A.O.; Akerman, A.; Milogiannakis, V.; Walker, G.; Rawlinson, W.; et al. Novel siRNA therapeutics demonstrate multi-variant efficacy against SARS-CoV-2. Antivir. Res. 2023, 217, 105677. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Park, J.H.; Sailor, M.J. Rekindling RNAi Therapy: Materials Design Requirements for In Vivo siRNA Delivery. Adv. Mater. 2019, 31, e1903637. [Google Scholar] [CrossRef] [PubMed]
- Kanasty, R.; Dorkin, J.R.; Vegas, A.; Anderson, D. Delivery materials for siRNA therapeutics. Nat. Mater. 2013, 12, 967–977. [Google Scholar] [CrossRef] [PubMed]
- Keam, S.J. Vutrisiran: First Approval. Drugs 2022, 82, 1419–1425. [Google Scholar] [CrossRef] [PubMed]
- Syed, Y.Y. Nedosiran: First Approval. Drugs 2023, 83, 1729–1733. [Google Scholar] [CrossRef]
- Scott, L.J. Givosiran: First Approval. Drugs 2020, 80, 335–339. [Google Scholar] [CrossRef] [PubMed]
- Lamb, Y.N. Inclisiran: First Approval. Drugs 2021, 81, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Scott, L.J.; Keam, S.J. Lumasiran: First Approval. Drugs 2021, 81, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Hoy, S.M. Patisiran: First Global Approval. Drugs 2018, 78, 1625–1631. [Google Scholar] [CrossRef] [PubMed]
- Afonin, K.A.; Viard, M.; Koyfman, A.Y.; Martins, A.N.; Kasprzak, W.K.; Panigaj, M.; Desai, R.; Santhanam, A.; Grabow, W.W.; Jaeger, L.; et al. Multifunctional RNA Nanoparticles. Nano Lett. 2014, 14, 5662–5671. [Google Scholar] [CrossRef] [PubMed]
- Afonin, K.A.; Kasprzak, W.K.; Bindewald, E.; Kireeva, M.; Viard, M.; Kashlev, M.; Shapiro, B.A. In Silico Design and Enzymatic Synthesis of Functional RNA Nanoparticles. Acc. Chem. Res. 2014, 47, 1731–1741. [Google Scholar] [CrossRef] [PubMed]
- Afonin, K.A.; Viard, M.; Kagiampakis, I.; Case, C.L.; Dobrovolskaia, M.A.; Hofmann, J.; Vrzak, A.; Kireeva, M.; Kasprzak, W.K.; KewalRamani, V.N.; et al. Triggering of RNA Interference with RNA–RNA, RNA–DNA, and DNA–RNA Nanoparticles. ACS Nano 2015, 9, 251–259. [Google Scholar] [CrossRef]
- Afonin, K.A.; Bindewald, E.; Yaghoubian, A.J.; Voss, N.; Jacovetty, E.; Shapiro, B.A.; Jaeger, L. In vitro assembly of cubic RNA-based scaffolds designed in silico. Nat. Nanotechnol. 2010, 5, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Gao, H.; Bao, G. Physical Principles of Nanoparticle Cellular Endocytosis. ACS Nano 2015, 9, 8655–8671. [Google Scholar] [CrossRef] [PubMed]
- Afonin, K.A.; Grabow, W.W.; Walker, F.M.; Bindewald, E.; Dobrovolskaia, M.A.; Shapiro, B.A.; Jaeger, L. Design and self-assembly of siRNA-functionalized RNA nanoparticles for use in automated nanomedicine. Nat. Protoc. 2011, 6, 2022–2034. [Google Scholar] [CrossRef] [PubMed]
- Ui-Tei, K.; Naito, Y.; Takahashi, F.; Haraguchi, T.; Ohki-Hamazaki, H.; Juni, A.; Ueda, R.; Saigo, K. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res. 2004, 32, 936–948. [Google Scholar] [CrossRef]
- Reynolds, A.; Leake, D.; Boese, Q.; Scaringe, S.; Marshall, W.S.; Khvorova, A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004, 22, 326–330. [Google Scholar] [CrossRef] [PubMed]
- Amarzguioui, M.; Prydz, H. An algorithm for selection of functional siRNA sequences. Biochem. Biophys. Res. Commun. 2004, 316, 1050–1058. [Google Scholar] [CrossRef] [PubMed]
- Rangan, R.; Zheludev, I.N.; Hagey, R.J.; Pham, E.A.; Wayment-Steele, H.K.; Glenn, J.S.; Das, R. RNA genome conservation and secondary structure in SARS-CoV-2 and SARS-related viruses: A first look. RNA 2020, 26, 937–959. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Akiyama, M.; Sakakibara, Y. RNA secondary structure prediction using deep learning with thermodynamic integration. Nat. Commun. 2021, 12, 941. [Google Scholar] [CrossRef] [PubMed]
- Welcome to the DuplexFold Web Server [EB/OL]. Available online: https://rna.urmc.rochester.edu/RNAstructureWeb/Servers/DuplexFold/DuplexFold.html (accessed on 5 January 2024).
