Structural Impact of the Interaction of the Influenza A Virus Nucleoprotein with Genomic RNA Segments
Abstract
1. Introduction
2. Materials and Methods
2.1. In Vitro Transcription of the M and NS vRNAs
2.2. Expression, Purification, and Characterization of the WSN NP Protein
2.3. Modification of the M and NS vRNAs
2.4. Reverse Transcription and cDNA Analysis
2.5. Data Analysis
3. Results
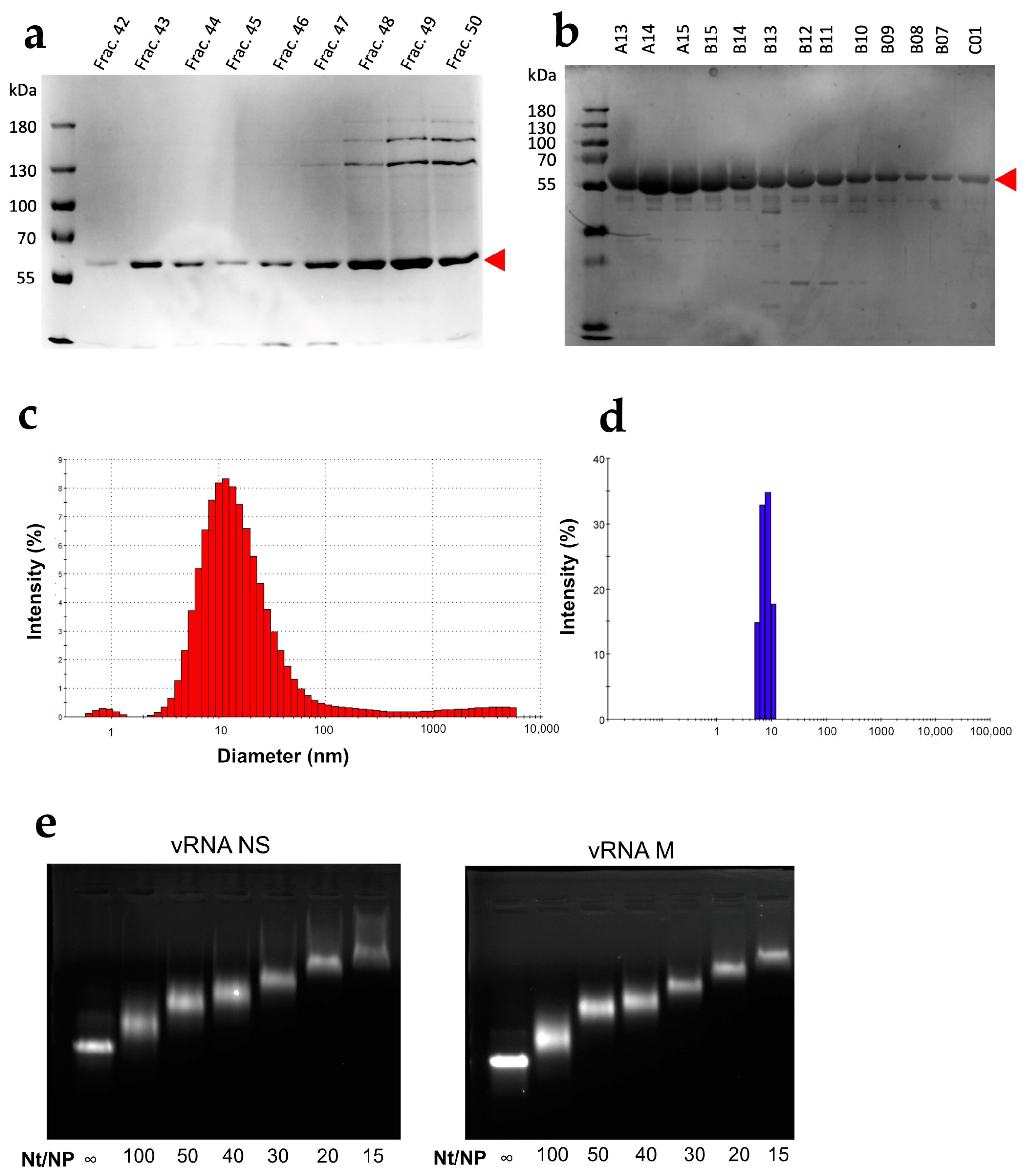
3.1. Characterization of the WSN NP Protein
3.2. NP Has Limited but Significant RNA Chaperone Activity
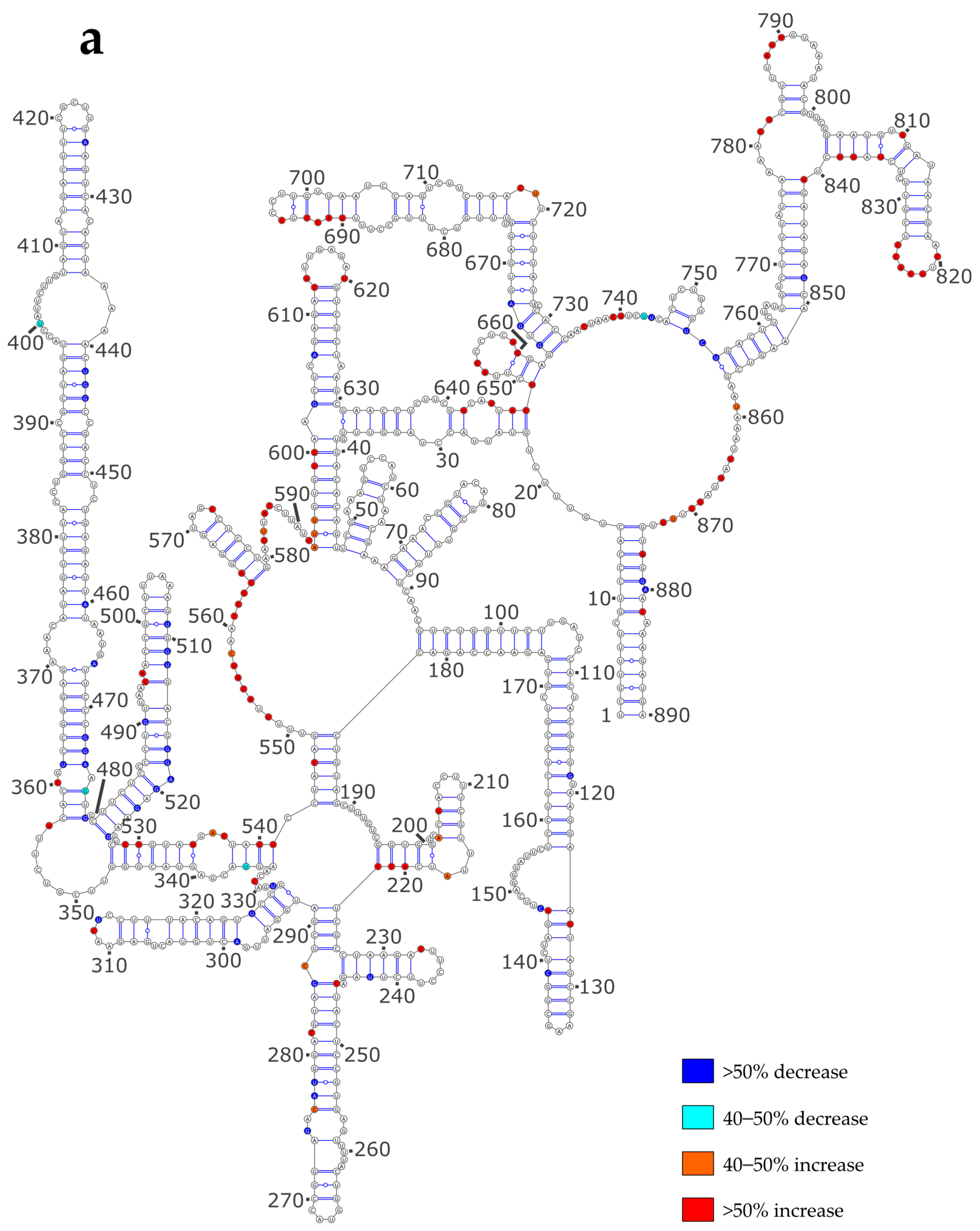
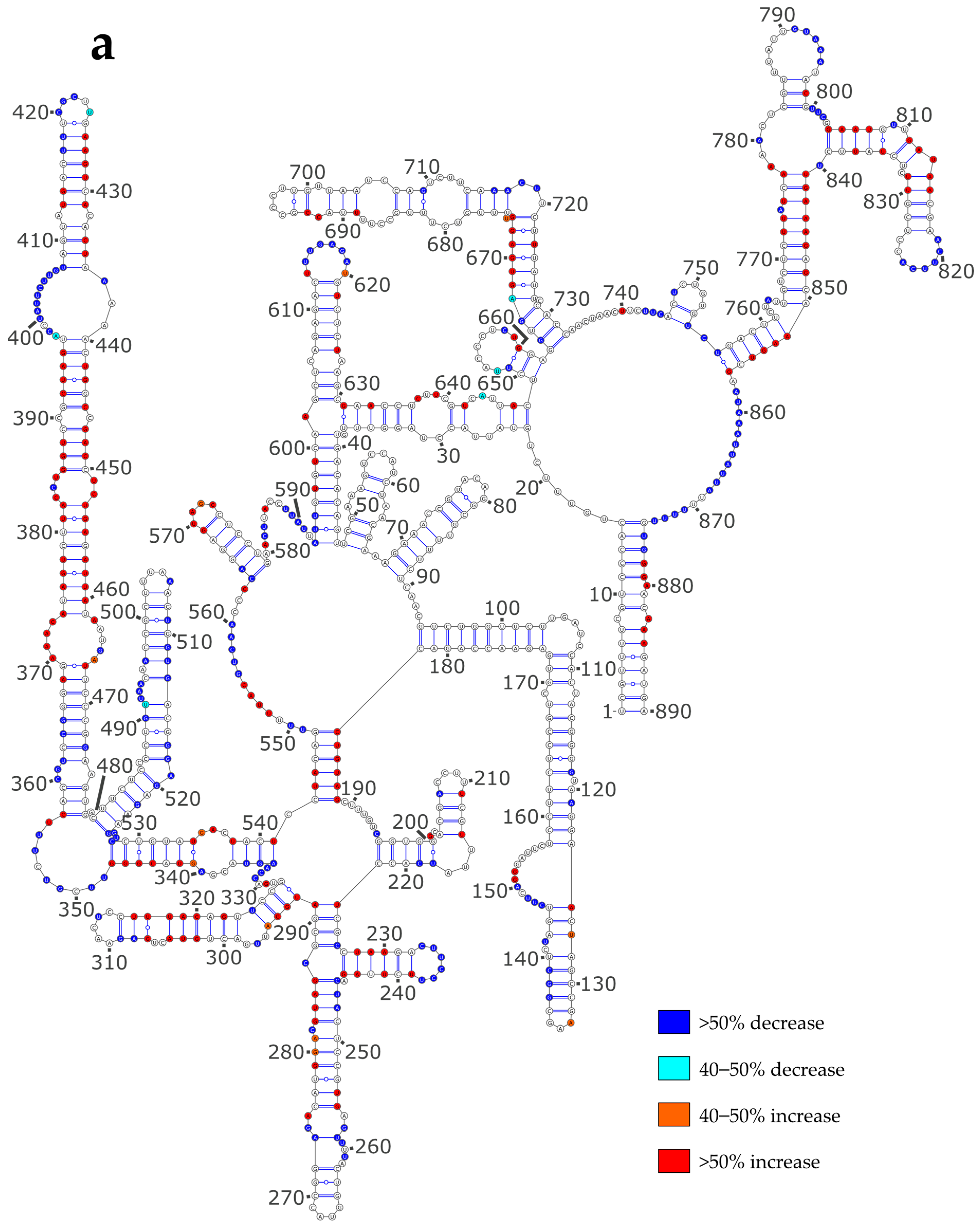
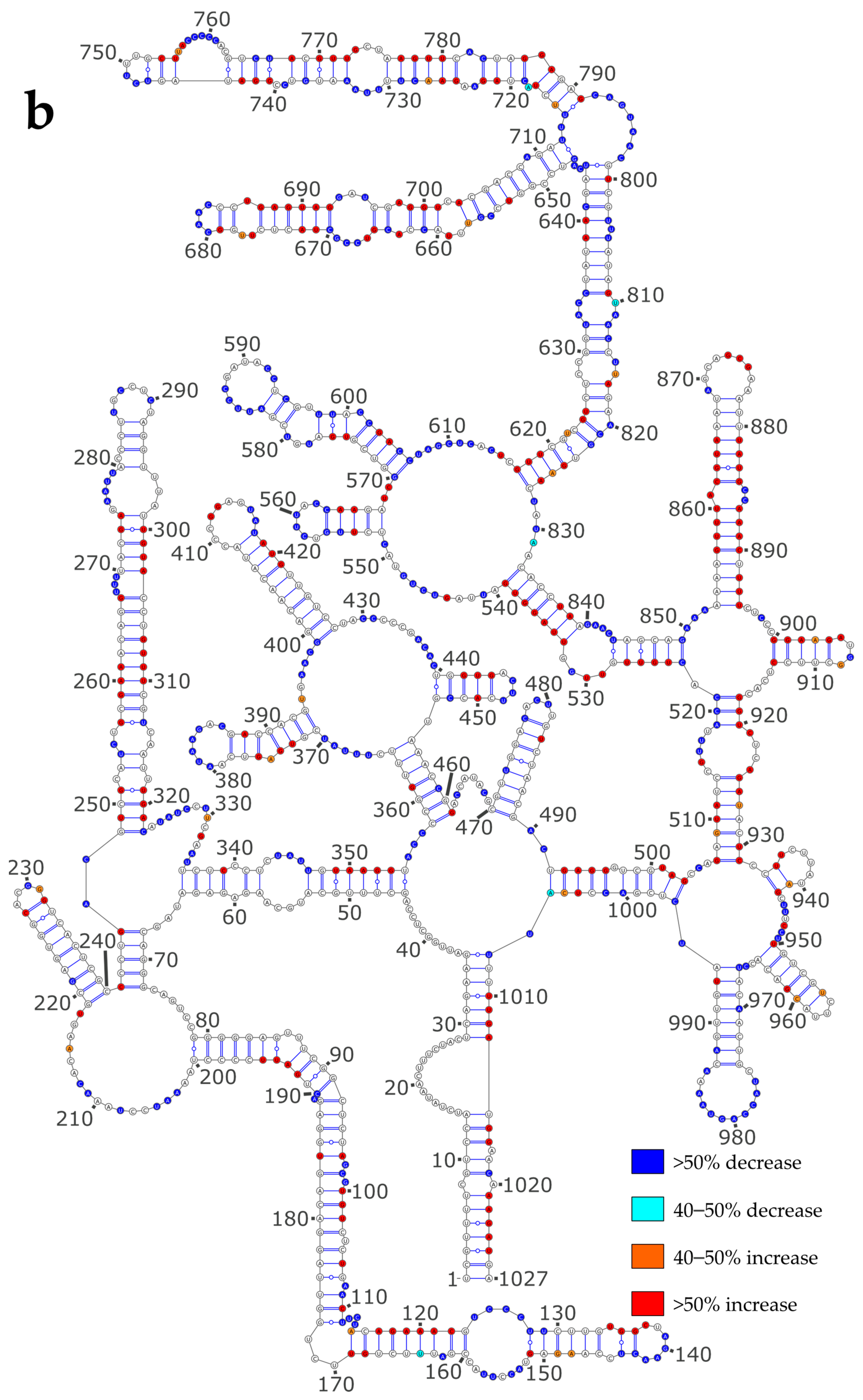
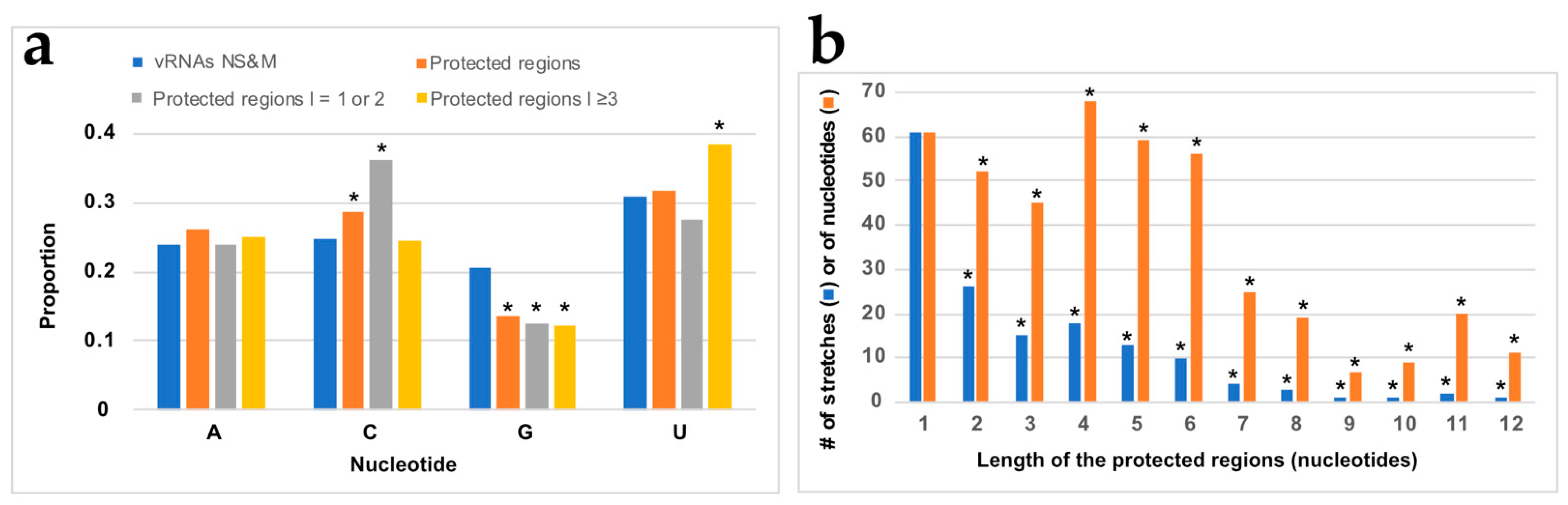
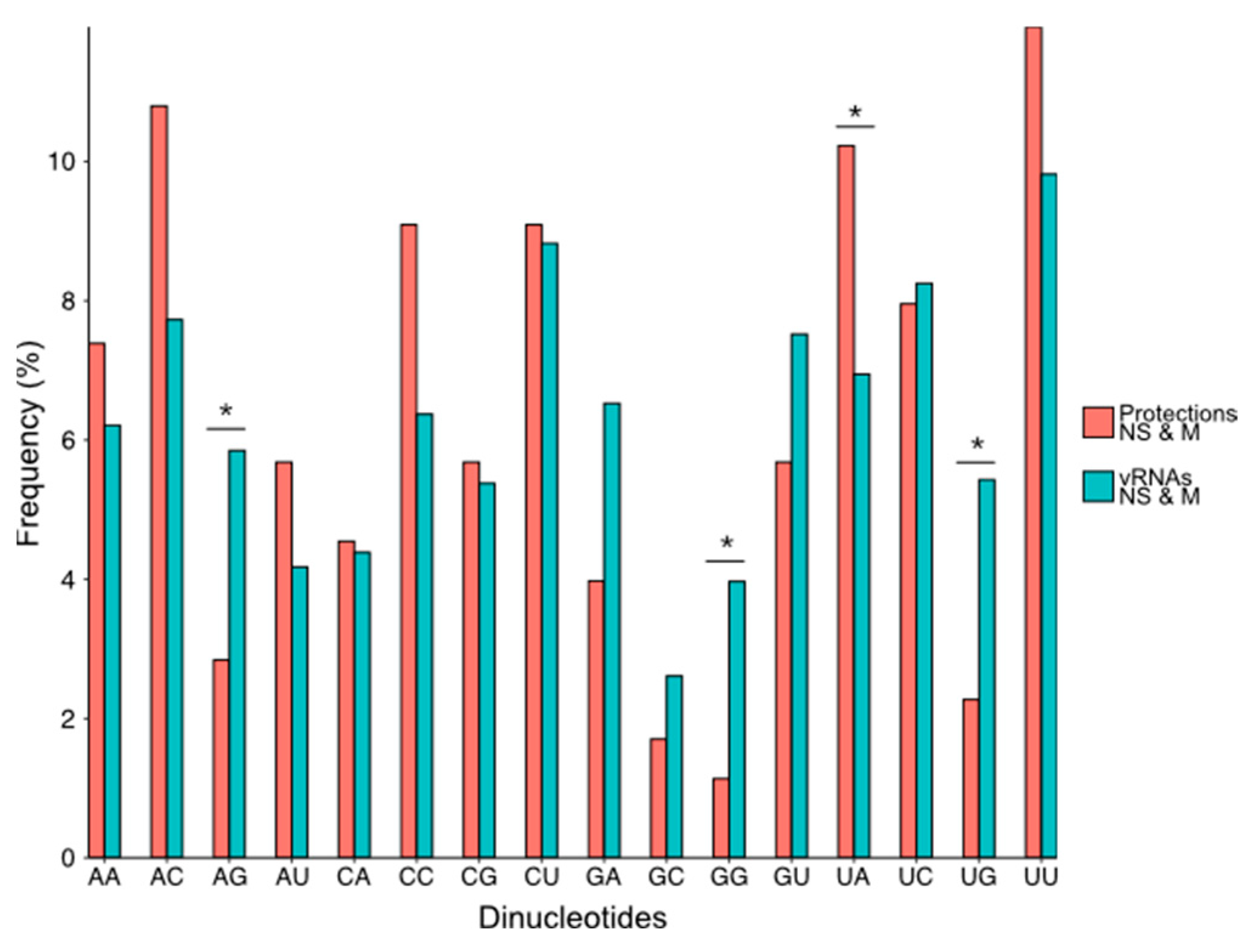
3.3. NP Has a Major Impact on the RNA Structures to Which It Binds
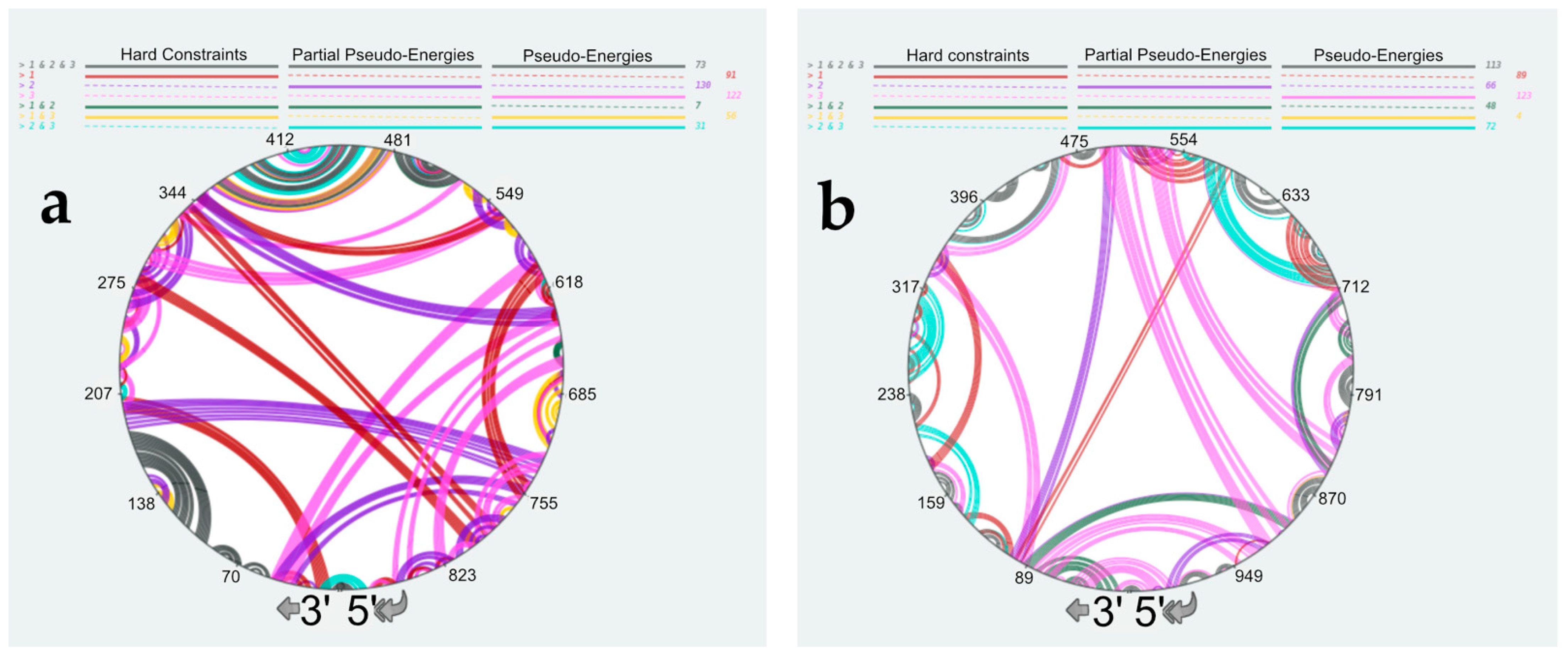
3.4. Using SHAPE Probing Data to Model the Secondary Structure of the vRNA/NP Complexes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Iuliano, A.D.; Roguski, K.M.; Chang, H.H.; Muscatello, D.J.; Palekar, R.; Tempia, S.; Cohen, C.; Gran, J.M.; Schanzer, D.; Cowling, B.J.; et al. Estimates of global seasonal influenza-associated respiratory mortality: A modelling study. Lancet 2018, 391, 1285–1300. [Google Scholar] [CrossRef] [PubMed]
