Abstract
Glycosylation, a dynamic modification prevalent in viruses and higher eukaryotes, is principally regulated by uridine diphosphate (UDP)-glycosyltransferases (UGTs) in plants. Although UGTs are involved in plant defense responses, their responses to most pathogens, especially plant viruses, remain unclear. Here, we aimed to identify UGTs in the whole genome of Nicotiana benthamiana (N. benthamiana) and to analyze their function in Chinese wheat mosaic virus (CWMV) infection. A total of 147 NbUGTs were identified in N. benthamiana. To conduct a phylogenetic analysis, the UGT protein sequences of N. benthamiana and Arabidopsis thaliana were aligned. The gene structure and conserved motifs of the UGTs were also analyzed. Additionally, the physicochemical properties and predictable subcellular localization were examined in detail. Analysis of cis-acting elements in the putative promoter revealed that NbUGTs were involved in temperature, defense, and hormone responses. The expression levels of 20 NbUGTs containing defense-related cis-acting elements were assessed in CWMV-infected N. benthamiana, revealing a significant upregulation of 8 NbUGTs. Subcellular localization analysis of three NbUGTs (NbUGT12, NbUGT16 and NbUGT17) revealed their predominant localization in the cytoplasm of N. benthamiana leaves, and NbUGT12 was also distributed in the chloroplasts. CWMV infection did not alter the subcellular localization of NbUGT12, NbUGT16, and NbUGT17. Transient overexpression of NbUGT12, NbUGT16, and NbUGT17 enhanced CWMV infection, whereas the knockdown of NbUGT12, NbUGT16 and NbUGT17 inhibited CWMV infection in N. benthamiana. These NbUGTs could serve as potential susceptibility genes to facilitate CWMV infection. Overall, the findings throw light on the evolution and function of NbUGTs.
1. Introduction
Plants produce several secondary metabolites that are critical for their interactions with the environment, reproductive strategies, and defense responses [1]. Glycosylation, hydroxylation, acylation, and methylation play important roles in the diversity and complexity of plant secondary metabolites [2]. Glycosylation, which is essential for maintaining cellular homeostasis by regulating the levels, activities, and locations of key cellular metabolites [3], is widespread from viruses to higher eukaryotes [4,5]. During glycosylation, the glycosyl group from the donor molecule is transferred to the recipient molecule by glycosyltransferases (GTs) to form a glucoside bond, which further becomes converted into more stable and inactive storage forms [6,7]. Glycosylation produces oligosaccharides, polysaccharides, glycoproteins and glycolipids, and other glycoside compounds [8,9]. The process involves various types of glycosyl donors, including nucleotide-activated sugars such as UDP-glucose, UDP-galactose, UDP-rhamnose, UDP-xylose, and UDP-glucuronic acid. Glycosyl receptors can be carbohydrates or non-saccharide compounds, such as proteins, antibiotics, and phytohormones [10,11]. Glycosylation can affect the homeostasis of these compounds by modifying their chemical activity, degradation, or localization [10,12]. According to the recently updated CAZy (CAZy, http://www.cazy.org, accessed on 3 December 2023), GTs from different species have been classified into 117 enzyme families based on their amino acid sequence similarity, catalytic mechanism, substrate specificity, and the presence of conserved sequence motifs. Among them, UGTs belong to the largest GT family associated with secondary metabolites, such as phytohormones, terpenoids, and sterols [9,13,14]. UGTs are abundant in the plant kingdom, they are a highly differentiated and multi-lineated multi-gene family that is widely involved in the glycosylation of plant secondary metabolites [1]. UGTs are responsible for the glycosylation of small-molecule compounds and participate in multiple plant growth and development processes as well as plant defense responses [15].
With the development of sequencing technologies, an increasing number of UGT genes have been identified at the whole genome in various plant species, including Arabidopsis thaliana, Brachypodium distachyon, Brassica rapa, Brassica oleracea, rice, wheat, maize, soybean, cotton, alfalfa, and peach. The C-terminus of UGTs contains a highly conserved plant secondary product, the glycosyltransferase box (PSPG), composed of 44 amino acids [16]. The entire fold and core regions of plant UGTs are suggested to be conserved, because the C-terminal domain of UGTs mainly recognizes the same or similar glycosyl donors, while the N-terminal domain mainly recognizes specific receptor substrates [17,18]. Hence, the PSPG motif plays a critical role in regulating the glycosylation of specific phytohormones, defense compounds, and other secondary metabolites in plants [19,20]. Phytohormones are the key endogenous factors mediating the plant stress response, which is the integration center of plants to cope with environmental stimuli and plays an important role in plant defense responses [15]. UGTs indirectly regulate biotic stress responses by glycosylating phytohormones, including jasmonic acid (JA) and salicylic acid (SA), or phytohormone-related compounds. For example, the overexpression of UGT74F2 in Arabidopsis thaliana reduced free SA levels and resistance to Pseudomonas syringae (P. syringae) infection; conversely, the knockout of UGT74F2 had the opposite effect [21,22]. The knockout of UGT74J1 in Oryza sativa resulted in increased SA levels and promoted resistance to Magnaporthe oryzae [23]. The disruption of the expression of methyl salicylate (MeSA)-specific UGT71C3 in Arabidopsis thaliana enhanced systemic acquired resistance to P. syringae by increasing MeSA and free SA levels [24]. UGTs with secondary metabolite activities are considered important for plant resistance to pathogens [15]. For instance, Arabidopsis thaliana UGT71C1 can glycosylate the secondary metabolite flavonoids, thus affecting the level of reactive oxygen species (ROS) and reducing the damage to plants caused by excessive ROS during pathogen invasion [25]. Silencing of Twi1, a homolog of TOGT in Nicotiana tabacum (N. tabacum), resulted in an increase in total scopoletin and decreased resistance to tomato spotted wilt virus in Solanum lycopersicum [26].
CWMV was discovered and identified at the end of the last century on winter wheat in Shandong Province, China [27,28,29]. It naturally infects wheat plants, leading to typical mosaic symptoms, and is one of the most common pathogens causing wheat yellow mosaic disease in China. CWMV is a member of the genus Furovirus and is spread by the plasmodiophorid Polymyxa graminis (P. graminis) [27,30]. According to the life history of P. graminis [31], its resting sporangium can survive in the residue or soil after wheat harvest. Under suitable environmental conditions, these spores can germinate and produce zoospores that colonize wheat roots. The CWMV particles carried by P. graminis can infect the roots during this stage, leading to proliferation and systemic infection. CWMV is comprised of two positive-sense single-stranded RNAs, RNA1 and RNA2. RNA1 possesses 7147 nucleotides and includes three major predicted open reading frames that encode the replication-associated protein, RNA-dependent RNA polymerase (RdRp), and a movement protein (MP), which are required for viral replication and movement. RNA2 contains 3563–3569 nucleotides according to different isolates and encodes four proteins, coat protein (CP), CP–read-through (CP-RT), N-terminal extension CP (N-CP), and cysteine-rich protein (CRP) [30]. CWMV and wheat yellow mosaic virus (WYMV) often co-infect wheat, posing a serious threat to grain production safety [29]. In laboratory experiments, it has been demonstrated that CWMV can infect N. benthamiana through mechanical inoculation and cause symptoms similar to those of CWMV-infected wheat, making it a common model system for investigating the interaction between CWMV and plants [32].
Some UGTs are involved in the regulation of host resistance to viruses in plants, such as the overexpression of Togt1 and Togt2 in N. tabacum, which increased resistance to tobacco mosaic virus and potato virus Y [33,34]. However, only a few UGT genes have been reported to regulate viral disease resistance in wheat. In this study, UGTs from N. benthamiana were identified at the whole-genome level. Subsequently, phylogenetic and gene structure analyses were conducted, followed by the prediction of subcellular localization and the analysis of cis-acting elements in the gene promoter. The expression levels of some UGTs were measured in the CWMV infection time cross, and UGTs responding to CWMV infection in N. benthamiana were screened. Finally, the regulatory effects of three UGTs on CWMV were investigated. This study provides a reference for the role of UGTs in regulating viral disease resistance in plants.
2. Materials and Method
2.1. Identification and Bioinformatics Analysis of NbUGTs
Protein sequences of 19 Arabidopsis thaliana UGT family members were downloaded from The Arabidopsis Information Resource (https://www.arabidopsis.org/, accessed on 10 September 2022) and used as templates to perform a BLASTP search for all UGTs in N. benthamiana. The possible genome of N. benthamiana was identified using the hidden Markov model (HMM) profile of the UDPGT superfamily domain (PF00201), which was downloaded from the Sol Genomics Network (https://solgenomics.net/, accessed on 12 September 2022). The PSPG domain was used as a further screening criterion, and after ruling out 65 genes, 147 NbUGTs were identified. Then, phylogenetic analysis was performed based on the full-length protein sequences of 19 Arabidopsis thaliana UGT family members and 147 NbUGTs using MEGA11.0 software [8,12] with the neighbor-joining (NJ) method, and a bootstrap test was carried out with 1000 iterations [35]. In addition, the coding sequence (CDS) lengths, isoelectric points (pIs), and molecular weights (MWs) of the NbUGTs were predicted using ExPASy [36], and the subcellular localizations of the NbUGTs were predicted using the tool in the website (http://cello.life.nctu.edu.tw/cello.html, accessed on 16 September 2022).
