Isolation and Characterization of a Lytic Bacteriophage RH-42-1 of Erwinia amylovora from Orchard Soil in China
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.1.1. Bacterial Strains and Culture Media
2.1.2. Soil Samples
2.2. Enrichment and Isolation
2.3. Purification, Concentration, and Titer Determination
2.4. Electron Microscopy
2.5. Characterization of Bacteriophage
2.5.1. Determination of Optimal Multiplicity of Infection (MOI)
2.5.2. One-Step Growth Curve Determination
2.5.3. Host Range
2.5.4. Tolerance to Temperature, pH, UV Light, and Chemicals
2.6. Whole Genome Sequencing
2.7. Statistical Analysis
3. Results and Analysis
3.1. Isolation, Purification, and Electron Microscopy
3.2. Optimal Multiplicity of Infection (MOI) for Bacteriophage RH-42-1
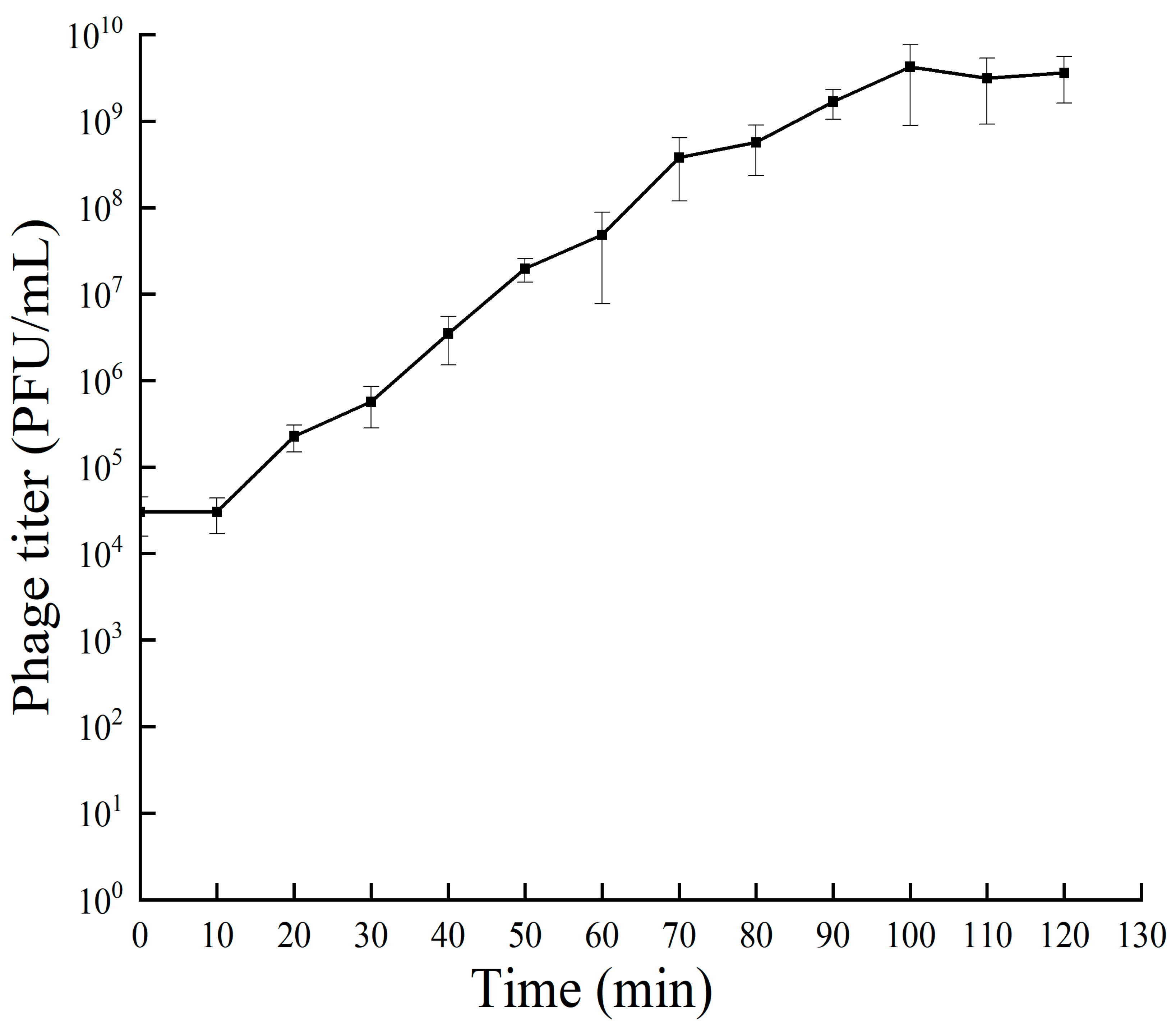
3.3. One-Step Growth Curve of Bacteriophage RH-42-1
3.4. Host Range of Bacteriophage RH-42-1
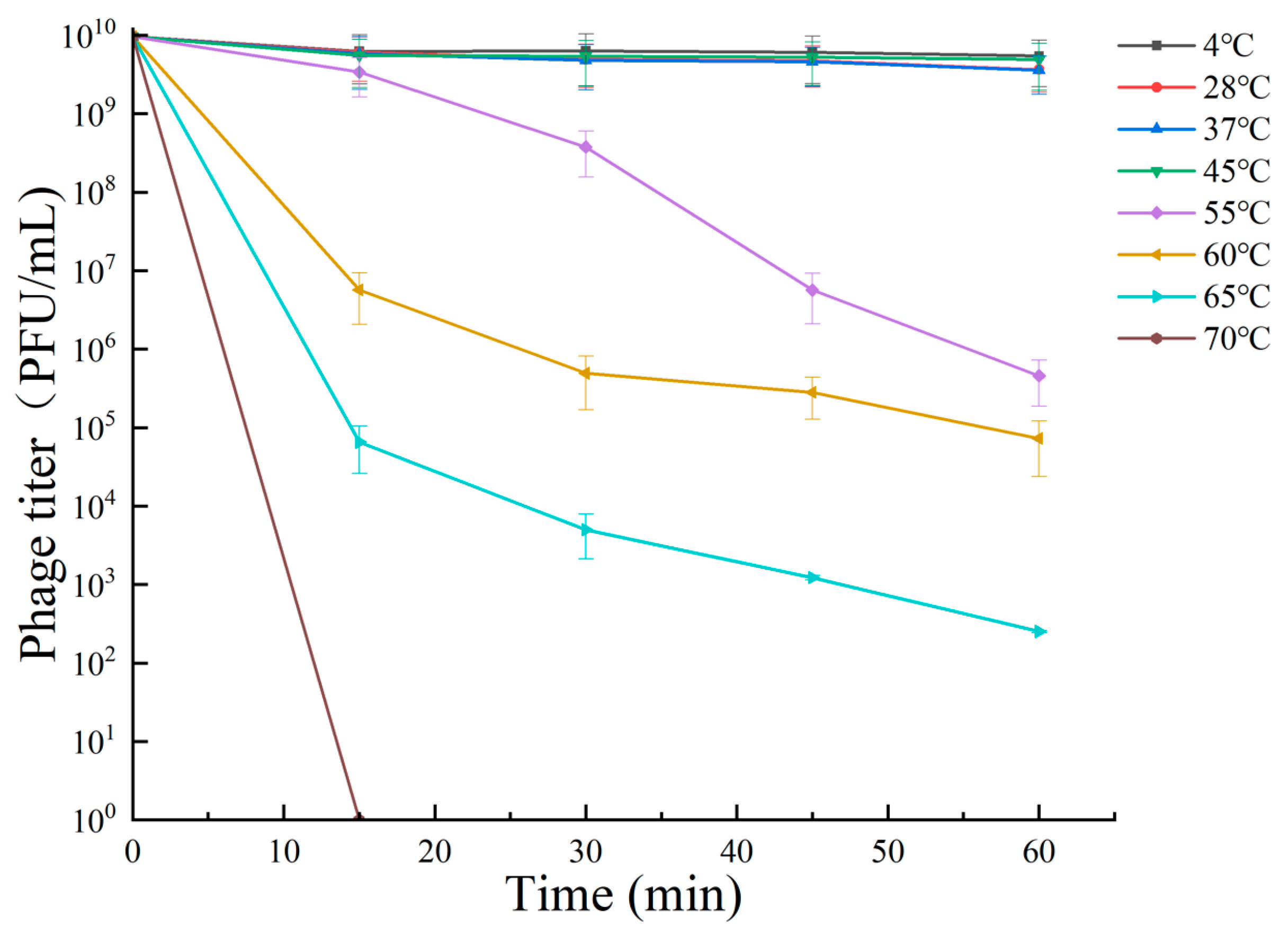
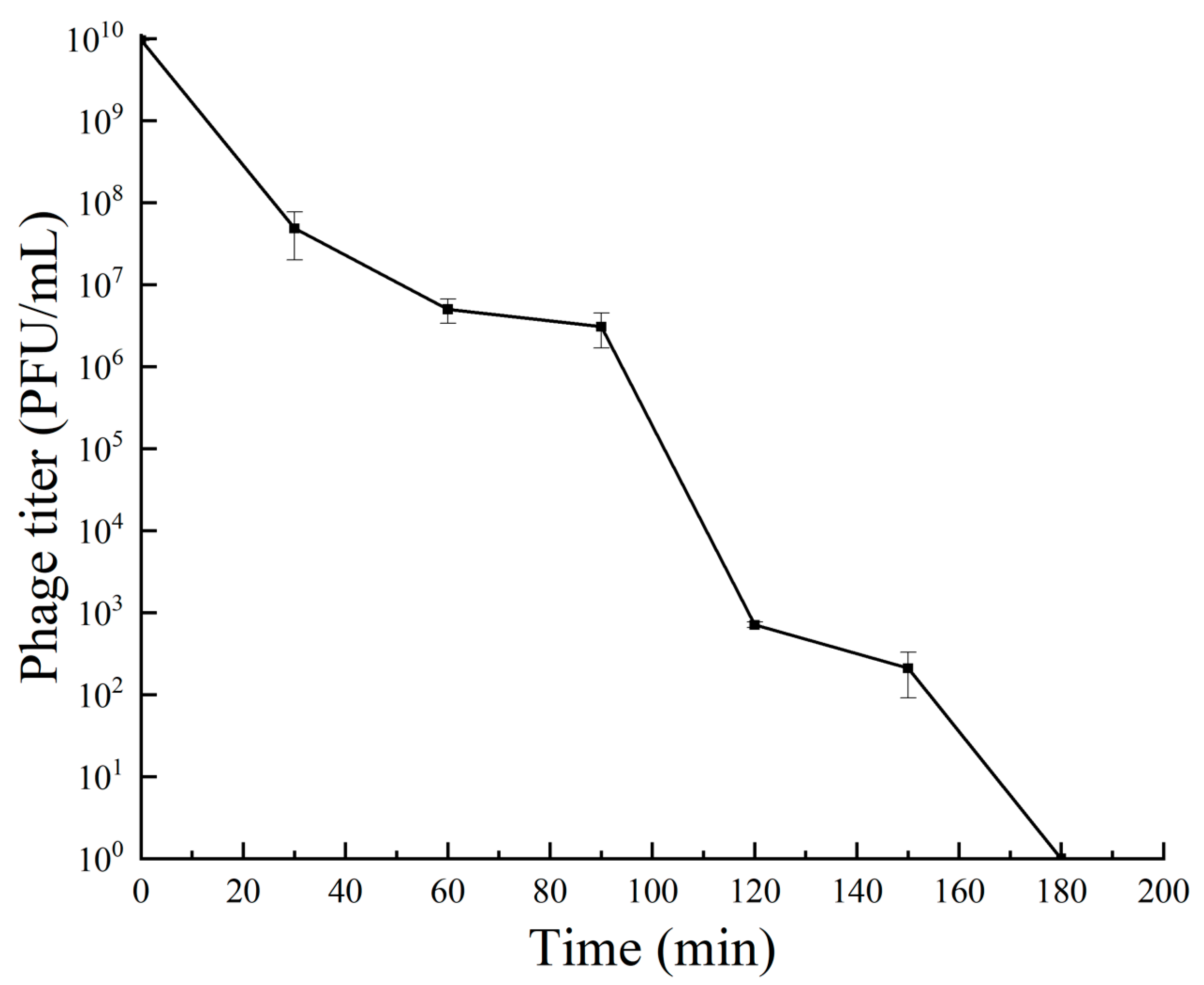
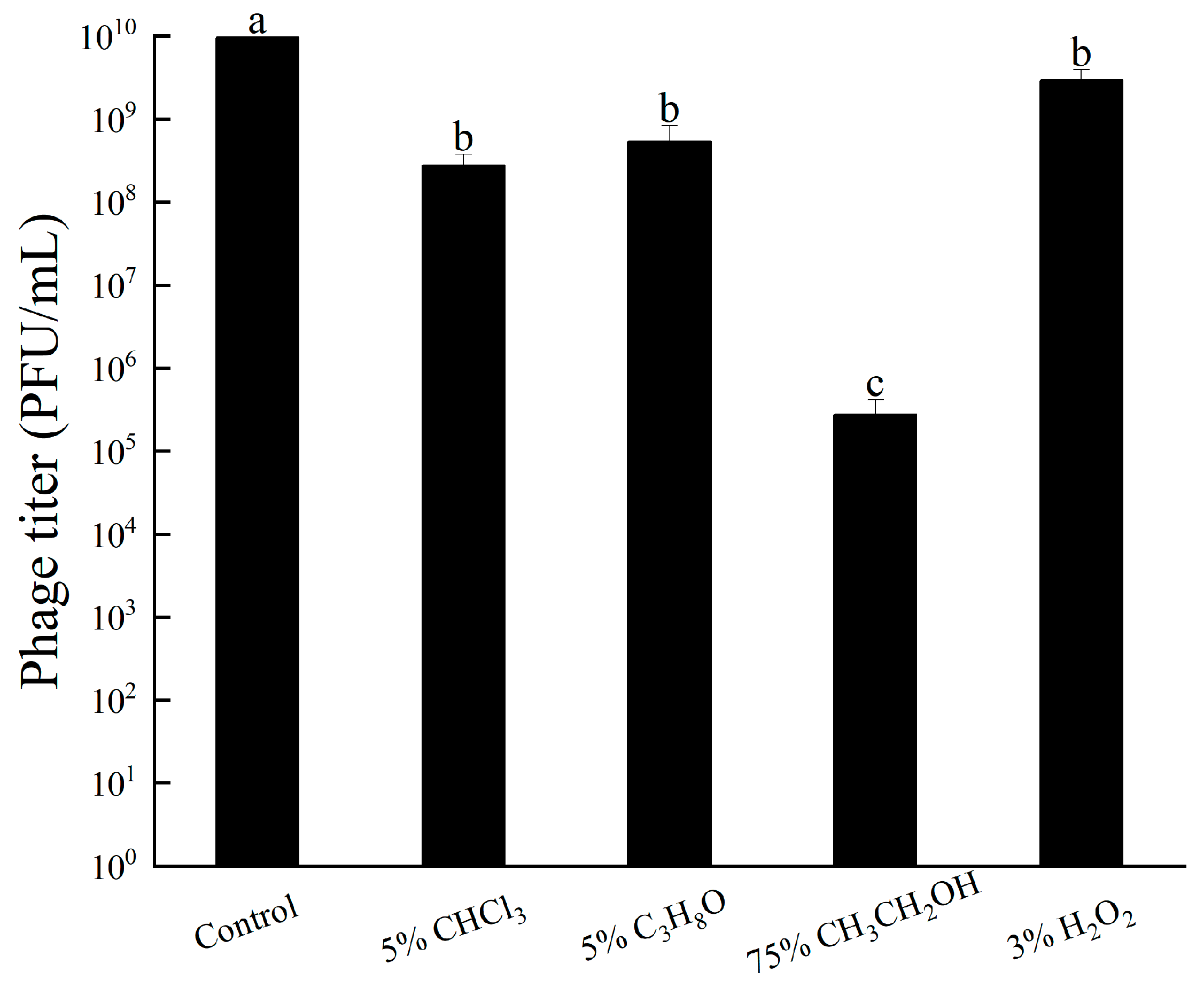
3.5. Tolerance of Bacteriophage RH-42-1
3.5.1. Temperature Tolerance
3.5.2. pH Tolerance
3.5.3. UV Radiation Tolerance
3.5.4. Other Chemicals Tolerance
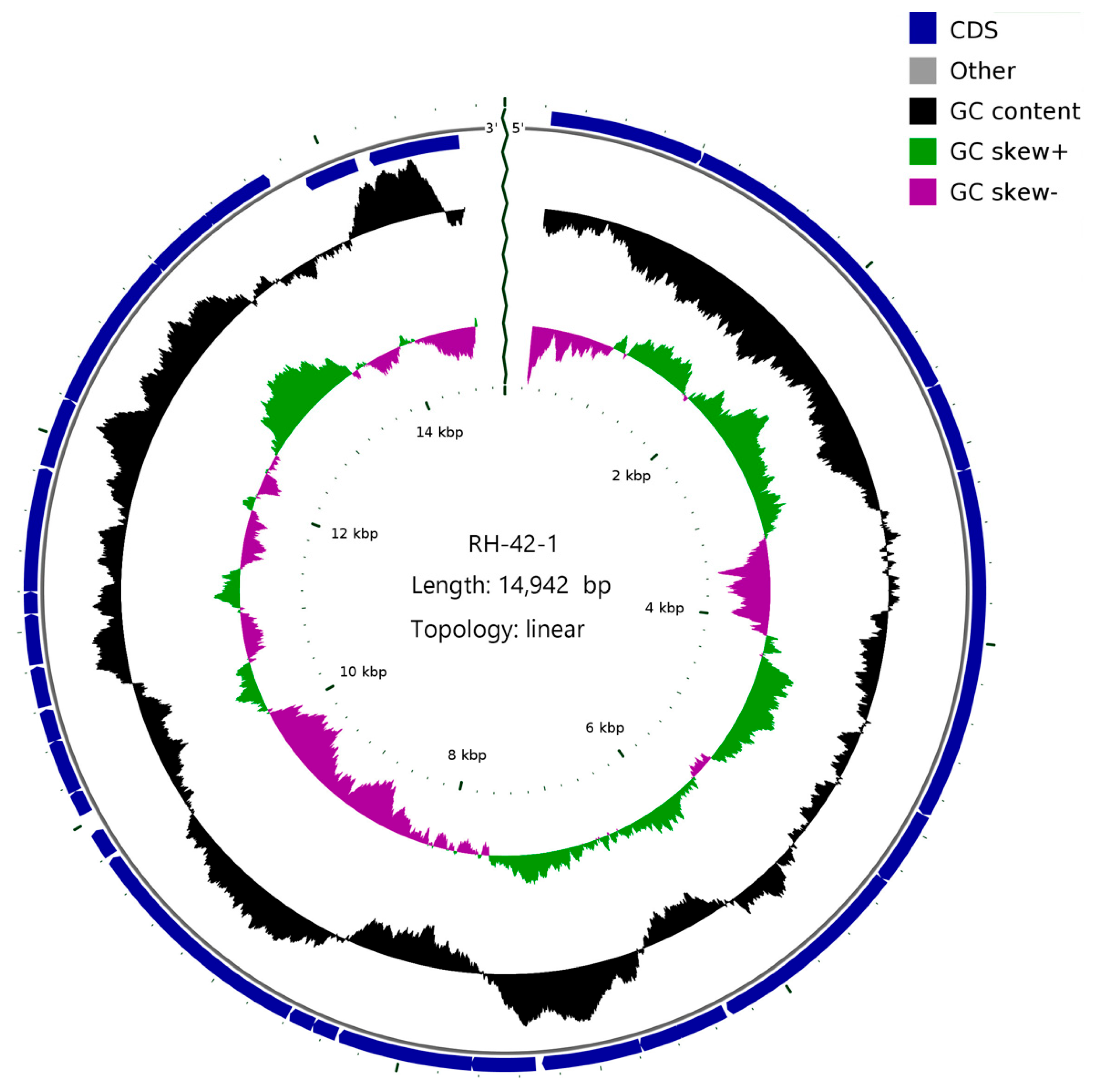
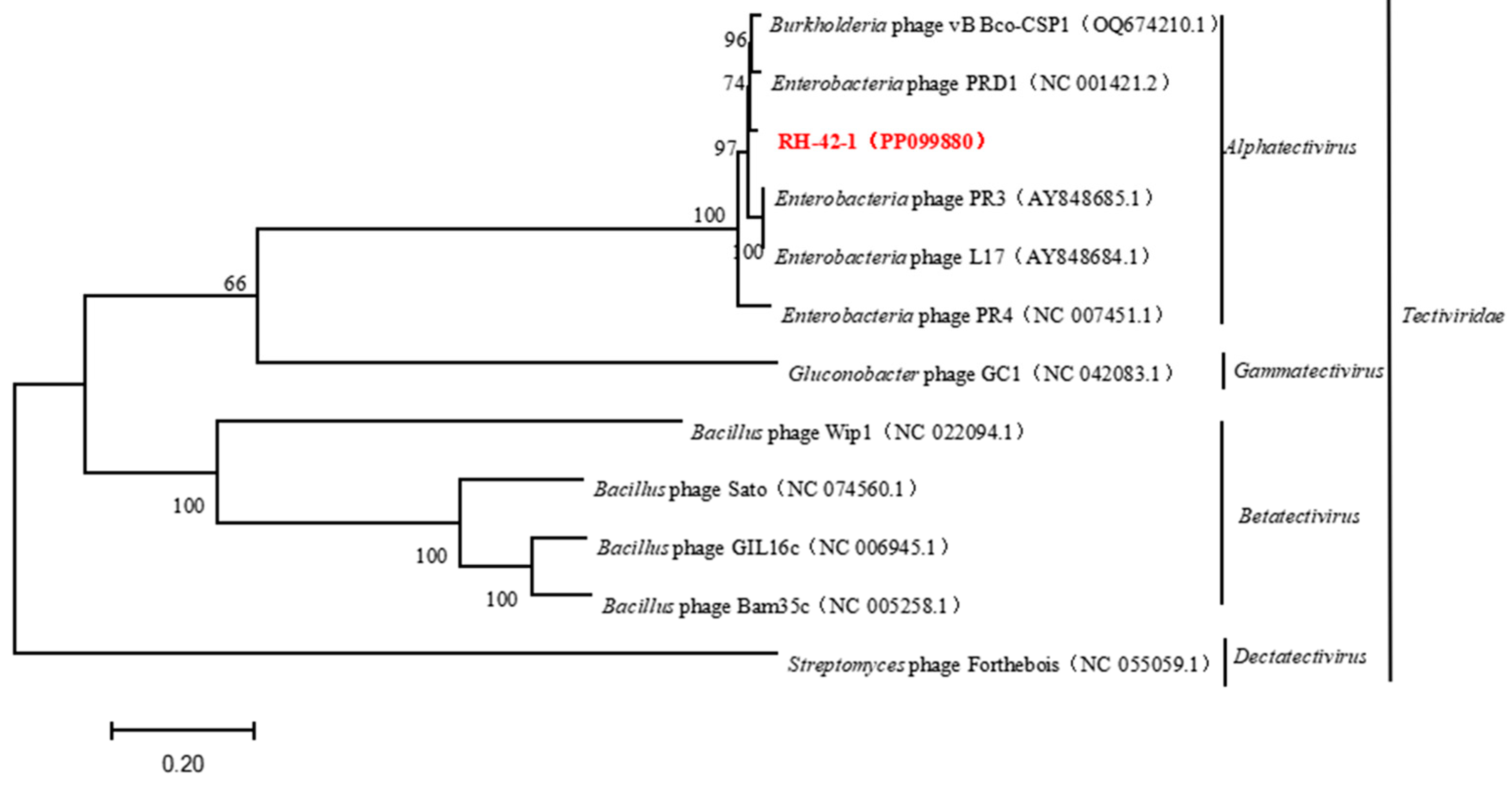
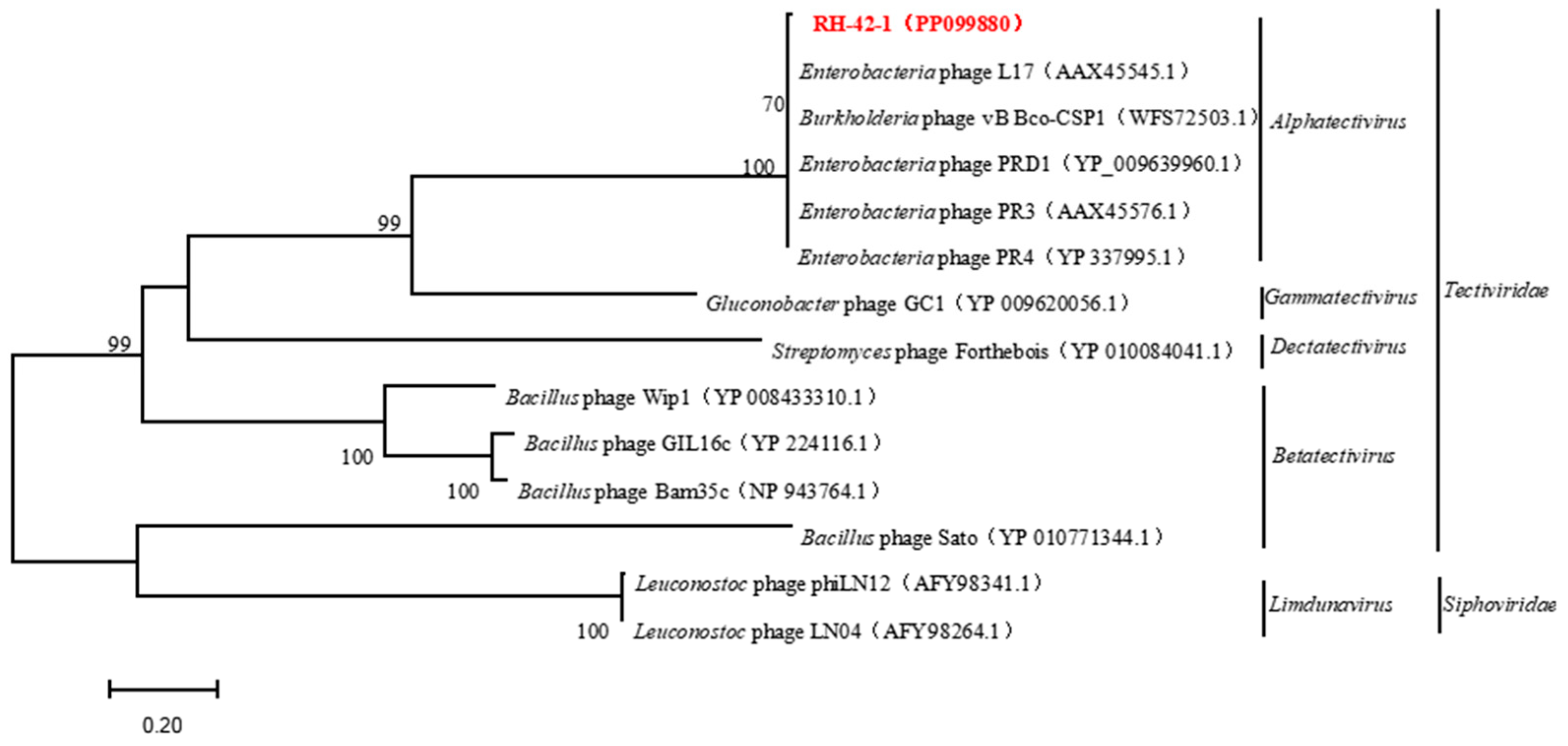
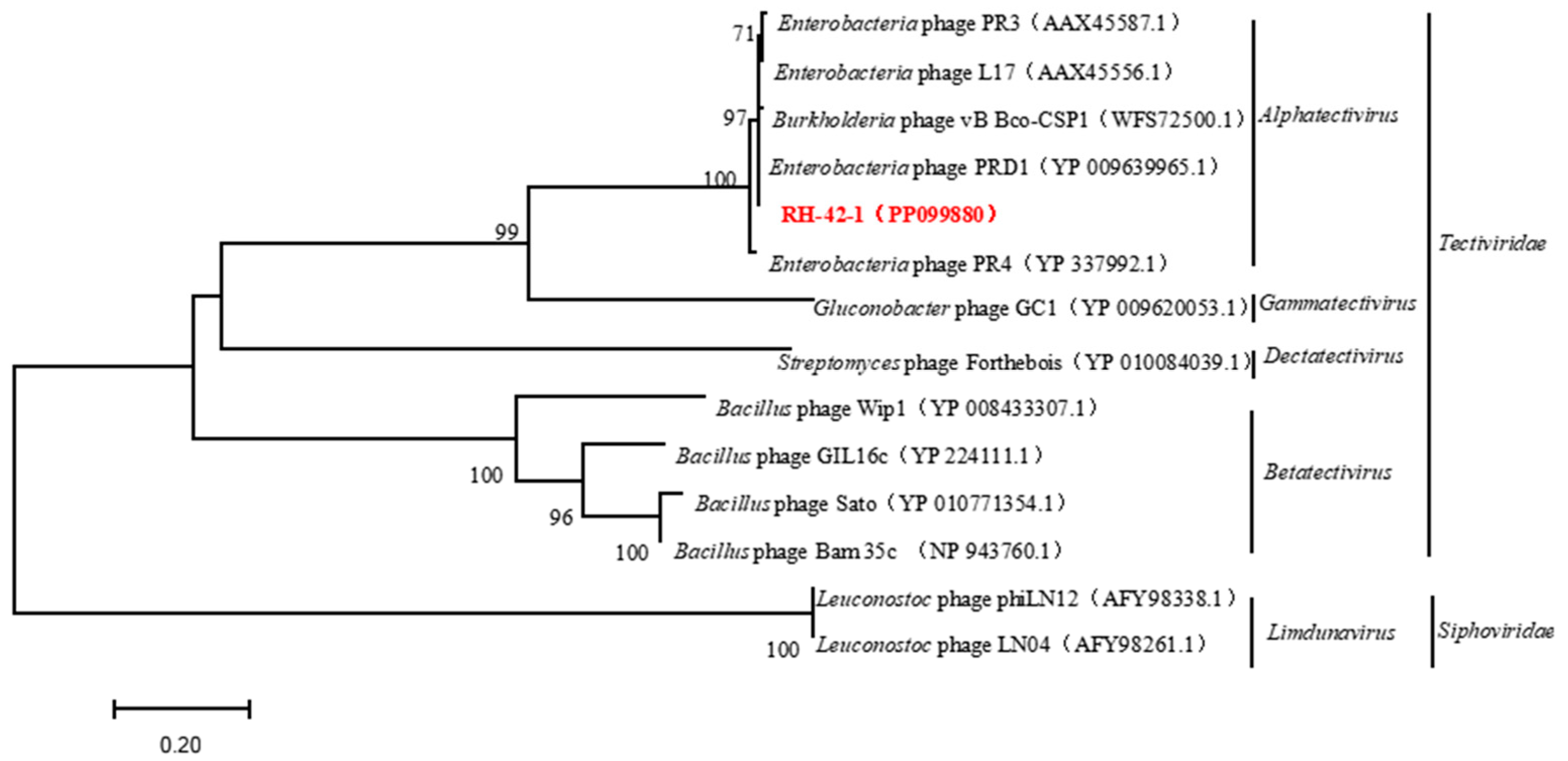
3.6. Genomic Features of Bacteriophage RH-42-1
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Mansfield, J.; Genin, S.; Magori, S.; Citovsky, V.; Sriariyanum, M.; Ronald, P.; Dow, M.; Verdier, V.; Beer, S.V.; Machado, M.A.; et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 614–629. [Google Scholar] [CrossRef] [PubMed]
- Eastgate, J.A. Erwinia amylovora: The molecular basis of fireblight disease. Mol. Plant Pathol. 2000, 1, 325–329. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Sheng, Q.; Luo, M.; Ma, D.; Zhang, C. Occurrence status of fire blight on Korla fragrant pear in Xinjiang and the control proposals. Plant Prot. Sin. 2022, 48, 207–213. [Google Scholar]