- Eisfeld, A.J.; Neumann, G.; Kawaoka, Y. At the centre: Influenza A virus ribonucleoproteins. Nat. Rev. Microbiol. 2015, 13, 28–41. [Google Scholar] [CrossRef] [PubMed]
- Arranz, R.; Coloma, R.; Chichon, F.J.; Conesa, J.J.; Carrascosa, J.L.; Valpuesta, J.M.; Ortin, J.; Martin-Benito, J. The Structure of Native Influenza Virion Ribonucleoproteins. Science 2012, 338, 1634–1637. [Google Scholar] [CrossRef] [PubMed]
- Hutchinson, E.C.; von Kirchbach, J.C.; Gog, J.R.; Digard, P. Genome packaging in influenza A virus. J. Gen. Virol. 2010, 91, 313–328. [Google Scholar] [CrossRef] [PubMed]
- Gerber, M.; Isel, C.; Moules, V.; Marquet, R. Selective packaging of the influenza A genome and consequences for genetic reassortment. Trends Microbiol. 2014, 22, 446–455. [Google Scholar] [CrossRef] [PubMed]
- Ferhadian, D.; Contrant, M.; Printz-Schweigert, A.; Smyth, R.P.; Paillart, J.-C.; Marquet, R. Structural and Functional Motifs in Influenza Virus RNAs. Front. Microbiol. 2018, 9, 559. [Google Scholar] [CrossRef]
- Compans, R.W.; Content, J.; Duesberg, P.H. Structure of the Ribonucleoprotein of Influenza Virus. J. Virol. 1972, 10, 795–800. [Google Scholar] [CrossRef]