2.2. Analysis of Conserved Motifs, Gene Structure, and Conserved Domains of NbUGTs
The conserved motifs of the NbUGTs were analyzed using the MEME program, with the maximum number of motifs set to 10 [37] (Table S1). The gene structures of the NbUGTs were analyzed and visualized using TBtools-II v1.108 with the N. benthamiana genome annotation file. The conserved domains of the genes were analyzed using the TBtools-II v1.108.
2.3. Prediction of Cis-Acting Elements in the Putative Promoter Regions
To investigate cis-acting elements in the putative promoter regions of the obtained NbUGTs, 2 kb genomic DNA sequences upstream of the initiation codon (ATG) of all NbUGTs were downloaded from the Sol Genomics Network (https://solgenomics.net/, accessed on 18 September 2022). Cis-regulatory elements in the putative promoter sequences were analyzed using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 18 September 2022) [38] and visualized by TBtools-II v1.108.
2.4. Plant Culture and Virus Inoculation
N. benthamiana needed to be cultured in a growth chamber at 15 ± 2 °C under a 16 h/8 h light/dark photoperiod when inoculated with CWMV, while the other plants were grown in soil inside a growth chamber maintained at 25 ± 2 °C with a 16 h/8 h light/dark cycle and 65 ± 5% relative humidity. To inoculate CWMV, briefly, plasmids pCB-35S-R1 and pCB-35S-R2, respectively, containing CWMV RNA1 and RNA2 full-length sequences were individually transformed into the Agrobacterium tumefaciens strain GV3101. The Agrobacterium cultures were grown individually overnight at 28 °C. The resulting Agrobacterium cultures were pelleted and resuspended in an infiltration buffer (10 mM MES, pH of 5.7, 10 mM MgCl2, 0.2 mM acetosyringone) at room temperature (OD600 = 0.6–0.8). Agrobacterium harboring pCB-35S-R1 was mixed with Agrobacterium harboring pCB-35S-R2 in a 1:1 ratio. Mixed cultures were individually infiltrated into N. benthamiana leaves. A TRV-based VIGS system in N. benthamiana was used in this study, while the plasmids pTRV1 and pTRV2 or pTRV2-NbUGT12, NbUGT16, and NbUGT17 were individually transformed into the Agrobacterium tumefaciens strain GV3101. The Agrobacterium cultures were grown individually overnight at 28 °C. Then, the cultures were pelleted and resuspended in the infiltration buffer at room temperature (OD600 = 0.3–0.6). Agrobacterium harboring pTRV1 was mixed with Agrobacterium harboring pTRV2 or pTRV2-NbUGT12, pTRV2-NbUGT16, and pTRV2-NbUGT17 in a 1:1 ratio. Mixed cultures were infiltrated into N. benthamiana leaves. Moreover, H2B-RFP transgenic N. benthamiana plants were also used in this study. These plants can express a nuclear marker H2B-RFP, and are always used for subcellular localization research [39,40].
2.5. Plasmid Construction
Firstly, NbUGT12, NbUGT16, and NbUGT17 were cloned from the cDNA of N. benthamiana. The cloned fragments were recombined into the Ti vector plasmid according to the instructions of the pEASY®-Blunt Zero Cloning Kit (TransGen Biotech, Beijing, China). Then, the Ti-NbUGT12, Ti-NbUGT16, and Ti-NbUGT17 plasmids were used as templates to amplify the full-length and silent fragments of the genes, respectively. Next, the NbUGT12, NbUGT16, and NbUGT17 CDS full-length fragments were sequentially inserted into the intermediate vector Donor 207 and the GFP-tagged PGWB505 vector according to the Gateway series construction method. Finally, the recombinant NbUGT12-GFP, NbUGT16-GFP, and NbUGT17-GFP plasmids were transferred into the Agrobacterium by electroporation, and the corresponding Agrobacterium with NbUGT12-GFP, NbUGT16-GFP, and NbUGT17-GFP plasmids were obtained. To silence NbUGT12, NbUGT16, and NbUGT17 in N. benthamiana, approximately 300 bp fragments of NbUGT12, NbUGT16, and NbUGT17 were inserted into the pTRV2 vector, respectively. The silenced fragments were designed on the Sol Genomics Network (https://solgenomics.net/, accessed on 15 October 2022) and their sequences are listed in Table S2. Two endonuclease BamHI and SmaI sequences were added when designing the primers for amplifying the fragments. Then, the amplified fragments and the pTRV2 vector plasmid were all digested with the two enzymes and recovered. Then, the pTRV2-NbUGT12, pTRV2-NbUGT16, and pTRV2-NbUGT17 fragments were ligated to the digested pTRV2 plasmid using T4 DNA Ligase (Thermo Scientific, Waltham, MA, USA), respectively. Finally, the recombinant plasmids were transferred into Agrobacterium.
2.6. RNA Extraction and Quantitative Reverse-Transcription PCR (qRT-PCR) Assay
The total RNA of the N. benthamiana tissue samples was extracted using the HiPure Plant RNA Mini Kit (Magen, Guangzhou, China). First-strand cDNA was synthesized using the First-Strand cDNA Synthesis Kit (Vazyme, Nanjing, China). The qRT-PCR was carried out using Hieff® qPCR SYBR Green Master Mix (Yeasen, Shanghai, China) on an Applied Biosystems Quantstudio 6 Flex system (Applied Biosystems, Foster City, CA, USA), and the relative expression levels of the assayed genes were calculated using the 2−ΔΔCt method. Each treatment had three biological replicates and three technical replicates. The primer sequences used in this study are listed in Table S3.
2.7. Western Blot (WB) Assay
The total protein was extracted from N. benthamiana leaves and homogenized in a lysis buffer containing 2% β-mercaptoethanol, 6% sodium dodecyl sulfate (SDS), and 100 mM Tris-HCl (pH of 8.8). The protein samples were individually mixed with SDS loading buffer and boiled for approximately 8 min. Protein samples were separated by SDS–polyacrylamide gel electrophoresis (PAGE) and transferred to nitrocellulose membranes. The blots were incubated in a blocking buffer (5% skim milk in 1×PBS) for 1 h, followed by detection using specific anti-GFP (TransGen Biotech, Beijing, China) or anti-CWMV CP primary antibody and then an HRP-conjugated anti-mouse or anti-rabbit secondary antibody (Abbkine Scientific, California, USA). The detection signal was visualized using an Amersham Imager 680 machine (GE Healthcare BioSciences, Pittsburgh, PA, USA). In addition, the specific primary antibody for detecting CWMV CP was prepared and preserved in our laboratory.
2.8. Confocal Fluorescence Microscope Observation
The confocal fluorescence microscope (TCS SP8 X) was used to observe the expression of fluorescently labeled genes, and the LAS X application was used for photographing and analysis. Briefly, the inoculation leaves of N. benthamiana were sampled with a puncher; then, the samples were placed on a glass slide and covered with a coverslip before being observed under the microscope. LAS X is equipped with channels to observe different fluorescence, such as GFP, RFP, and YFP, and it also has a special chloroplast channel that can be used to observe whether the gene can express in the chloroplasts [41]. When detecting the fluorescence signal, the excitation and emission wavelengths of the GFP fluorescence were 488 nm and 500–555 nm, respectively, the RFP fluorescence excitation and emission wavelengths were 552 nm and 565–620 nm, respectively, while those of the chloroplast fluorescence excitation and emission wavelengths were 552 nm and 670–730 nm, respectively.
2.9. Chloroplast Extraction
The chloroplasts were extracted using the MinuteTM Chloroplast Isolation Kit (Invent Biotechnologies, Beijing, China) according to the manufacturer’s instructions. Subsequently, the resulting samples were analyzed using WB analysis [41].
3. Results
3.1. Identification and Phylogenetic Analysis of NbUGTs
The genome sequences of N. benthamiana were downloaded from the Sol Genomics Network (https://solgenomics.net/, accessed on 12 September 2022). Initially, 212 possible NbUGTs were identified using the HMM profile of the UDPGT superfamily domain (PF00201) in N. benthamiana. After removing 19 sequences that lacked the PSPG domain, 193 NbUGTs were obtained. Furthermore, truncated UGTs and genes with less than 50% identity with the 44 amino acids of the PSPG motif were removed. In total, 147 UGT genes were identified in N. benthamiana.
To determine the evolutionary relationships between the UGT family genes in N. benthamiana and Arabidopsis thaliana, a phylogenetic tree was constructed based on the amino acid sequence similarity of 19 UGTs in Arabidopsis thaliana that were distributed in 14 different groups from A–N and 147 NbUGTs using MEGA11.0 [8,12]. The results showed that the 147 NbUGTs were classified into 16 major phylogenetic groups, while all 14 conserved evolutionary groups (A–N) originally described in Arabidopsis thaliana were included in N. benthamiana (Figure 1). The number of NbUGTs in each group varied, with the largest one, Group A, containing 25 NbUGTs, and the smallest one, Group M, having only 2 members (Figure 1). In addition, two new evolutionary groups not found in Arabidopsis thaliana were identified in N. benthamiana: Group O with 24 members and Group P with 12 members (Figure 1).