- Mikiciński, A.; Puławska, J.; Molzhigitova, A. Bacterial species recognized for the first time for its biocontrol activity against fire blight (Erwinia amylovora). Eur. J. Plant Pathol. 2020, 156, 257–272. [Google Scholar] [CrossRef]
- Sun, W.; Gong, P.; Zhao, Y.; Ming, L.; Zeng, Q.; Liu, F. Current Situation of Fire Blight in China. Phytopathology 2023, 113, 2143–2151. [Google Scholar] [CrossRef] [PubMed]
- Jurgens, A.G.; Babadoost, M. Sensitivity of Erwinia amylovora in Illinois Apple Orchards to Streptomycin, Oxytetracyline, Kasugamycin, and Copper. Plant Dis. 2013, 97, 1484–1490. [Google Scholar] [CrossRef] [PubMed]
- Stockwell, V.O.; Johnson, K.B.; Sugar, D.; Loper, J.E. Control of fire blight by Pseudomonas fluorescens A506 and Pantoea vagans C9-1 applied as single strains and mixed inocula. Phytopathology 2010, 100, 1330–1339. [Google Scholar] [CrossRef] [PubMed]
- Bahadou, S.A.; Ouijja, A.; Boukhari, M. Development of field strategies for fire blight control integrating biocontrol agents and plant defense activators in Morocco. J. Plant Pathol. 2017, 99, 51–58. [Google Scholar]
- Broggini, G.A.L.; Duffy, B.; Holliger, E.; Schärer, H.J.; Gessler, C.; Patocchi, A. Detection of the fire blight biocontrol agent Bacillus subtilis BD170 (Biopro®) in a Swiss apple orchard. Eur. J. Plant Pathol. 2005, 111, 93–100. [Google Scholar] [CrossRef]
- Cui, Z.; Hu, L.; Zeng, L.; Meng, W.; Guo, D.; Sun, L. Isolation and characterization of Priestia megaterium KD7 for the biological control of pear fire blight. Front. Microbiol. 2023, 14, 1099664. [Google Scholar] [CrossRef]
- Colavecchio, A.; Cadieux, B.; Lo, A.; Goodridge, L.D. Bacteriophages Contribute to the Spread of Antibiotic Resistance Genes among Foodborne Pathogens of the Enterobacteriaceae Family—A Review. Front. Microbiol. 2017, 8, 1108. [Google Scholar] [CrossRef] [PubMed]