- Kasprzak, W.; Bindewald, E.; Kim, T.J.; Jaeger, L.; Shapiro, B.A. Use of RNA structure flexibility data in nanostructure modeling. Methods 2011, 54, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Ju, X.; Zhu, Y.; Wang, Y.; Li, J.; Zhang, J.; Gong, M.; Ren, W.; Li, S.; Zhong, J.; Zhang, L.; et al. A novel cell culture system modeling the SARS-CoV-2 life cycle. PLOS Pathog. 2021, 17, e1009439. [Google Scholar] [CrossRef] [PubMed]
- Rennick, J.J.; Johnston, A.P.R.; Parton, R.G. Key principles and methods for studying the endocytosis of biological and nanoparticle therapeutics. Nat. Nanotechnol. 2021, 16, 266–276. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhu, W.; Fan, M.; Zhang, J.; Peng, Y.; Huang, F.; Wang, N.; He, L.; Zhang, L.; Holmdahl, R.; et al. Dependence of SARS-CoV-2 infection on cholesterol-rich lipid raft and endosomal acidification. Comput. Struct. Biotechnol. J. 2021, 19, 1933–1943. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Liu, D.X.; Tam, J.P. Lipid rafts are involved in SARS-CoV entry into Vero E6 cells. Biochem. Biophys. Res. Commun. 2008, 369, 344–349. [Google Scholar] [CrossRef]
- Toussi, S.S.; Hammond, J.L.; Gerstenberger, B.S.; Anderson, A.S. Therapeutics for COVID-19. Nat. Microbiol. 2023, 8, 771–786. [Google Scholar] [CrossRef] [PubMed]
- Abbasian, M.H.; Mahmanzar, M.; Rahimian, K.; Mahdavi, B.; Tokhanbigli, S.; Moradi, B.; Sisakht, M.M.; Deng, Y. Global landscape of SARS-CoV-2 mutations and conserved regions. J. Transl. Med. 2023, 21, 152. [Google Scholar] [CrossRef] [PubMed]
- Moazzam, M.; Zhang, M.; Hussain, A.; Yu, X.; Huang, J.; Huang, Y. The landscape of nanoparticle-based siRNA delivery and therapeutic development. Mol. Ther. 2024, 32, 284–312. [Google Scholar] [CrossRef] [PubMed]
- Otter, C.J.; Bracci, N.; Parenti, N.A.; Ye, C.; Asthana, A.; Blomqvist, E.K.; Tan, L.H.; Pfannenstiel, J.J.; Jackson, N.; Fehr, A.R.; et al. SARS-CoV-2 nsp15 endoribonuclease antagonizes dsRNA-induced antiviral signaling. Proc. Natl. Acad. Sci. USA 2024, 121, e2320194121. [Google Scholar] [CrossRef] [PubMed]
- Simeoni, M.; Cavinato, T.; Rodriguez, D.; Gatfield, D. I(nsp1)ecting SARS-CoV-2–ribosome interactions. Commun. Biol. 2021, 4, 715. [Google Scholar] [CrossRef] [PubMed]
- Calleja, D.J.; Lessene, G.; Komander, D. Inhibitors of SARS-CoV-2 PLpro. Front. Chem. 2022, 10, 876212. [Google Scholar] [CrossRef]
- Corona, A.; Madia, V.N.; De Santis, R.; Manelfi, C.; Emmolo, R.; Ialongo, D.; Patacchini, E.; Messore, A.; Amatore, D.; Faggioni, G.; et al. Diketo acid inhibitors of nsp13 of SARS-CoV-2 block viral replication. Antivir. Res. 2023, 217, 105697. [Google Scholar] [CrossRef]
- Afonin, K.A.; Dobrovolskaia, M.A.; Ke, W.; Grodzinski, P.; Bathe, M. Critical review of nucleic acid nanotechnology to identify gaps and inform a strategy for accelerated clinical translation. Adv. Drug Deliv. Rev. 2022, 181, 114081. [Google Scholar] [CrossRef] [PubMed]
- Kanarskaya, M.A.; Pyshnyi, D.V.; Lomzov, A.A. Diversity of Self-Assembled RNA Complexes: From Nanoarchitecture to Nanomachines. Molecules 2023, 29, 10. [Google Scholar] [CrossRef] [PubMed]