- Ortega, J.; Martin-Benito, J.; Zurcher, T.; Valpuesta, J.M.; Carrascosa, J.L.; Ortin, J. Ultrastructural and functional analyses of recombinant influenza virus ribonucleoproteins suggest dimerization of nucleoprotein during virus amplification. J. Virol. 2000, 74, 156–163. [Google Scholar] [CrossRef]
- Williams, G.D.; Townsend, D.; Wylie, K.M.; Kim, P.J.; Amarasinghe, G.K.; Kutluay, S.B.; Boon, A.C.M. Nucleotide resolution mapping of influenza A virus nucleoprotein-RNA interactions reveals RNA features required for replication. Nat. Commun. 2018, 9, 465. [Google Scholar] [CrossRef]
- Lee, N.; Le Sage, V.; Nanni, A.V.; Snyder, D.J.; Cooper, V.S.; Lakdawala, S.S. Genome-wide analysis of influenza viral RNA and nucleoprotein association. Nucleic Acids Res. 2017, 45, 8968–8977. [Google Scholar] [CrossRef]
- Le Sage, V.; Nanni, A.V.; Bhagwat, A.R.; Snyder, D.J.; Cooper, V.S.; Lakdawala, S.S.; Lee, N. Non-Uniform and Non-Random Binding of Nucleoprotein to Influenza A and B Viral RNA. Viruses 2018, 10, 522. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Krug, R.M.; Tao, Y.J. The mechanism by which influenza A virus nucleoprotein forms oligomers and binds RNA. Nature 2006, 444, 1078–1082. [Google Scholar] [CrossRef] [PubMed]