- Buttimer, C.; McAuliffe, O.; Ross, R.P.; Hill, C.; O’Mahony, J.; Coffey, A. Bacteriophages and Bacterial Plant Diseases. Front. Microbiol. 2017, 8, 34. [Google Scholar] [CrossRef] [PubMed]
- Sabri, M.; Benkirane, R.; Habbadi, K.; Sadik, S.; Ou-Zine, M.; Diouri, M.; Achbani, E.H. Phages as a Potential Biocontrol of Phytobacteria. Arch. Phytopathol. Plant Prot. 2021, 54, 1277–1291. [Google Scholar] [CrossRef]
- Akremi, I.; Holtappels, D.; Brabra, W.; Jlidi, M.; Hadj Ibrahim, A.; Ben Ali, M.; Fortuna, K.; Ahmed, M.; Meerbeek, B.V.; Rhouma, A.; et al. First Report of Filamentous Phages Isolated from Tunisian Orchards to Control Erwinia amylovora. Microorganisms 2020, 8, 1762. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yang, Y. Preliminary study on bacteriophage of fire blight. Plant Quar. 2001, 1, 414–417. [Google Scholar]
- Kropinski, A.M.; Mazzocco, A.; Waddell, T.E.; Lingohr, E.; Johnson, R.P. Enumeration of bacteriophages by double agar overlay plaque assay. Methods Mol. Biol. 2009, 501, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Wen, C.; Ai, C.; Lu, S.; Yang, Q.; Liao, H.; Zhou, S. Isolation and Characterization of the Lytic Pseudoxanthomonas kaohsiungensi Phage PW916. Viruses 2022, 14, 1709. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Wang, Y.; Hong, B.; Li, Y.; Ma, Y.; Wang, J. Isolation and Characterization of a Lytic Vibrio parahaemolyticus Phage vB_VpaP_GHSM17 from Sewage Samples. Viruses 2022, 14, 1601. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, M.; Hu, R.; Bai, J.; He, X.; Jin, Y. Isolation of the Novel Phage PHB09 and Its Potential Use against the Plant Pathogen Pseudomonas syringae pv. actinidiae. Viruses 2021, 13, 2275. [Google Scholar] [CrossRef]
- Coil, D.; Jospin, G.; Darling, A.E. A5-miseq: An updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 2015, 31, 587–589. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef] [PubMed]
- Besemer, J.; Lomsadze, A.; Borodovsky, M. GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001, 29, 2607–2618. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Stothard, P.; Wishart, D.S. Circular genome visualization and exploration using CGView. Bioinformatics 2005, 21, 537–539. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Morello, E.; Saussereau, E.; Maura, D.; Huerre, M.; Touqui, L.; Debarbieux, L. Pulmonary bacteriophage therapy on Pseudomonas aeruginosa cystic fibrosis strains: First steps towards treatment and prevention. PLoS ONE 2011, 6, e16963. [Google Scholar] [CrossRef] [PubMed]
- Malnoy, M.; Martens, S.; Norelli, J.L.; Barny, M.A.; Sundin, G.W.; Smits, T.H.; Duffy, B. Fire blight: Applied genomic insights of the pathogen and host. Annu. Rev. Phytopathol. 2012, 50, 475–494. [Google Scholar] [CrossRef]
- Meczker, K.; Domotor, D.; Vass, J.; Rakhely, G.; Schneider, G.; Kovacs, T. The genome of the Erwinia amylovora phage PhiEaH1 reveals greater diversity and broadens the applicability of phages for the treatment of fire blight. FEMS Microbiol. Lett. 2014, 350, 25–27. [Google Scholar] [CrossRef]
- Tang, Z.; Liu, J. Exploration and functional identification of novel Xanthomonas oryzae pv. oryzae (Xoo) phages. Biol. Resour. 2018, 40, 18–23. [Google Scholar] [CrossRef]
- Balogh, B.; Dickstein, E.R.; Jones, J.B.; Canteros, B.I. Narrow host range phages associated with citrus canker lesions in Florida and Argentina. Eur. J. Plant Pathol. 2013, 135, 253–264. [Google Scholar] [CrossRef]
- Besarab, N.V.; Letarov, A.V.; Kulikov, E.E.; Babenko, V.V.; Belalov, I.S.; Lagonenko, A.L.; Golomidova, A.K.; Evtushenkov, A.N. Two novel Erwinia amylovora bacteriophages, Loshitsa2 and Micant, isolated in Belarus. Arch. Virol. 2022, 167, 2633–2642. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Shi, J.; Ma, W.; Li, Z.; Wang, J.; Li, J.; Wang, X. Isolation, characterization, and application of a novel specific Salmonella bacteriophage in different food matrices. Food Res. Int. 2018, 111, 631–641. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.G.; Roh, E.; Park, J.; Giri, S.S.; Kwon, J.; Kim, S.W.; Kang, J.W.; Lee, S.B.; Jung, W.J.; Lee, Y.M.; et al. The Bacteriophage pEp_SNUABM_08 Is a Novel Singleton Siphovirus with High Host Specificity for Erwinia pyrifoliae. Viruses 2021, 13, 1231. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Lee, G.M.; Kim, D.; Park, D.H.; Oh, C.S. Characterization of the Lytic Bacteriophage phiEaP-8 Effective against Both Erwinia amylovora and Erwinia pyrifoliae Causing Severe Diseases in Apple and Pear. Plant Pathol. J. 2018, 34, 445–450. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.J.; Kim, S.G.; Lee, Y.M.; Giri, S.S.; Kang, J.W.; Lee, S.B.; Jung, W.J.; Hwang, M.H.; Park, J.; Cheng, C.; et al. Evaluation of the Antimicrobial Potential and Characterization of Novel T7-Like Erwinia Bacteriophages. Biology 2023, 12, 180. [Google Scholar] [CrossRef] [PubMed]
- Choe, J.; Kim, B.; Park, M.K.; Roh, E. Biological and Genetic Characterizations of a Novel Lytic PhiFifi106 against Indigenous Erwinia amylovora and Evaluation of the Control of Fire Blight in Apple Plants. Biology 2023, 12, 1060. [Google Scholar] [CrossRef] [PubMed]
- Muller, I.; Lurz, R.; Kube, M.; Quedenau, C.; Jelkmann, W.; Geider, K. Molecular and physiological properties of bacteriophages from North America and Germany affecting the fire blight pathogen Erwinia amylovora. Microb. Biotechnol. 2011, 4, 735–745. [Google Scholar] [CrossRef]
- Sabri, M.; El Handi, K.; Valentini, F.; De Stradis, A.; Achbani, E.H.; Benkirane, R.; Resch, G.; Elbeaino, T. Identification and Characterization of Erwinia Phage IT22: A New Bacteriophage-Based Biocontrol against Erwinia amylovora. Viruses 2022, 14, 2455. [Google Scholar] [CrossRef]
- Besarab, N.V.; Akhremchuk, A.E.; Zlatohurska, M.A.; Romaniuk, L.V.; Valentovich, L.N.; Tovkach, F.I.; Lagonenko, A.L.; Evtushenkov, A.N. Isolation and characterization of Hena1—A novel Erwinia amylovora bacteriophage. FEMS Microbiol. Lett. 2020, 367. [Google Scholar] [CrossRef]
- Tayyarcan, E.K.; Boyaci, I.H. Isolation, characterization, and application of bacteriophage cocktails for the biocontrol of Pseudomonas fluorescens group strains in whole and skimmed milk. Braz. J. Microbiol. 2023, 54, 3061–3071. [Google Scholar] [CrossRef] [PubMed]
- Jalasvuori, M.; Koskinen, K. Extending the hosts of Tectiviridae into four additional genera of Gram-positive bacteria and more diverse Bacillus species. Virology 2018, 518, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Ravantti, J.J.; Gaidelyte, A.; Bamford, D.H.; Bamford, J.K. Comparative analysis of bacterial viruses Bam35, infecting a gram-positive host, and PRD1, infecting gram-negative hosts, demonstrates a viral lineage. Virology 2003, 313, 401–414. [Google Scholar] [CrossRef] [PubMed]
- Bamford, D.H.; Rouhiainen, L.; Takkinen, K.; Soderlund, H. Comparison of the lipid-containing bacteriophages PRD1, PR3, PR4, PR5 and L17. J. Gen. Virol. 1981, 57, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Olsen, R.H.; Siak, J.S.; Gray, R.H. Characteristics of PRD1, a plasmid-dependent broad host range DNA bacteriophage. J. Virol. 1974, 14, 689–699. [Google Scholar] [CrossRef] [PubMed]
- Llop, P.; Barbé, S.; López, M.M. Functions and origin of plasmids in Erwinia species that are pathogenic to or epiphytically associated with pome fruit trees. Trees 2012, 26, 31–46. [Google Scholar] [CrossRef][Green Version]


| Strain Number | Origin Tissues | Places |
|---|---|---|
| Ea 001 | Apple branches | Khorgas city, Xinjiang |
| X24 | Fragrant pear leaf | Korla city, Xinjing |
| X26 | Fragrant pear branch | Bugur county, Xinjing |
| X36 | Fragrant pear leaf | Korla city, Xinjing |
| Y85 | Fragrant pear core | Korla city, Xinjing |
| Y126 | Hawthorn leaf | Bugur county, Xinjing |
| Y134 | Quince branch | Bugur county, Xinjing |
| Y137 | Fragrant pear branch | Korla city, Xinjing |
| FK-1 | Apple branches | Fukang city, Xinjing |
| KEL-1 | Fragrant pear leaf | Yuli county, Xinjing |
| KEL-2 | Fragrant pear branch | Yuli county, Xinjing |
| TC-1 | Apple branches | Tacheng city, Xinjing |
| TC-2 | Apple branches | Tacheng city, Xinjing |
| TC-3 | Hawthorn branch | Tacheng city, Xinjing |
| MOI | Bacterial Population (CFU/mL) | Phage Concentration (PFU/mL) | Phage Titer (PFU/mL) |
|---|---|---|---|
| 100:1 | 107 | 109 | (2.7 ± 0.15) × 107 |
| 10:1 | 107 | 108 | (4.4 ± 0.2) × 107 |
| 1:1 | 107 | 107 | (9.6 ± 0.1) × 109 |
| 0.1:1 | 107 | 106 | (1.54 ± 0.03) × 108 |
| 0.01:1 | 107 | 105 | (6.26 ± 0.15) × 107 |
| 0.001:1 | 107 | 104 | (4.18 ± 0.03) × 106 |
| Host Species | Strains | Lysis Result |
|---|---|---|
| E. amylovora | FK-1, KEL-1, KEL-2, TC-1, TC-2 | + |
| Ea001, TC-3, X24, X26, X36, Y85, Y126, Y134, Y137 | − | |
| P. syringae pv. syringage | D1 | − |
| Escherichia coli | B13 | − |
| B. velezensis | FN12, F34, F44 | − |
| B. amyloliquefaciens | FX74 | − |
| Functional Category | ORF Order | Position of ORF | Accession No. | Annotation |
|---|---|---|---|---|
| Structure and packaging | 5 | 4908−5288 | YP_009639959.1 | major spike protein |
| 6 | 5288−6310 | AAX45552.1 | spike | |
| 7 | 6329−6589 | YP_009639961.1 | protein P17 | |
| 8 | 6579−6785 | AAR99748.1 | protein P33 | |
| 9 | 6785−7285 | YP_337990.1 | minor head protein | |
| 10 | 7318−7641 | UDY80295.1 | protein P10 | |
| 11 | 7638−8321 | YP_009639965.1 | terminase large subunit | |
| 13 | 8461−8589 | YP_009639967.1 | packaging protein | |
| 14 | 8596−9783 | YP_337995.1 | major capsid protein | |
| 20 | 10,834−11,088 | YP_009639975.1 | minor capsid protein | |
| DNA replication and regulation | 1 | 234−1013 | YP_009639955.1 | terminal protein |
| 2 | 1017−2678 | AAR99740.1 | DNA polymerase | |
| 4 | 3129−4904 | AAX45635.1 | receptor binding | |
| 15 | 9802−9945 | YP_337996.1 | DNA packaging | |
| 17 | 10,169−10,441 | YP_009639972.1 | DNA delivery | |
| 18 | 10,441−10,605 | YP_009639973.1 | membrane DNA delivery | |
| 19 | 10,618−10,824 | YP_009639974.1 | membrane DNA delivery | |
| 22 | 11,203−11,826 | YP_009639977.1 | DNA delivery | |
| 23 | 11,837−12,190 | AAR99763.1 | infectivity protein | |
| 24 | 12,191−13,000 | AAX45554.1 | transclycosylase | |
| 26 | 13,343−13,705 | YP_009639982.1 | hypothetical protein | |
| 27 | 13,861−14,145 | AAR99769.1 | ssDNA binding protein | |
| 28 | 14,218−14,700 | YP_009639985.1 | single strand DNA binding protein | |
| Cracking | 3 | 2680−3129 | YP_338008.1 | endolysin |
| 25 | 12,997−13,350 | AAX45661.1 | holin | |
| Unknown | 12 | 8333−8461 | YP_009639966.1 | hypothetical protein |
| 16 | 10,045−10,167 | YP_009639971.1 | hypothetical protein | |
| 21 | 11,091−11,201 | YP_009639976.1 | hypothetical protein |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xi, H.; Fu, B.; Sheng, Q.; Luo, M.; Sun, L. Isolation and Characterization of a Lytic Bacteriophage RH-42-1 of Erwinia amylovora from Orchard Soil in China. Viruses 2024, 16, 509. https://doi.org/10.3390/v16040509
Xi H, Fu B, Sheng Q, Luo M, Sun L. Isolation and Characterization of a Lytic Bacteriophage RH-42-1 of Erwinia amylovora from Orchard Soil in China. Viruses. 2024; 16(4):509. https://doi.org/10.3390/v16040509
Chicago/Turabian StyleXi, Haishen, Benzhong Fu, Qiang Sheng, Ming Luo, and Liying Sun. 2024. "Isolation and Characterization of a Lytic Bacteriophage RH-42-1 of Erwinia amylovora from Orchard Soil in China" Viruses 16, no. 4: 509. https://doi.org/10.3390/v16040509
APA StyleXi, H., Fu, B., Sheng, Q., Luo, M., & Sun, L. (2024). Isolation and Characterization of a Lytic Bacteriophage RH-42-1 of Erwinia amylovora from Orchard Soil in China. Viruses, 16(4), 509. https://doi.org/10.3390/v16040509