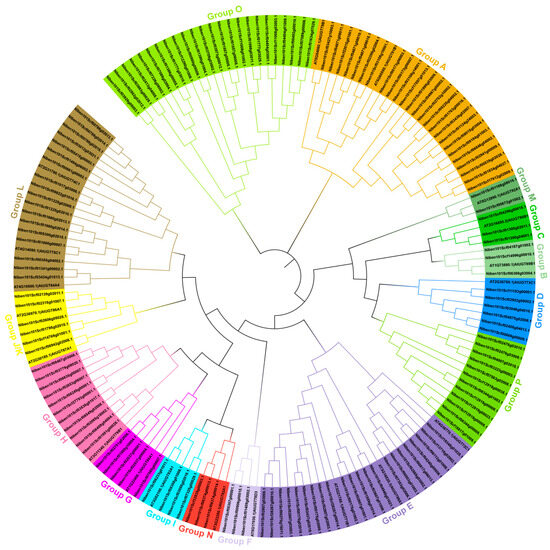
Figure 1.
Phylogenetic analysis of NbUGT gene family. The MUSCLE and MEGA11.0 software were used for the sequence alignment and construction of the phylogenetic tree using the full-length sequences of 147 NbUGTs and 19 Arabidopsis thaliana UGT genes.
3.2. Prediction of the Subcellular Localization and Physicochemical Properties of NbUGTs
The NbUGTs’ physicochemical properties, including the MW, protein length (PL), pI, CDS length, and intron number (IN), were determined using ExPASy (Table 1). The MW of these NbUGTs varied from 30.55219 kDa (Niben101Scf04967g00002.1) to 91.45533 kDa (Niben101Scf03046g04015.1), with a range of 270 to 801 amino acids (aa), respectively. The CDS lengths of the NbUGTs ranged from 813 bp (Niben101Scf04967g00002.1) to 2406 bp (Niben101Scf03046g04015.1). A significant number (132) of the NbUGTs were acidic proteins (pI < 7). Out of these NbUGTs, 81.63% (120) either lacked or contained only one intron. The subcellular localizations predicted on the website (http://cello.life.nctu.edu.tw/cello.html, accessed on 16 September 2022) are shown in Table 1. The results showed that the NbUGTs were most likely localized in the cytoplasm, followed by the plasma membrane, chloroplasts, nucleus, and outer membrane (Table 1).

Table 1.
List of physicochemical properties and subcellular localization of NbUGTs.
3.3. Gene Structure and Conserved Motifs Analysis of NbUGTs
Ten conserved motifs were identified among the 147 NbUGTs using online MEME analysis. The majority of members contained five to nine motifs and always started with motif three and ended with motif two. Only a few genes were different, for example, Niben101Scf03929g05003.1, which contained only four motifs (Figure 2a,b). Additionally, most of the 147 members did not have any introns or contained only one intron (Figure 2c).
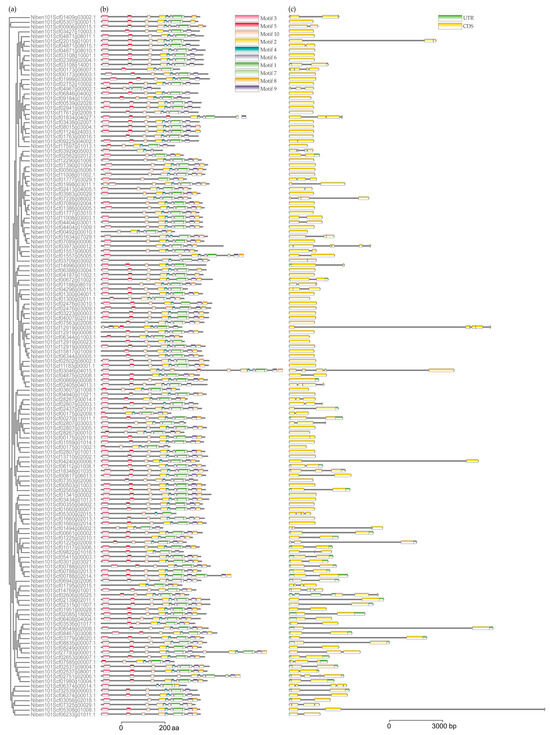
Figure 2.
Analysis of conserved motifs and gene structure of NbUGTs. (a) Phylogenetic tree constructed using the NbUGT protein sequences. (b) Ten types of conserved motifs were predicted in the NbUGT protein sequences. Different motifs are shown in different color boxes. The sequence information for each motif is provided in Supplementary Table S1. (c) The gene structure of NbUGTs (untranslated regions, exons, and introns are shown as light green boxes, yellow boxes, and horizontal lines, respectively).
3.4. Identification of Conserved Domains and Cis-Acting Elements in the Putative Promoter of NbUGTs
Among the 147 NbUGTs, 90.48% (133) contained the Glycosyltransferase_GTB-type superfamily domain, 9.52% (14) had a GT1_Gtf-like domain, and only 1 NbUGT (Niben101Scf06942g02006.1) contained the PLN02448 domain. This indicated a high level of conservation among the NbUGTs (Figure 3a,b). To investigate the possible regulatory mechanisms of the NbUGTs, the PlantCARE web server was used to search for possible cis-acting elements with the 2000 bp putative promoter regions of the NbUGTs. A total of 17 types of cis-acting elements were identified. These elements were associated with environmental stress, hormonal responses, development, and defense responses (Figure 3c).
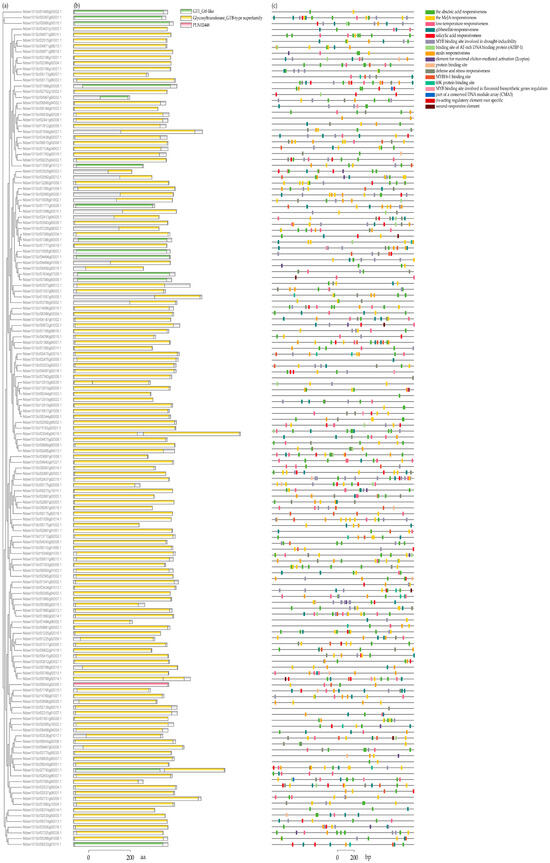
Figure 3.
Predicted conserved domains and cis-acting elements of NbUGTs. (a) Phylogenetic tree constructed using the NbUGT protein sequences. (b) The conserved domains of the NbUGTs were analyzed through TBtools-II v1.108 software. (c) The type, quantity, and position of predicted cis-acting elements in the putative promoter regions of NbUGTs. Different elements are shown in different color boxes.
3.5. Expression Patterns of NbUGTs during CWMV Infection
To further clarify the functions of the NbUGTs in response to CWMV infection, the expression patterns of the partial NbUGTs were analyzed following CWMV infection in N. benthamiana. The plants were inoculated with CWMV and cultured in an artificial climate incubator at 15 °C. Then, the total RNA was extracted from their systemic leaves on 0, 7, 14, 21, and 28 days post-infection (dpi), respectively, followed by qRT-PCR analysis of 20 NbUGTs containing defense-related cis-acting elements (the corresponding gene names are listed in Table S4). At 21 dpi, there was a significant increase in CWMV CP accumulation. After an additional 7 days, the accumulation level of CWMV CP increased by nearly 70 times compared to that on day 7 (Figure 4a). Through the qRT-PCR assay, 8 NbUGTs were identified as potential responders to CWMV infection (Figure 4), whereas the expression levels of the remaining 12 NbUGT genes did not show significant changes during CWMV infection (Figure S1). The expression patterns of NbUGT1, NbUGT12, NbUGT16, and NbUGT24 were similar, displaying a minor, non-significant difference at 0–21 dpi but showing a significant increase at 28 dpi (Figure 4b–d,h). However, the expression profiles of NbUGT17, NbUGT21, and NbUGT25 showed an initial increase, followed by a decrease, and then an increase again (Figure 4e,g,i). NbUGT19 exhibited a significant increase in expression over time, reaching a level 26 times higher than that on day 0 at 28 dpi (Figure 4f). In summary, eight NbUGTs were significantly upregulated during CWMV infection, suggesting their potential involvement in the interaction between CWMV infection and the defense response of N. benthamiana. Subsequently, according to the expression patterns, the eight significantly upregulated genes were divided into the following three groups: the high upregulation, medium upregulation, and low upregulation categories containing three, three, and two genes, respectively. We then randomly selected one in each of the groups for functional studies. Consequently, NbUGT12, NbUGT16, and NbUGT17 were picked for further investigation. In addition, it is worth noting that NbUGT12, NbUGT16, and NbUGT17 all contain one intron (Figure S2), which is consistent with the predicted results (Table 1).