- Poppleton, E.; Urbanek, N.; Chakraborty, T.; Griffo, A.; Monari, L.; Göpfrich, K. RNA origami: Design, simulation and application. RNA Biol. 2023, 20, 510–524. [Google Scholar] [CrossRef]
- Afonin, K.A.; Dobrovolskaia, M.A.; Church, G.; Bathe, M. Opportunities, Barriers, and a Strategy for Overcoming Translational Challenges to Therapeutic Nucleic Acid Nanotechnology. ACS Nano 2020, 14, 9221–9227. [Google Scholar] [CrossRef] [PubMed]
- Kumar, H.; Mishra, G.; Sharma, A.K.; Gothwal, A.; Kesharwani, P.; Gupta, U. Intranasal Drug Delivery: A Non-Invasive Approach for the Better Delivery of Neurotherapeutics. Pharm. Nanotechnol. 2017, 5, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Song, X.J.; Chen, X.; Wang, J.W.; Cui, Y.L. Advances and future perspectives of intranasal drug delivery: A scientometric review. J. Control. Release 2024, 367, 366–384. [Google Scholar] [CrossRef] [PubMed]
| ID | Strand | Sequence (5′-3′) | Secondary Structure | Coverage | Target Genes |
|---|---|---|---|---|---|
| siRNA1 | guide | UGUGUUUUCUCGUUGAAACCA | ((.(((((......))))))) (−0.8) | 99.83% | NSP1_N |
| passenger | GUUUCAACGAGAAAACACACG | (((((.....)))).)..... (−3.3) | |||
| siRNA2 | guide | UAUUACCGUUCUUACGAAGAA | ..((..(((....)))..)). (−2) | 99.69% | NSP1_C |
| passenger | CUUCGUAAGAACGGUAAUAAA | ((((....))).)........ (−2.3) | |||
| siRNA3 | guide | UAUUAUUGGGUAAACCUUGGG | (......((.....))....) (−0.5) | 99.36% | Nucleocapsid |
| passenger | CAAGGUUUACCCAAUAAUACU | ..((..(((.....)))..)) (−4) | |||
| siRNA4 | guide | UUGUAUAUGCGAAAAGUGCAU | .(((((.........))))). (−0.1) | 99.52% | RdRp |
| passenger | GCACUUUUCGCAUAUACAAAA | ((.......)).......... (−2.9) | |||
| siRNA5 | guide | AAUAAACACGCCAAGUAGGAG | ..........((.....)).. (−1.8) | 99.90% | Spike_S2 |
| passenger | CCUACUUGGCGUGUUUAUUCU | ((.....))...(......). (−2.8) | |||
| siRNA6 | guide | GAAUUCCAAGCUAUAACGCAG | .........((......)).. (−3.9) | 99.69% | Spike_RBD |
| passenger | GCGUUAUAGCUUGGAAUUCUA | ((......))........... (−2.3) | |||
| siRNA7 | guide | UUUCAACGUACACUUUGUUUC | ....((((.......)))).. (−4.7) | 99.77% | Spike_NTD |
| passenger | AACAAAGUGUACGUUGAAAUC | ............(.......) (−4.9) | |||
| siRNA8 | guide | GUAGCUUUGAGCGUUUCUGCU | ((((............)))). (−0.5) | 99.77% | NSP13_Stem |
| passenger | CAGAAACGCUCAAAGCUACUG | (((.(..(((...)))).))) (−0.8) | |||
| siRNA9 | guide | UUAGUUACACGAUAACCAGUA | (..((((.....))))....) (−2.8) | 99.93% | NSP13_1B |
| passenger | CUGGUUAUCGUGUAACUAAAA | .(((((((...)))))))... (−0.7) | |||
| siRNA10 | guide | ACAUCAUGCGUGAUAACACCC | ..((((....))))....... (−2.6) | 98.89% | NSP13_helicase |
| passenger | GUGUUAUCACGCAUGAUGUUU | (...(((((....)))))..) (−0.6) | |||
| siRNA11 | guide | UAAGAAUGGUCUACGUAUGCA | (..(.(((.....))).)..) (−2.7) | 99.46% | NSP13_ZBD |
| passenger | CAUACGUAGACCAUUCUUAUG | ((((...(((....))))))) (−0.4) | |||
| siRNA12 | guide | AGCUUUAGGGUUACCAAUGUC | .((....((....))...)). (−0.5) | 99.87% | NSP14 |
| passenger | CAUUGGUAACCCUAAAGCUAU | ...((((.........)))). (−3.3) | |||