- Tarus, B.; Bakowiez, O.; Chenavas, S.; Duchemin, L.; Estrozi, L.F.; Bourdieu, C.; Lejal, N.; Bernard, J.; Moudjou, M.; Chevalier, C.; et al. Oligomerization paths of the nucleoprotein of influenza A virus. Biochimie 2012, 94, 776–785. [Google Scholar] [CrossRef] [PubMed]
- Labaronne, A.; Swale, C.; Monod, A.; Schoehn, G.; Crépin, T.; Ruigrok, R.W.H. Binding of RNA by the Nucleoproteins of Influenza Viruses A and B. Viruses 2016, 8, 247. [Google Scholar] [CrossRef]
- Ng, A.K.; Zhang, H.; Tan, K.; Li, Z.; Liu, J.H.; Chan, P.K.; Li, S.M.; Chan, W.Y.; Au, S.W.; Joachimiak, A.; et al. Structure of the influenza virus A H5N1 nucleoprotein: Implications for RNA binding, oligomerization, and vaccine design. Faseb J. 2008, 22, 3638–3647. [Google Scholar] [CrossRef] [PubMed]
- Chenavas, S.; Estrozi, L.F.; Slama-Schwok, A.; Delmas, B.; Di Primo, C.; Baudin, F.; Li, X.; Crépin, T.; Ruigrok, R.W.H. Monomeric nucleoprotein of influenza A virus. PLoS Pathog. 2013, 9, e1003275. [Google Scholar] [CrossRef]
- Liu, C.-L.; Hung, H.-C.; Lo, S.-C.; Chiang, C.-H.; Chen, I.-J.; Hsu, J.T.-A.; Hou, M.-H. Using mutagenesis to explore conserved residues in the RNA-binding groove of influenza A virus nucleoprotein for antiviral drug development. Sci. Rep. 2016, 6, 21662. [Google Scholar] [CrossRef] [PubMed]