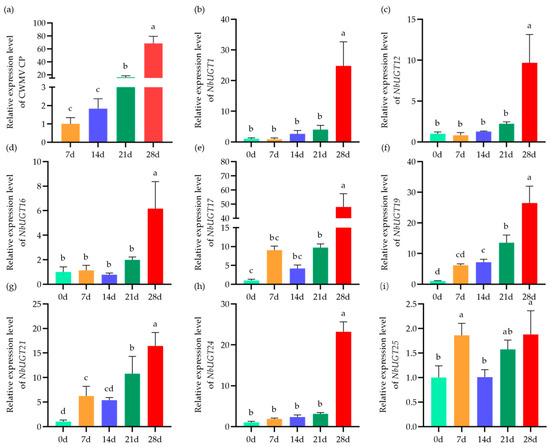
Figure 4.
The expression patterns of 8 NbUGTs in CWMV-infected N. benthamiana plants. (a) The accumulation of CWMV CP in the assayed systemic leaves of N. benthamiana infected with CWMV at 7 to 28 days. (b–i) Relative expression levels of NbUGT1, NbUGT12, NbUGT16, NbUGT17, NbUGT19, NbUGT21, NbUGT24, and NbUGT25 in CWMV-infected N. benthamiana plants at 0 to 28 dpi. Each treatment had three biological replicates; the data presented are the means ± SD, determined using Student’s t-test. Different letters show statistically significant differences (p < 0.05, Tukey’s test).
3.6. Subcellular Localization of NbUGT12, NbUGT16, and NbUGT17
The protein functions are closely related to their subcellular localization. To analyze the characteristics and effects of the identified NbUGTs, their subcellular localizations were investigated using a website (http://cello.life.nctu.edu.tw/cello.html, accessed on 16 September 2022). Based on the predictions, NbUGT12 and NbUGT16 are most likely localized in the cytoplasm, while NbUGT17 has the potential localization in the cell membrane and cytoplasm (Table 1). Here, NbUGT12-GFP, NbUGT16-GFP, and NbUGT17-GFP were expressed in the transgenic N. benthamiana leaves (expressing H2B-RFP, a nuclear marker) via agroinfiltration. Non-fused GFP was used as a control. The inoculated leaves were collected at 72 h post-infiltration (hpi) to observe subcellular localization under a confocal microscope. The results revealed that NbUGT12-GFP was located in the cytoplasm and chloroplasts, NbUGT16-GFP was mainly located in the cytoplasm, whereas NbUGT17-GFP was present in the cytoplasm and nucleus (Figure 5a). To analyze the relationship between NbUGTs and CWMV, we tested whether CWMV affected the subcellular localization of the three proteins. Consequently, NbUGT12-GFP, NbUGT16-GFP, and NbUGT17-GFP were co-injected with CWMV in H2B-RFP transgenic N. benthamiana leaves using agroinfiltration. In brief, Agrobacterium individually harboring pCB-35S-R1 and pCB-35S-R2 were mixed with Agrobacterium harboring NbUGT12-GFP, NbUGT16-GFP, and NbUGT17-GFP in a 1:1:1 ratio, respectively. Mixed cultures were individually infiltrated into the plants leaves. At 7 dpi, the inoculated leaves were observed by confocal microscopy. The results showed no significant changes (Figure 5b). In addition, to determine whether these three proteins can express in chloroplasts, the chloroplast protein was extracted and analyzed using the WB assay. The results showed that NbUGT12 was indeed localized in the chloroplasts, while the control proteins, NbUGT16 and NbUGT17, did not express in the chloroplasts, regardless of CWMV infection (Figure 5c,d). In general, NbUGT12, NbUGT16, and NbUGT17 were primarily expressed in the cytoplasm, although NbUGT12 was also distributed in the chloroplasts and did not exhibit changes during CWMV infection.
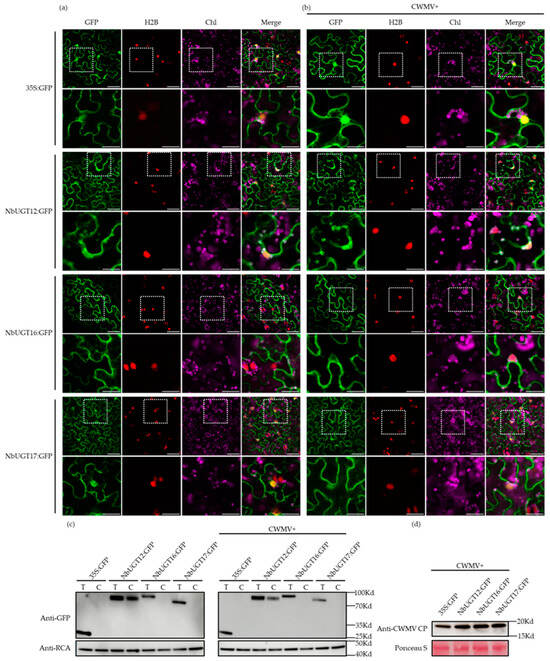
Figure 5.
Analyzing the subcellular localization of NbUGT12, NbUGT16, and NbUGT17. (a) Subcellular localization of NbUGT12-GFP, NbUGT16-GFP, and NbUGT17-GFP in H2B-RFP transgenic N. benthamiana epidermal cells. Confocal images were taken at 72 hpi. Scale bar = 50 μm. The corresponding region in the white box was magnified below it. Scale bar = 25 μm, Chl (chloroplast). (b) Subcellular localization of NbUGT12-GFP, NbUGT16-GFP, and NbUGT17-GFP at 15 °C in H2B-RFP transgenic N. benthamiana leaves infected with CWMV at 7 dpi. Scale bar = 50 μm. The corresponding region in the white box was magnified below it. Scale bar = 25 μm. (c) The chloroplast proteins of N. benthamiana-inoculated leaves were analyzed by the WB assay and RCA and were used as a chloroplast maker. T (total protein); C (chloroplast). (d) The accumulation of CWMV CP in the assayed N. benthamiana-inoculated leaves was analyzed by the WB assay using CWMV CP-specific antibody. Ponceau S was used to visualize the sample loadings.
3.7. NbUGT12, NbUGT16, and NbUGT17 Positively Regulate CWMV Infection in N. benthamiana
To investigate the role of NbUGT12, NbUGT16, and NbUGT17 in CWMV infection, NbUGT12-GFP, NbUGT16-GFP, and NbUGT17-GFP were co-injected with CWMV into N. benthamiana, respectively. Plants inoculated with GFP and CWMV were used as controls. After confirming the expression of these proteins using a confocal fluorescence microscope at 7 dpi, the total protein of the inoculated leaves was extracted. Then, the accumulation levels of NbUGT12-GFP, NbUGT16-GFP, and NbUGT17-GFP, and GFP and CWMV CP were detected by the WB assay. The results indicated that the accumulation levels of NbUGT12-GFP, NbUGT16-GFP, and NbUGT17-GFP were comparable to their respective controls, while the accumulation level of CWMV CP significantly increased in NbUGT12-GFP+CWMV-, NbUGT16-GFP+CWMV-, and NbUGT17-GFP+CWMV-inoculated plants compared to that in the control plants (Figure 6a).
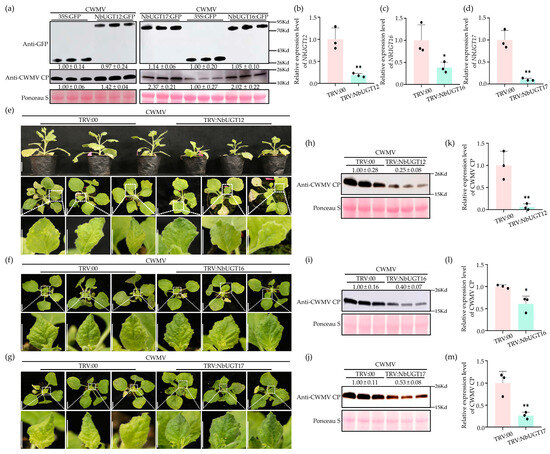
Figure 6.