| siRNA13 | guide | UAAACGAUAUGUUCGAAGGCA | ....(((.....)))...... (−2.5) | 99.79% | NSP15_NendoU domain |
| passenger | CCUUCGAACAUAUCGUUUAUG | (...(((.....))).....) (−1.8) | |||
| siRNA14 | guide | UCUACUUGACCAUCAACUCUA | (....((((...))))....) (−3) | 99.66% | NSP15_Middle domain |
| passenger | GAGUUGAUGGUCAAGUAGACU | .((((.((......)).)))) (−2.3) | |||
| siRNA15 | guide | AAAAUCUAGCACCAUAAUCAA | ..................... (−1.9) | 99.85% | NSP3 _Single-stranded poly(A) binding domain |
| passenger | GAUUAUGGUGCUAGAUUUUAC | (...((........))....) (−3.5) | |||
| siRNA16 | guide | ACAAACACGGUUUAAACACCG | .......((((......)))) (−2) | 99.76% | NSP3 _PLpro |
| passenger | GUGUUUAAACCGUGUUUGUAC | (((..(((((...)))))))) (−0.1) | |||
| siRNA17 | guide | AAUUACAACCGUCUACAACAU | ..................... (−2.9) | 99.84% | NSP3_C |
| passenger | GUUGUAGACGGUUGUAAUUCA | (...((.(....).))...). (−3.3) | |||
| siRNA18 | guide | GUAAACUACGUCAUCAAGCCA | ..................... (−2.6) | 99.54% | NSP5 |
| passenger | GCUUGAUGACGUAGUUUACUG | (........).(((....))) (−3.1) |
| siRNA Nanoparticles | siRNA | RNA Motif | Length | Target Gene |
|---|---|---|---|---|
| siRNA_nano_1 | siRNA1 | CUCCUACUUGGCGUGUUUAUUCCUGUCAAUCAUGGCAAGUGUGUUUUCUCGUUGAAACCA | 60 | NSP1_N |
| siRNA2 | UGGUUUCAACGAGAAAACACACUUGUCAUGUGUAUGUUGCCUUAUUACCGUUCUUACGAAGAA | 63 | NSP1_C | |
| siRNA5 | UUCUUCGUAAGAACGGUAAUAAGGCACAUACUUUGUUGAUAGGAAUAAACACGCCAAGUAGGAG | 64 | Spike_S2 | |
| siRNA_nano_2 | siRNA8 | AGCAGAGACGCUCGGAGCUGCUGUUUUGGUCUACUUGACCAUCAACUCUA | 50 | NSP15_Middle domain |
| siRNA14 | UAGAGUUGGUGGUCGAGUAGGCCUUUUGCAAAAUCUAGCACCAUAAUCAA | 50 | NSP3 _Single-stranded poly(A) binding domain | |
| siRNA15 | UUGAUUAUGGUGCUAGGUUUUGCUUUUCAGUAGCUUUGAGCGUUUCUGCU | 50 | NSP13_Stem |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, J.; Lu, S.; Xiao, J.; Xu, N.; Li, Y.; Xu, J.; Deng, M.; Xuanyuan, H.; Zhang, Y.; Wu, F.; et al. Inhibition of SARS-CoV-2 Replication by Self-Assembled siRNA Nanoparticles Targeting Multiple Highly Conserved Viral Sequences. Viruses 2024, 16, 1072. https://doi.org/10.3390/v16071072
Sun J, Lu S, Xiao J, Xu N, Li Y, Xu J, Deng M, Xuanyuan H, Zhang Y, Wu F, et al. Inhibition of SARS-CoV-2 Replication by Self-Assembled siRNA Nanoparticles Targeting Multiple Highly Conserved Viral Sequences. Viruses. 2024; 16(7):1072. https://doi.org/10.3390/v16071072
Chicago/Turabian StyleSun, Jianan, Siya Lu, Jizhen Xiao, Nuo Xu, Yingbin Li, Jinfeng Xu, Maohua Deng, Hanlu Xuanyuan, Yushi Zhang, Fangli Wu, and et al. 2024. "Inhibition of SARS-CoV-2 Replication by Self-Assembled siRNA Nanoparticles Targeting Multiple Highly Conserved Viral Sequences" Viruses 16, no. 7: 1072. https://doi.org/10.3390/v16071072
APA StyleSun, J., Lu, S., Xiao, J., Xu, N., Li, Y., Xu, J., Deng, M., Xuanyuan, H., Zhang, Y., Wu, F., Jin, W., & Liu, K. (2024). Inhibition of SARS-CoV-2 Replication by Self-Assembled siRNA Nanoparticles Targeting Multiple Highly Conserved Viral Sequences. Viruses, 16(7), 1072. https://doi.org/10.3390/v16071072