- Baudin, F.; Bach, C.; Cusack, S.; Ruigrok, R.W. Structure of influenza virus RNP. I. Influenza virus nucleoprotein melts secondary structure in panhandle RNA and exposes the bases to the solvent. Embo J. 1994, 13, 3158–3165. [Google Scholar] [CrossRef]
- Zheng, W.; Olson, J.; Vakharia, V.; Tao, Y.J. The crystal structure and RNA-binding of an orthomyxovirus nucleoprotein. PLoS Pathog. 2013, 9, e1003624. [Google Scholar] [CrossRef]
- Chenavier, F.; Estrozi, L.F.; Teulon, J.-M.; Zarkadas, E.; Freslon, L.-L.; Pellequer, J.-L.; Ruigrok, R.W.H.; Schoehn, G.; Ballandras-Colas, A.; Crépin, T. Cryo-EM structure of influenza helical nucleocapsid reveals NP-NP and NP-RNA interactions as a model for the genome encapsidation. Sci. Adv. 2023, 9, eadj9974. [Google Scholar] [CrossRef]
- Jakob, C.; Paul-Stansilaus, R.; Schwemmle, M.; Marquet, R.; Bolte, H. The influenza A virus genome packaging network-complex, flexible and yet unsolved. Nucleic Acids Res. 2022, 50, 9023–9038. [Google Scholar] [CrossRef]
- Fournier, E.; Moules, V.; Essere, B.; Paillart, J.C.; Sirbat, J.D.; Isel, C.; Cavalier, A.; Rolland, J.P.; Thomas, D.; Lina, B.; et al. A supramolecular assembly formed by influenza A virus genomic RNA segments. Nucleic Acids Res. 2012, 40, 2197–2209. [Google Scholar] [CrossRef]
- Fournier, E.; Moules, V.; Essere, B.; Paillart, J.C.; Sirbat, J.D.; Cavalier, A.; Rolland, J.P.; Thomas, D.; Lina, B.; Isel, C.; et al. Interaction network linking the human H3N2 influenza A virus genomic RNA segments. Vaccine 2012, 30, 7359–7367. [Google Scholar] [CrossRef]
- Noda, T.; Sugita, Y.; Aoyama, K.; Hirase, A.; Kawakami, E.; Miyazawa, A.; Sagara, H.; Kawaoka, Y. Three-dimensional analysis of ribonucleoprotein complexes in influenza A virus. Nat. Commun. 2012, 3, 639. [Google Scholar] [CrossRef]
- Gavazzi, C.; Isel, C.; Fournier, E.; Moules, V.; Cavalier, A.; Thomas, D.; Lina, B.; Marquet, R. An in vitro network of intermolecular interactions between viral RNA segments of an avian H5N2 influenza A virus: Comparison with a human H3N2 virus. Nucleic Acids Res. 2013, 41, 1241–1254. [Google Scholar] [CrossRef] [PubMed]
- Gavazzi, C.; Yver, M.; Isel, C.; Smyth, R.P.; Rosa-Calatrava, M.; Lina, B.; Moulès, V.; Marquet, R. A functional sequence-specific interaction between influenza A virus genomic RNA segments. Proc. Natl. Acad. Sci. USA 2013, 110, 16604–16609. [Google Scholar] [CrossRef] [PubMed]
- Dadonaite, B.; Gilbertson, B.; Knight, M.L.; Trifkovic, S.; Rockman, S.; Laederach, A.; Brown, L.E.; Fodor, E.; Bauer, D.L.V. The structure of the influenza A virus genome. Nat. Microbiol. 2019, 4, 1781–1789. [Google Scholar] [CrossRef] [PubMed]
- Le Sage, V.; Kanarek, J.P.; Snyder, D.J.; Cooper, V.S.; Lakdawala, S.S.; Lee, N. Mapping of Influenza Virus RNA-RNA Interactions Reveals a Flexible Network. Cell Rep. 2020, 31, 107823. [Google Scholar] [CrossRef] [PubMed]
- Moreira, É.A.; Weber, A.; Bolte, H.; Kolesnikova, L.; Giese, S.; Lakdawala, S.; Beer, M.; Zimmer, G.; García-Sastre, A.; Schwemmle, M.; et al. A conserved influenza A virus nucleoprotein code controls specific viral genome packaging. Nat. Commun. 2016, 7, 12861. [Google Scholar] [CrossRef] [PubMed]
- Klumpp, K.; Ruigrok, R.W.; Baudin, F. Roles of the influenza virus polymerase and nucleoprotein in forming a functional RNP structure. Embo J. 1997, 16, 1248–1257. [Google Scholar] [CrossRef] [PubMed]
- Godet, J.; Boudier, C.; Humbert, N.; Ivanyi-Nagy, R.; Darlix, J.-L.; Mély, Y. Comparative nucleic acid chaperone properties of the nucleocapsid protein NCp7 and Tat protein of HIV-1. Virus. Res. 2012, 169, 349–360. [Google Scholar] [CrossRef] [PubMed]
- Henriet, S.; Sinck, L.; Bec, G.; Gorelick, R.J.; Marquet, R.; Paillart, J.-C. Vif is a RNA chaperone that could temporally regulate RNA dimerization and the early steps of HIV-1 reverse transcription. Nucleic Acids Res. 2007, 35, 5141–5153. [Google Scholar] [CrossRef]
- Ivanyi-Nagy, R.; Lavergne, J.-P.; Gabus, C.; Ficheux, D.; Darlix, J.-L. RNA chaperoning and intrinsic disorder in the core proteins of Flaviviridae. Nucleic Acids Res. 2008, 36, 712–725. [Google Scholar] [CrossRef] [PubMed]
- Pitchai, F.N.N.; Chameettachal, A.; Vivet-Boudou, V.; Ali, L.M.; Pillai, V.N.; Krishnan, A.; Bernacchi, S.; Mustafa, F.; Marquet, R.; Rizvi, T.A. Identification of Pr78Gag Binding Sites on the Mason-Pfizer Monkey Virus Genomic RNA Packaging Determinants. J. Mol. Biol. 2021, 433, 166923. [Google Scholar] [CrossRef]
- Karabiber, F.; McGinnis, J.L.; Favorov, O.V.; Weeks, K.M. QuShape: Rapid, accurate, and best-practices quantification of nucleic acid probing information, resolved by capillary electrophoresis. RNA 2013, 19, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Reuter, J.S.; Mathews, D.H. RNAstructure: Software for RNA secondary structure prediction and analysis. BMC Bioinform. 2010, 11, 129. [Google Scholar] [CrossRef]
- Hajdin, C.E.; Bellaousov, S.; Huggins, W.; Leonard, C.W.; Mathews, D.H.; