Sequences Related to Chimay Rhabdovirus Are Widely Distributed in Ixodes ricinus Ticks across England and Wales
Abstract
1. Introduction
2. Materials and Methods
3. Results
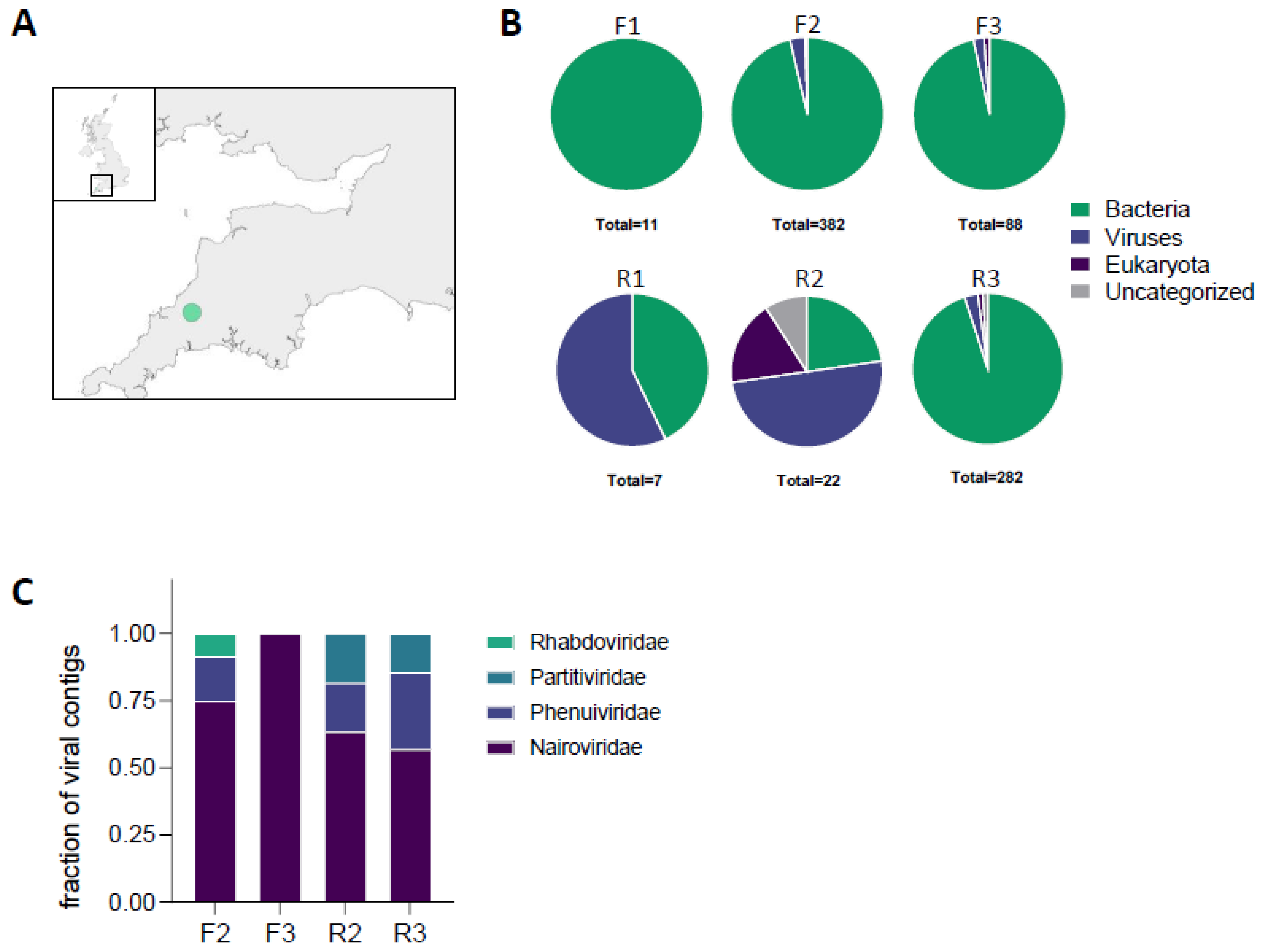
3.1. Microbiomes of Ixodes ricinus Collected in a Single Location Show High Diversity
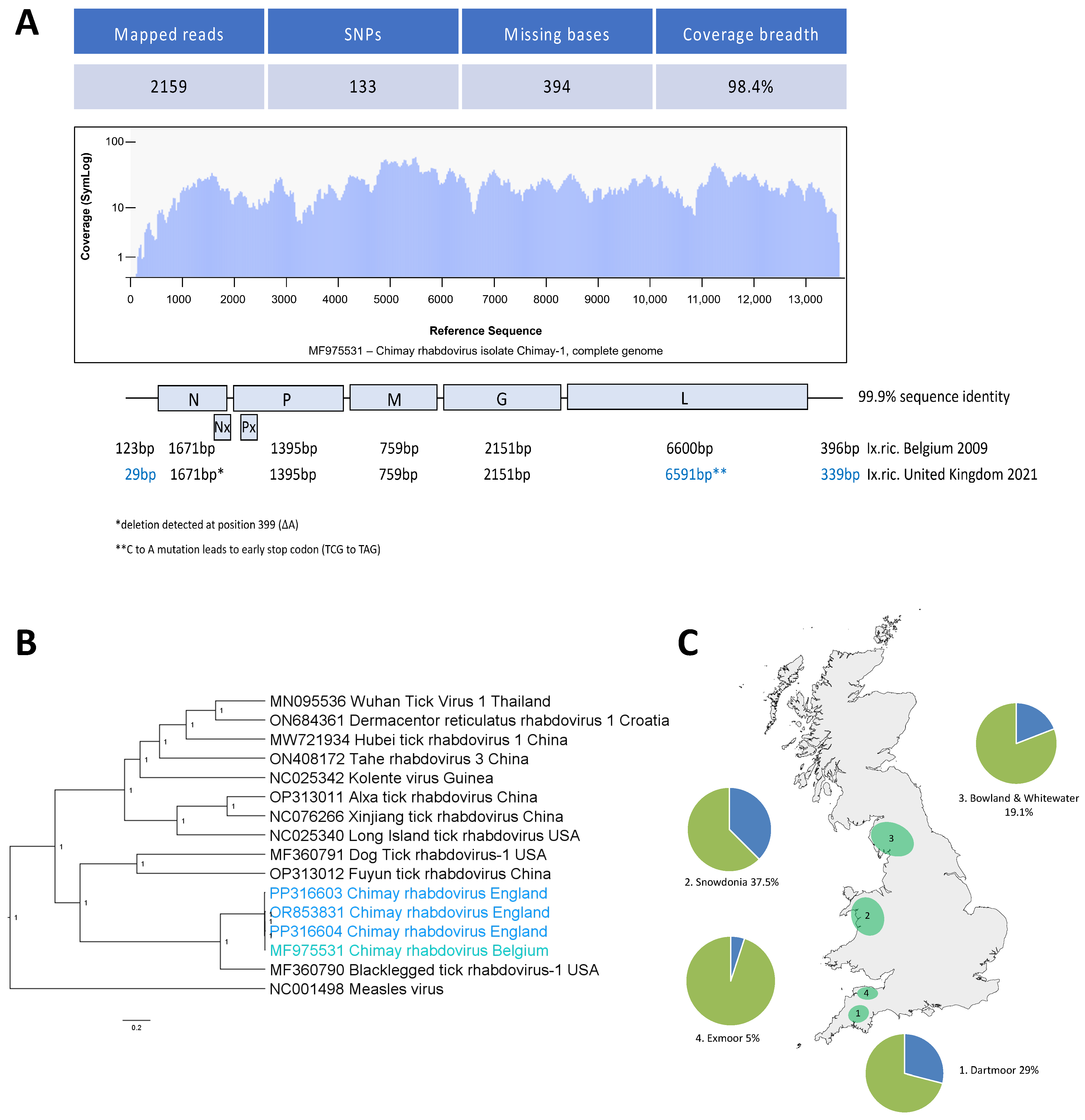
3.2. Sequences Related to Chimay Rhabdovirus Are Widely Distributed in Ticks across England and Wales
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shi, J.; Hu, Z.; Deng, F.; Shen, S. Tick-Borne Viruses. Virol. Sin. 2018, 33, 21–43. [Google Scholar] [CrossRef]
- Estrada-Peña, A.; de la Fuente, J. Species interactions in occurrence data for a community of tick-transmitted pathogens. Sci. Data 2016, 3, 160056. [Google Scholar] [CrossRef] [PubMed]
- Hubálek, Z.; Rudolf, I. Tick-borne viruses in Europe. Parasitol. Res. 2012, 111, 9–36. [Google Scholar] [CrossRef]
- Holding, M.; Dowall, S.D.; Medlock, J.M.; Carter, D.P.; Pullan, S.T.; Lewis, J.; Vipond, R.; Rocchi, M.S.; Baylis, M.; Hewson, R. Tick-Borne Encephalitis Virus, United Kingdom. Emerg. Infect. Dis. 2020, 26, 90–96. [Google Scholar] [CrossRef]
- Jahfari, S.; de Vries, A.; Rijks, J.M.; Van Gucht, S.; Vennema, H.; Sprong, H.; Rockx, B. Tick-Borne Encephalitis Virus in Ticks and Roe Deer, the Netherlands. Emerg. Infect. Dis. 2017, 23, 1028–1030. [Google Scholar] [CrossRef] [PubMed]
- Moutailler, S.; Popovici, I.; Devillers, E.; Vayssier-Taussat, M.; Eloit, M. Diversity of viruses in Ixodes ricinus, and characterization of a neurotropic strain of Eyach virus. New Microbes New Infect. 2016, 11, 71–81. [Google Scholar] [CrossRef]