NbUGT12, NbUGT16, and NbUGT17 positively regulate CWMV infection in N. benthamiana. (a) NbUGT12-GFP, NbUGT16-GFP, and NbUGT17-GFP were co-injected with CWMV in N. benthamiana leaves, respectively. Plants inoculated with GFP + CWMV were used as controls. The protein expression levels of NbUGT12-GFP, NbUGT16-GFP, and NbUGT17-GFP were detected by anti-GFP immunoblots. The accumulation levels of CWMV CP in the assayed plants were determined by WB analysis using a CWMV CP-specific antibody. Gray analysis was performed on the WB bands using ImageJ 1.8.0, each treatment had three biological replicates, and the data presented are the means ± SD. Ponceau S was used to visualize sample loadings. (b–d) qRT-PCR assay was used to determine the relative expression levels of NbUGT12, NbUGT16, and NbUGT17 in TRV: NbUGT12-, TRV: NbUGT16-, TRV: NbUGT17-, and TRV: 00-inoculated N. benthamiana plants. (e–g) Phenotypes of N. benthamiana plants inoculated with TRV: NbUGT12+CWMV, TRV: NbUGT16+CWMV, and TRV: NbUGT17+CWMV at 30 dpi. Plants inoculated with TRV: 00+CWMV were used as controls. Scale bar = 2 cm. (h–j) The accumulation of CWMV CP in TRV: NbUGT12+CWMV-, TRV: NbUGT16+CWMV-, and TRV: NbUGT17+CWMV-inoculated assayed plants were analyzed by WB. Plants inoculated with TRV: 00+CWMV acted as controls. Gray analysis was performed on the WB bands using ImageJ 1.8.0, each treatment had three biological replicates, and the data presented are the means ± SD. Ponceau S was used to show the protein loadings. (k–m) Transcriptional levels of CWMV in TRV: NbUGT12+CWMV-, TRV: NbUGT16+CWMV-, and TRV: NbUGT17+CWMV-inoculated assayed plants were detected by qRT-PCR assay. Plants inoculated with TRV: 00+CWMV acted as controls. Each treatment had three biological replicates, and the data presented are the means ± SD, determined using Student’s t-test. * p < 0.05, ** p < 0.01.
NbUGT12, NbUGT16, and NbUGT17 were also silenced in N. benthamiana by the TRV-mediated VIGS assay. Firstly, the infectious clones of TRV-NbUGT12, TRV-NbUGT16, and TRV-NbUGT17 were inoculated in N. benthamiana, respectively, with TRV: 00 serving as the control. The total RNA was extracted from the systemic leaves at 7 dpi, and qRT-PCR analysis was performed. The results showed that the expression levels of NbUGT12, NbUGT16, and NbUGT17 were reduced by approximately 80%, 60%, and 90% in TRV: NbUGT12-, TRV: NbUGT16-, and TRV: NbUGT17-infected plants, respectively, compared to those in TRV: 00-infected plants (Figure 6b–d). Subsequently, the plants successfully silenced NbUGT12, NbUGT16, and NbUGT17 were inoculated with CWMV, respectively. After another 30 dpi, the TRV: NbUGT12+CWMV-, TRV: NbUGT16+CWMV-, and TRV: NbUGT17+CWMV-inoculated plants exhibited milder CWMV disease symptoms than their controls (Figure 6e–g). In addition, TRV: NbUGT12+CWMV-inoculated plants were shorter than the controls, indicating that NbUGT12 may also play a significant role in plant growth (Figure 6e). The total RNA and protein were extracted from the systemic leaves. Then, the accumulation levels of CWMV CP in these plants were analyzed by WB and qRT-PCR assays. The WB assay revealed a lower accumulation of CWMV CP in TRV: NbUGT12+CWMV-, TRV: NbUGT16+CWMV-, and TRV: NbUGT17+CWMV-inoculated plants than in TRV: 00+CWMV-inoculated plants (Figure 6h–j). Moreover, qRT-PCR analysis revealed that the transcriptional levels of CWMV CP were downregulated by approximately 95%, 40%, and 75% in the TRV: NbUGT12+CWMV-, TRV: NbUGT16+CWMV-, and TRV: NbUGT17+CWMV-inoculated plants, respectively, compared those in the control plants (Figure 6k–m). In summary, these findings suggested that NbUGT12, NbUGT16, and NbUGT17 positively regulate CWMV infection in N. benthamiana.
4. Discussion
Glycosylation, a type of post-translational modification (PTM) of protein, is catalyzed by GTs and can affect several cellular processes and metabolic pathways in plants, including host–pathogen interactions [42,43,44]. GTs are a class of highly differentiated multi-membered metabolic enzymes belonging to the multi-gene transferase family. Based on the similarity of amino acid sequences and substrate specificity, they are divided into 117 families (CAZy, http://www.cazy.org, accessed on 3 December 2023), among which the GT1 family has the most members and is the most functionally important. The GT1 family mainly catalyzes UDP-glucides to specific receptors, so it is also commonly referred to as the UGT family [45]. The crystal structures of plant UGTs exhibit GT-B folding, which is a common structural folding of nucleotide sugar-dependent enzymes [6,46]. The GT-B folding consists of two flexibly connected β/α/β Roseman domains. The gap between these two domains is critical for the catalytic activity [6,47]. The two domains are referred to as the C-terminal and N-terminal domains. Its C-terminal domain recognizes and binds UDP-glucides through a highly conserved 44 amino acid motif known as the PSPG motif, which is the basis of this study; the N-terminal domain binds aglycones loosely, resulting in substrate structural diversity [48,49,50].
In addition, the implementation of the Human Genome Project (HGP) and the rapid development of high-throughput sequencing technology have provided a vast amount of data for bioinformatics development. UGT genes have been identified and analyzed in many plants, such as maize, cotton, and tomato [51,52,53]. N. benthamiana is a heterotetraploid species with 19 chromosomes. It is known for small plants, luxuriant leaves, and is easy to cultivate. Moreover, N. benthamiana is susceptible to many pathogenic microorganisms, particularly viruses, and is subjected to gene expression regulation and PTMs in vivo. Therefore, it is commonly used as a model plant in biology. This study presented a genome-wide analysis of the UGT family in N. benthamiana and investigated the response of some NbUGT genes to CWMV. Based on the PSPG motif, 147 UGT genes have been identified in N. benthamiana. These genes were divided into 16 groups using cluster analysis. Among them, Group A had the largest number of genes, with 25 genes accounting for 17.00% of the UGT genes in N. benthamiana. In addition, there were two new groups, Group O and Group P, which were specific to Arabidopsis thaliana and included 24 and 12 genes, respectively (Figure 1). These two groups have been found in wheat, as well [54]. In N. benthamiana, 81.63% of the NbUGTs did not contain any introns or contained only one intron, which is consistent with the number of UGT gene introns in tomato, rice, and cotton [51,53,55]. In this study, the website (http://cello.life.nctu.edu.tw/cello.html, accessed on 16 September 2022) was used to predict that these NbUGT genes were primarily located in the cytoplasm, whereas the UGT genes of wheat were mostly located in the cytoplasm, cell membrane, and chloroplast [54], and the UGT genes of cotton were mostly located in the cytosol and chloroplast [56]. Out of the 147 UGT genes, 92.52% had CDS lengths between 1000 and 1500 bp, and 89.80% were acidic proteins (pI < 7). Moreover, three genes, NbUGT12, NbUGT16, and NbUGT17, were found to be acidic proteins containing only one intron (Table 1).
Bronze1, the first gene in the plant UGT family, was accidentally discovered in 1977. The protein encoded by Bronze1 is an enzyme with UGT activity that synthesizes flavonoid glycosides and regulates melanin accumulation in maize grains [57]. Since then, numerous studies have been conducted on plant UGT genes, particularly in food crops, medicinal plants, fruits, and flavonoid-rich plants such as Rhodiola sachalinensis, Ginkgo biloba, and strawberry. However, most reports focused on the identification of UGT genes and their role in plant growth and development, fruit quality, or adaptation to abiotic stress [58,59,60,61,62,63,64]. In-depth studies have been conducted on some model crops, such as rice, Arabidopsis thaliana, tomato, and the phenomenon and mechanism of some UGT genes responding to pathogens have been elucidated [55,65,66]. However, the relationship between most UGT genes in plants and pathogens, especially viruses, remains unclear.
The expression levels of eight NbUGTs were upregulated significantly during CWMV infection (Figure 4). These genes may play a role in the interaction between N. benthamiana and CWMV, which could be necessary for CWMV infection or the host response to it. Further analysis revealed that NbUGT12, NbUGT16, and NbUGT17 were susceptible genes that positively regulated CWMV invasion (Figure 6). Interestingly, NbUGT12-silenced N. benthamiana plants showed a significant dwarf phenotype, suggesting the potential role that NbUGT12 may play in CWMV–host interactions, as well as plant growth and development. A similar phenomenon has been observed in Arabidopsis thaliana, where UGT73C7 contributes to plant disease resistance, but the overexpression lines of UGT73C7 show a dwarf phenotype [66].
The subcellular localization of NbUGT12, NbUGT16, and NbUGT17 remained unchanged during CWMV infection (Figure 5), indicating that they should not affect viral infection through changes in localization. Many UGT genes are involved in the phenylpropanoid metabolism in plants, which can regulate the accumulation of metabolites such as hydroxycinnamic acids, coumarins, flavonoids, phenols, and tannins [66,67,68,69,70]. Flavonoid glycosylation is typically mediated by UGTs [1]. It provides the core flavonoid skeleton with complexity, enhances molecular stability and solubility, alters its chemical properties, and affects subcellular transport and biological activity [10,70,71]. The phenylpropanoid metabolism also regulates plant growth and development [72,73]. Therefore, it is possible that these three UGT genes, particularly NbUGT12, are involved in the flavonoid metabolism or other links in the phenylpropanoid metabolic pathway, which could affect plant growth and susceptibility to CWMV infection. Some UGTs are also involved in hormone pathways, such as JA, SA, and abscisic acid (ABA) [23,74,75,76], which are also related to plant resistance. For instance, it was discovered that aspirin (acetyl-SA) was resistant to tobacco mosaic virus in 1979, marking the first report of SA’s involvement in plant immunity [77]. Subsequently, numerous studies have demonstrated that SA is a defense-related hormone [78,79,80]. Additionally, plant immunity is highly dependent on the interaction between SA and other hormones [81,82]. The balance between plant growth and immunity is also influenced by SA [83]. Whether NbUGT12, NbUGT16, and NbUGT17 are associated with these hormone pathways or the phenylpropanoid metabolism, and whether they participate in singular or multiple pathways to synergistically affect the interaction between N. benthamiana and CWMV, are directions for future studies.
5. Conclusions
Accumulating evidence suggests that glycosylation, as a common PTM, is essential for growth, development, and immunity in eukaryotes. However, to date, there have been very few instances concerning the study of the functions and mechanisms of glycosylation in the interaction between pathogens, especially viruses and their hosts in plants. In this study, we identified and analyzed 147 UGT genes in N. benthamiana via bioinformatics methods. Bioinformatics analysis, such as the physicochemical information, gene structure, conserved motifs, and conserved domains, showed that the UGT gene family is highly conserved in N. benthamiana. More importantly, the expression patterns showed that eight NbUGT genes may be involved in CWMV infection. The function of three genes, NbUGT12, NbUGT16, and NbUGT17, which positively regulate CWMV infection has been initially confirmed by transient overexpression and VIGS assays. These findings could facilitate the investigation of the molecular mechanisms of the interplay between CWMV and its natural host wheat, which can contribute to breeding resistant varieties to CWMV.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/v16040489/s1, Table S1: Amino acid sequences of ten conserved motifs; Table S2: Nucleotide sequences of silent fragments for NbUGT12, NbUGT16, and NbUGT17; Table S3: All primer sequences used in this study; Table S4: The corresponding genes’ names in this study; Figure S1: The expression patterns of 12 NbUGTs without significant changes under CWMV infection; Figure S2: Detection of the presence of introns for NbUGT12, NbUGT16, and NbUGT17.
Author Contributions
J.L. and W.L. conceived the project and designed the experiments; X.W., Y.Z. and Y.L. performed the experiments and collected the data; J.L., J.Y., H.H., T.Y. and X.W. participated in the analysis and discussion of the data; J.L., J.Y. and X.W. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by National Key R&D Program of China (2023YFD1400800), the China Agriculture Research System from the Ministry of Agriculture of the China (CARS-03), and the China Postdoctoral Science Foundation (2022M721731).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article and Supplementary Materials.
Conflicts of Interest
The authors state that there are no conflicts of interest.
References
- Caputi, L.; Malnoy, M.; Goremykin, V.; Nikiforova, S.; Martens, S. A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. Plant J. 2012, 69, 1030–1042. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, P.; Sangwan, R.S.; Sangwan, N.S. Plant secondary metabolism linked glycosyltransferases: An update on expanding knowledge and scopes. Biotechnol. Adv. 2016, 34, 714–739. [Google Scholar] [CrossRef] [PubMed]
- Lim, E.K.; Bowles, D.J. A class of plant glycosyltransferases involved in cellular homeostasis. EMBO J. 2004, 23, 2912–2955. [Google Scholar] [CrossRef] [PubMed]
- Rehman, H.M.; Nawaz, M.A.; Shah, Z.H.; Ludwig-Müller, J.; Chung, G.; Ahmad, M.Q.; Yang, S.H.; Lee, S.I. Comparative genomic and transcriptomic analyses of Family-1 UDP glycosyltransferase in three Brassica species and Arabidopsis indicates stress-responsive regulation. Sci. Rep. 2018, 8, 1875. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Härtl, K.; McGraphery, K.; Hoffmann, T.; Schwab, W. Attractive but Toxic: Emerging Roles of Glycosidically Bound Volatiles and Glycosyltransferases Involved in Their Formation. Mol. Plant 2018, 11, 1225–1236. [Google Scholar] [CrossRef] [PubMed]
- Lairson, L.L.; Henrissat, B.; Davies, G.J.; Withers, S.G. Glycosyltransferases: Structures, functions, and mechanisms. Annu. Rev. Biochem. 2008, 77, 521–555. [Google Scholar] [CrossRef]
- Xiao, X.; Lu, Q.; Liu, R.; Gong, J.; Gong, W.; Liu, A.; Ge, Q.; Li, J.; Shang, H.; Li, P.; et al. Genome-wide characterization of the UDP-glycosyltransferase gene family in upland cotton. 3 Biotech 2019, 9, 453. [Google Scholar] [CrossRef]
- Ross, J.; Li, Y.; Lim, E.; Bowles, D.J. Higher plant glycosyltransferases. Genome Biol. 2001, 2, 3004.1–3004.6. [Google Scholar] [CrossRef]
- Vogt, T.; Jones, P. Glycosyltransferases in plant natural product synthesis: Characterization of a supergene family. Trends Plant Sci. 2000, 5, 380–386. [Google Scholar] [CrossRef]
- Bowles, D.; Isayenkova, J.; Lim, E.K.; Poppenberger, B. Glycosyltransferases: Managers of small molecules. Curr. Opin. Plant Biol. 2005, 8, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Zhang, Z.; Zhang, L.; Wang, J.; Wu, C. Glycosyltransferase GT1 family: Phylogenetic distribution, substrates coverage, and representative structural features. Comput. Struct. Biotechnol. J. 2020, 18, 1383–1390. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Baldauf, S.; Lim, E.K.; Bowles, D.J. Phylogenetic analysis of the UDP-glycosyltransferase multigene family of Arabidopsis thaliana. J. Biol. Chem. 2001, 276, 4338–4343. [Google Scholar] [CrossRef]
- Gachon, C.M.; Langlois-Meurinne, M.; Saindrenan, P. Plant secondary metabolism glycosyltransferases: The emerging functional analysis. Trends Plant Sci. 2005, 10, 542–549. [Google Scholar] [CrossRef] [PubMed]
- Nagatoshi, M.; Terasaka, K.; Nagatsu, A.; Mizukami, H. Iridoid-specific glucosyltransferase from Gardenia jasminoides. J. Biol. Chem. 2011, 286, 32866–32874. [Google Scholar] [CrossRef] [PubMed]
- Gharabli, H.; Della Gala, V.; Welner, D.H. The function of UDP-glycosyltransferases in plants and their possible use in crop protection. Biotechnol. Adv. 2023, 67, 108182. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Zhang, J.; Shao, Y.; Xu, Y.; Ge, H.; Yu, B.; Wang, W. Enzyme-Catalyzed Glycosylation of Curcumin and Its Analogues by Glycosyltransferases from Bacillus subtilis ATCC 6633. Catalysts 2019, 9, 734. [Google Scholar] [CrossRef]
- Osmani, S.A.; Bak, S.; Imberty, A.; Olsen, C.E.; Møller, B.L. Catalytic key amino acids and UDP-sugar donor specificity of a plant glucuronosyltransferase, UGT94B1: Molecular modeling substantiated by site-specific mutagenesis and biochemical analyses. Plant Physiol. 2008, 148, 1295–1308. [Google Scholar] [CrossRef]
- Rahimi, S.; Kim, J.; Mijakovic, I.; Jung, K.H.; Choi, G.; Kim, S.C.; Kim, Y.J. Triterpenoid-biosynthetic UDP-glycosyltransferases from plants. Biotechnol. Adv. 2019, 37, 107394. [Google Scholar] [CrossRef]
- Wang, D.; Wang, J.; Shi, Y.; Li, R.; Fan, F.; Huang, Y.; Li, W.; Chen, N.; Huang, L.; Dai, Z.; et al. Elucidation of the complete biosynthetic pathway of the main triterpene glycosylation products of Panax notoginseng using a synthetic biology platform. Metab. Eng. 2020, 61, 131–140. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhuo, X.; Yan, X.; Zhang, Q. Comparative Genomic and Transcriptomic Analyses of Family-1 UDP Glycosyltransferase in Prunus Mume. Int. J. Mol. Sci. 2018, 19, 3382. [Google Scholar] [CrossRef]
- Boachon, B.; Gamir, J.; Pastor, V.; Erb, M.; Dean, J.V.; Flors, V.; Mauch-Mani, B. Role of two UDP-Glycosyltransferases from the L group of arabidopsis in resistance against pseudomonas syringae. Eur. J. Plant Pathol. 2014, 139, 707–720. [Google Scholar] [CrossRef]
- Song, J.T.; Koo, Y.J.; Seo, H.S.; Kim, M.C.; Choi, Y.D.; Kim, J.H. Overexpression of AtSGT1, an Arabidopsis salicylic acid glucosyltransferase, leads to increased susceptibility to Pseudomonas syringae. Phytochemistry 2008, 69, 1128–1134. [Google Scholar] [CrossRef] [PubMed]
- Tezuka, D.; Matsuura, H.; Saburi, W.; Mori, H.; Imai, R. A Ubiquitously Expressed UDP-Glucosyltransferase, UGT74J1, Controls Basal Salicylic Acid Levels in Rice. Plants 2021, 10, 1875. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wang, W.S.; Wang, T.; Meng, X.F.; Chen, T.T.; Huang, X.X.; Li, Y.J.; Hou, B.K. Methyl Salicylate Glucosylation Regulates Plant Defense Signaling and Systemic Acquired Resistance. Plant Physiol. 2019, 180, 2167–2181. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.E.; Choi, J.N.; Kim, I.A.; Lee, S.A.; Hwang, Y.S.; Lee, C.H.; Lim, J. Improved resistance to oxidative stress by a loss-of-function mutation in the Arabidopsis UGT71C1 gene. Mol. Cells 2008, 25, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Campos, L.; López-Gresa, M.P.; Fuertes, D.; Bellés, J.M.; Rodrigo, I.; Lisón, P. Tomato glycosyltransferase Twi1 plays a role in flavonoid glycosylation and defence against virus. BMC Plant Biol. 2019, 19, 450. [Google Scholar] [CrossRef] [PubMed]
- Diao, A.; Chen, J.; Ye, R.; Zheng, T.; Yu, S.; Antoniw, J.F.; Adams, M.J. Complete sequence and genome properties of Chinese wheat mosaic virus, a new furovirus from China. J. Gen. Virol. 1999, 80, 1141–1145. [Google Scholar] [CrossRef]
- Chen, J. Occurrence of fungally transmitted wheat mosaic viruses in China. Ann. Appl. Biol. 1993, 123, 55–61. [Google Scholar]
- Ye, R.; Zheng, T.; Chen, J.; Diao, A.; Adams, M.J.; Yu, S.; Antoniw, F. Characterization and partial sequence of a new furovirus of wheat in China. Plant Pathol. 1999, 48, 379–387. [Google Scholar] [CrossRef]
- Guo, L.M.; He, J.; Li, J.; Chen, J.P.; Zhang, H.M. Chinese wheat mosaic virus: A long-term threat to wheat in China. J. Integr. Agric. 2019, 18, 821–829. [Google Scholar] [CrossRef]
- Kanyuka, K.; Ward, E.; Adams, M.J. Polymyxa graminis and the cereal viruses it transmits: A research challenge. Mol. Plant Pathol. 2003, 4, 393–406. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, F.; Xie, L.; Song, X.J.; Li, J.; Chen, J.P.; Zhang, H.M. Functional identification of two minor capsid proteins from Chinese wheat mosaic virus using its infectious full-length cDNA clones. J. Gen. Virol. 2016, 97, 2441–2450. [Google Scholar] [CrossRef]
- Chong, J.; Baltz, R.; Schmitt, C.; Beffa, R.; Fritig, B.; Saindrenan, P. Downregulation of a pathogen-responsive tobacco UDP-Glc: Phenylpropanoid glucosyltransferase reduces scopoletin glucoside accumulation, enhances oxidative stress, and weakens virus resistance. Plant Cell 2002, 14, 1093–1107. [Google Scholar] [CrossRef] [PubMed]
- Matros, A.; Mock, H.P. Ectopic Expression of a UDP-Glucose: Phenylpropanoid Glucosyltransferase Leads to Increased Resistance of Transgenic Tobacco Plants against Infection with Potato Virus Y. Plant Cell Physiol. 2004, 45, 1185–1193. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Li, J.; Feng, H.; Liu, S.; Liu, P.; Chen, X.; Yang, J.; He, L.; Yang, J.; Chen, J. Phosphorylated viral protein evades plant immunity through interfering the function of RNA-binding protein. PLoS Pathog. 2022, 18, e1010412. [Google Scholar] [CrossRef]
- Zhang, T.; Shi, C.; Hu, H.; Zhang, Z.; Wang, Z.; Chen, Z.; Feng, H.; Liu, P.; Guo, J.; Lu, Q.; et al. N6-methyladenosine RNA modification promotes viral genomic RNA stability and infection. Nat. Commun. 2022, 13, 6576. [Google Scholar] [CrossRef]
- Yang, J.; Chen, L.; Zhang, J.; Liu, P.; Chen, M.; Chen, Z.; Zhong, K.; Liu, J.; Chen, J.; Yang, J. TaTHI2 interacts with Ca2+-dependent protein kinase TaCPK5 to suppress virus infection by regulating ROS accumulation. Plant Biotechnol. J. 2023. [Google Scholar] [CrossRef]
- Liu, C.; Talbot, N.J.; Chen, X.L. Protein glycosylation during infection by plant pathogenic fungi. New Phytol. 2021, 230, 1329–1335. [Google Scholar] [CrossRef]
- Shumilina, J.; Kusnetsova, A.; Tsarev, A.; Janse van Rensburg, H.C.; Medvedev, S.; Demidchik, V.; Van den Ende, W.; Frolov, A. Glycation of Plant Proteins: Regulatory Roles and Interplay with Sugar Signalling? Int. J. Mol. Sci. 2019, 20, 2366. [Google Scholar] [CrossRef]
- Wenjin, Z.; Sheng, W.; Jian, Y.; Chuanzhi, K.; Luqi, H.; Lanping, G. Glycosylation of plant secondary metabolites: Regulating from chaos to harmony. Environ. Exp. Bot. 2021, 194, 104703. [Google Scholar]
- Yonekura-Sakakibara, K.; Hanada, K. An evolutionary view of functional diversity in family 1 glycosyltransferases. Plant J. 2011, 66, 182–193. [Google Scholar] [CrossRef]
- Chang, A.; Singh, S.; Phillips, G.N., Jr.; Thorson, J.S. Glycosyltransferase structural biology and its role in the design of catalysts for glycosylation. Curr. Opin. Biotechnol. 2011, 22, 800–808. [Google Scholar] [CrossRef]
- Vrielink, A.; Rüger, W.; Driessen, H.P.; Freemont, P.S. Crystal structure of the DNA modifying enzyme beta-glucosyltransferase in the presence and absence of the substrate uridine diphosphoglucose. Embo J. 1994, 13, 3413–3422. [Google Scholar] [CrossRef]
- Blanchard, S.; Thorson, J.S. Enzymatic tools for engineering natural product glycosylation. Curr. Opin. Chem. Biol. 2006, 10, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, P.I.; Owens, I.S.; Burchell, B.; Bock, K.W.; Bairoch, A.; Bélanger, A.; Fournel-Gigleux, S.; Green, M.; Hum, D.W.; Iyanagi, T.; et al. The UDP glycosyltransferase gene superfamily: Recommended nomenclature update based on evolutionary divergence. Pharmacogenetics 1997, 7, 255–269. [Google Scholar] [CrossRef] [PubMed]
- Meech, R.; Mackenzie, P.I. Structure and function of uridine diphosphate glucuronosyltransferases. Clin. Exp. Pharmacol. Physiol. 1997, 24, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Fu, M.; Li, H.; Wang, L.; Liu, R.; Liu, Z. Genome-wide characterization of the UDP-glycosyltransferase gene family reveals their potential roles in leaf senescence in cotton. Int. J. Biol. Macromol. 2022, 222, 2648–2660. [Google Scholar] [CrossRef]
- Li, Y.; Li, P.; Wang, Y.; Dong, R.; Yu, H.; Hou, B. Genome-wide identification and phylogenetic analysis of Family-1 UDP glycosyltransferases in maize (Zea mays). Planta 2014, 239, 1265–1279. [Google Scholar] [CrossRef]
- Yu, G.; Chen, Q.; Chen, F.; Liu, H.; Lin, J.; Chen, R.; Ren, C.; Wei, J.; Zhang, Y.; Yang, F.; et al. Glutathione Promotes Degradation and Metabolism of Residual Fungicides by Inducing UDP-Glycosyltransferase Genes in Tomato. Front. Plant Sci. 2022, 13, 893508. [Google Scholar] [CrossRef]
- He, Y.; Ahmad, D.; Zhang, X.; Zhang, Y.; Wu, L.; Jiang, P.; Ma, H. Genome-wide analysis of family-1 UDP glycosyltransferases (UGT) and identification of UGT genes for FHB resistance in wheat (Triticum aestivum L.). BMC Plant Biol. 2018, 18, 67. [Google Scholar] [CrossRef]
- Dauda, W.P.; Shanmugam, V.; Tyagi, A.; Solanke, A.U.; Kumar, V.; Krishnan, S.G.; Bashyal, B.M.; Aggarwal, R. Genome-Wide Identification and Characterisation of Cytokinin-O-Glucosyltransferase (CGT) Genes of Rice Specific to Potential Pathogens. Plants 2022, 11, 917. [Google Scholar] [CrossRef]
- Elasad, M.; Wei, H.; Wang, H.; Su, J.; Ondati, E.; Yu, S. Genome-Wide Analysis and Characterization of the TRX Gene Family in Upland Cotton. Trop. Plant Biol. 2018, 11, 119–130. [Google Scholar] [CrossRef]
- Dooner, H.K.; Nelson, O.E. Controlling element-induced alterations in UDPglucose: Flavonoid glucosyltransferase, the enzyme specified by the bronze locus in maize. Proc. Natl. Acad. Sci. USA 1977, 74, 5623–5627. [Google Scholar] [CrossRef] [PubMed]
- Ao, B.; Han, Y.; Wang, S.; Wu, F.; Zhang, J. Genome-Wide Analysis and Profile of UDP-Glycosyltransferases Family in Alfalfa (Medicago sativa L.) under Drought Stress. Int. J. Mol. Sci. 2022, 23, 7243. [Google Scholar] [CrossRef]
- Dong, L.; Tang, Z.; Yang, T.; Hao, F.; Deng, X. Genome-Wide Analysis of UGT Genes in Petunia and Identification of PhUGT51 Involved in the Regulation of Salt Resistance. Plants 2022, 11, 2434. [Google Scholar] [CrossRef]
- He, Q.; Yin, H.; Jiang, J.; Bai, Y.; Chen, N.; Liu, S.; Zhuang, Y.; Liu, T. Fermentative Production of Phenolic Glucosides by Escherichia coli with an Engineered Glucosyltransferase from Rhodiola sachalinensis. J. Agric. Food Chem. 2017, 65, 4691–4697. [Google Scholar] [CrossRef]
- Zheng, S.W.; Chen, Z.F.; Liu, T.T.; Zhao, Z.Y.; Li, T.M.; Xing, G.M. Identification and Characterization of the Tomato UGT Gene Family and Effects of GAME 17 Overexpression on Plants and Growth and Development under High-CO2 Conditions. Agronomy 2022, 12, 1998. [Google Scholar] [CrossRef]
- Song, C.; Hong, X.; Zhao, S.; Liu, J.; Schulenburg, K.; Huang, F.C.; Franz-Oberdorf, K.; Schwab, W. Glucosylation of 4-Hydroxy-2,5-Dimethyl-3(2H)-Furanone, the Key Strawberry Flavor Compound in Strawberry Fruit. Plant Physiol. 2016, 171, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Shen, G.; Di, S.; Dixon, R.A.; Pang, Y. Characterization of UGT716A1 as a Multi-substrate UDP: Flavonoid Glucosyltransferase Gene in Ginkgo biloba. Front. Plant Sci. 2017, 8, 2085. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Li, C.; Wei, W.; Wei, Y.; Liu, Q.; Zhao, G.; Yue, J.; Yan, X.; Wang, P.; Zhou, Z. The unprecedented diversity of UGT94-family UDP-glycosyltransferases in Panax plants and their contribution to ginsenoside biosynthesis. Sci. Rep. 2020, 10, 15394. [Google Scholar] [CrossRef] [PubMed]
- Holmes, E.C.; Chen, Y.C.; Mudgett, M.B.; Sattely, E.S. Arabidopsis UGT76B1 glycosylates N-hydroxy-pipecolic acid and inactivates systemic acquired resistance in tomato. Plant Cell 2021, 33, 750–765. [Google Scholar] [CrossRef]
- Huang, X.X.; Wang, Y.; Lin, J.S.; Chen, L.; Li, Y.J.; Liu, Q.; Wang, G.F.; Xu, F.; Liu, L.; Hou, B.K. The novel pathogen-responsive glycosyltransferase UGT73C7 mediates the redirection of phenylpropanoid metabolism and promotes SNC1-dependent Arabidopsis immunity. Plant J. 2021, 107, 149–165. [Google Scholar] [CrossRef] [PubMed]
- Dare, A.P.; Yauk, Y.K.; Tomes, S.; McGhie, T.K.; Rebstock, R.S.; Cooney, J.M.; Atkinson, R.G. Silencing a phloretin-specific glycosyltransferase perturbs both general phenylpropanoid biosynthesis and plant development. Plant J. 2017, 91, 237–250. [Google Scholar] [CrossRef] [PubMed]
- Hettwer, K.; Böttcher, C.; Frolov, A.; Mittasch, J.; Albert, A.; von Roepenack-Lahaye, E.; Strack, D.; Milkowski, C. Dynamic metabolic changes in seeds and seedlings of Brassica napus (oilseed rape) suppressing UGT84A9 reveal plasticity and molecular regulation of the phenylpropanoid pathway. Phytochemistry 2016, 124, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Saxe, H.J.; Horibe, T.; Balan, B.; Butterfield, T.S.; Feinberg, N.G.; Zabaneh, C.M.; Jacobson, A.E.; Dandekar, A.M. Two UGT84A Family Glycosyltransferases Regulate Phenol, Flavonoid, and Tannin Metabolism in Juglans regia (English Walnut). Front. Plant Sci. 2021, 12, 626483. [Google Scholar] [CrossRef]
- Bowles, D.; Lim, E.K.; Poppenberger, B.; Vaistij, F.E. Glycosyltransferases of lipophilic small molecules. Annu. Rev. Plant Biol. 2006, 57, 567–597. [Google Scholar] [CrossRef]
- Fukuchi-Mizutani, M.; Okuhara, H.; Fukui, Y.; Nakao, M.; Katsumoto, Y.; Yonekura-Sakakibara, K.; Kusumi, T.; Hase, T.; Tanaka, Y. Biochemical and molecular characterization of a novel UDP-glucose: Anthocyanin 3′-O-glucosyltransferase, a key enzyme for blue anthocyanin biosynthesis, from gentian. Plant Physiol. 2003, 132, 1652–1663. [Google Scholar] [CrossRef]
- Dong, N.Q.; Lin, H.X. Contribution of phenylpropanoid metabolism to plant development and plant-environment interactions. J. Integr. Plant Biol. 2021, 63, 180–209. [Google Scholar] [CrossRef]
- Muro-Villanueva, F.; Mao, X.; Chapple, C. Linking phenylpropanoid metabolism, lignin deposition, and plant growth inhibition. Curr. Opin. Biotechnol. 2019, 56, 202–208. [Google Scholar] [CrossRef]
- Wang, T.; Ma, Y.Q.; Huang, X.X.; Mu, T.J.; Li, Y.J.; Li, X.K.; Liu, X.; Hou, B.K. Overexpression of OsUGT3 enhances drought and salt tolerance through modulating ABA synthesis and scavenging ROS in rice. Environ. Exp. Bot. 2021, 192, 104653. [Google Scholar] [CrossRef]
- Haroth, S.; Feussner, K.; Kelly, A.A.; Zienkiewicz, K.; Shaikhqasem, A.; Herrfurth, C.; Feussner, I. The glycosyltransferase UGT76E1 significantly contributes to 12-O-glucopyranosyl-jasmonic acid formation in wounded Arabidopsis thaliana leaves. J. Biol. Chem. 2019, 294, 9858–9872. [Google Scholar] [CrossRef] [PubMed]
- Mohnike, L.; Rekhter, D.; Huang, W.; Feussner, K.; Tian, H.; Herrfurth, C.; Zhang, Y.; Feussner, I. The glycosyltransferase UGT76B1 modulates N-hydroxy-pipecolic acid homeostasis and plant immunity. Plant Cell 2021, 33, 735–749. [Google Scholar] [CrossRef]
- White, R.F. Acetylsalicylic acid (aspirin) induces resistance to tobacco mosaic virus in tobacco. Virology 1979, 99, 410–412. [Google Scholar] [CrossRef]
- An, C.; Mou, Z. Salicylic acid and its function in plant immunity. J. Integr. Plant Biol. 2011, 53, 412–428. [Google Scholar] [CrossRef]
- Fu, Z.Q.; Dong, X. Systemic acquired resistance: Turning local infection into global defense. Annu. Rev. Plant Biol. 2013, 64, 839–863. [Google Scholar] [CrossRef] [PubMed]
- Ryals, J.; Uknes, S.; Ward, E. Systemic Acquired Resistance. Plant Physiol. 1994, 104, 1109–1112. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Sonbol, F.M.; Huot, B.; Gu, Y.; Withers, J.; Mwimba, M.; Yao, J.; He, S.Y.; Dong, X. Salicylic acid receptors activate jasmonic acid signalling through a non-canonical pathway to promote effector-triggered immunity. Nat. Commun. 2016, 7, 13099. [Google Scholar] [CrossRef] [PubMed]
- Vos, I.A.; Moritz, L.; Pieterse, C.M.; Van Wees, S.C. Impact of hormonal crosstalk on plant resistance and fitness under multi-attacker conditions. Front. Plant Sci. 2015, 6, 639. [Google Scholar] [CrossRef] [PubMed]
- Van Butselaar, T.; Van den Ackerveken, G. Salicylic Acid Steers the Growth-Immunity Tradeoff. Trends Plant Sci. 2020, 25, 566–576. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).