- Pettersson, J.H.-O.; Shi, M.; Bohlin, J.; Eldholm, V.; Brynildsrud, O.B.; Paulsen, K.M.; Andreassen, Å.; Holmes, E.C. Characterizing the virome of Ixodes ricinus ticks from northern Europe. Sci. Rep. 2017, 7, 10870. [Google Scholar] [CrossRef]
- Vanmechelen, B.; Merino, M.; Vergote, V.; Laenen, L.; Thijssen, M.; Martí-Carreras, J.; Claerebout, E.; Maes, P. Exploration of the Ixodes ricinus virosphere unveils an extensive virus diversity including novel coltiviruses and other reoviruses. Virus Evol. 2021, 7, veab066. [Google Scholar] [CrossRef] [PubMed]
- Lihou, K.; Vineer, H.R.; Wall, R. Distribution and prevalence of ticks and tick-borne disease on sheep and cattle farms in Great Britain. Parasites Vectors 2020, 13, 406. [Google Scholar] [CrossRef]
- McFadzean, H.; Johnson, N.; Phipps, L.P.; Hobbs, R.L. High morbidity associated with an outbreak of tick-borne disease in a dairy herd, Cornwall. Vet. Rec. Case Rep. 2021, 9, e171. [Google Scholar] [CrossRef]
- Cull, B.; Hansford, K.M.; McGinley, L.; Gillingham, E.L.; Vaux AG, C.; Smith, R.; Medlock, J.M. A nationwide study on Borrelia burgdorferi s.l. infection rates in questing Ixodes ricinus: A six-year snapshot study in protected recreational areas in England and Wales. Med. Vet. Entomol. 2021, 35, 352–360. [Google Scholar] [CrossRef] [PubMed]
- Kalantar, K.L.; Carvalho, T.; A de Bourcy, C.F.; Dimitrov, B.; Dingle, G.; Egger, R.; Han, J.; Holmes, O.B.; Juan, Y.-F.; King, R.; et al. IDseq—An open source cloud-based pipeline and analysis service for metagenomic pathogen detection and monitoring. GigaScience 2020, 9, giaa111. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Ohlendorf, V.; Marklewitz, M.; Kopp, A.; Yordanov, S.; Drosten, C.; Junglen, S. Huge diversity of phleboviruses in ticks from Strandja Nature Park, Bulgaria. Ticks Tick-Borne Dis. 2019, 10, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Prinz, M.; Fuchs, J.; Ehrmann, S.; Scherer-Lorenzen, M.; Kochs, G.; Panning, M. Molecular identification of novel phlebovirus sequences in European ticks. Ticks Tick-Borne Dis. 2017, 8, 795–798. [Google Scholar] [CrossRef] [PubMed]
- Vanmechelen, B.; Laenen, L.; Vergote, V.; Maes, P. Grotenhout Virus, a Novel Nairovirus Found in Ixodes ricinus in Belgium. Genome Announc. 2017, 5, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Ergunay, K.; Bourke, B.P.; Reinbold-Wasson, D.D.; Nikolich, M.P.; Nelson, S.P.; Caicedo-Quiroga, L.; Vaydayko, N.; Kirkitadze, G.; Chunashvili, T.; Long, L.S.; et al. The expanding range of emerging tick-borne viruses in Eastern Europe and the Black Sea Region. Sci. Rep. 2023, 13, 19824. [Google Scholar] [CrossRef]
- Walker, P.J.; Kongsuwan, K. Deduced structural model for animal rhabdovirus glycoproteins. J. Gen. Virol. 1999, 80, 1211–1220. [Google Scholar] [CrossRef][Green Version]
- Roche, S.; Bressanelli, S.; Rey, F.A.; Gaudin, Y. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science 2006, 313, 187–191. [Google Scholar] [CrossRef] [PubMed]
- Longdon, B.; Day, J.P.; Schulz, N.; Leftwich, P.T.; de Jong, M.A.; Breuker, C.J.; Gibbs, M.; Obbard, D.J.; Wilfert, L.; Smith, S.C.; et al. Vertically transmitted rhabdoviruses are found across three insect families and have dynamic interactions with their hosts. Proc. Biol. Sci. 2017, 284, 20162381. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schilling, M.; Golding, M.; Jones, B.P.; Mansfield, K.L.; Gandy, S.; Medlock, J.; Johnson, N. Sequences Related to Chimay Rhabdovirus Are Widely Distributed in Ixodes ricinus Ticks across England and Wales. Viruses 2024, 16, 504. https://doi.org/10.3390/v16040504
Schilling M, Golding M, Jones BP, Mansfield KL, Gandy S, Medlock J, Johnson N. Sequences Related to Chimay Rhabdovirus Are Widely Distributed in Ixodes ricinus Ticks across England and Wales. Viruses. 2024; 16(4):504. https://doi.org/10.3390/v16040504
Chicago/Turabian StyleSchilling, Mirjam, Megan Golding, Ben P. Jones, Karen L. Mansfield, Sara Gandy, Jolyon Medlock, and Nicholas Johnson. 2024. "Sequences Related to Chimay Rhabdovirus Are Widely Distributed in Ixodes ricinus Ticks across England and Wales" Viruses 16, no. 4: 504. https://doi.org/10.3390/v16040504
APA StyleSchilling, M., Golding, M., Jones, B. P., Mansfield, K. L., Gandy, S., Medlock, J., & Johnson, N. (2024). Sequences Related to Chimay Rhabdovirus Are Widely Distributed in Ixodes ricinus Ticks across England and Wales. Viruses, 16(4), 504. https://doi.org/10.3390/v16040504





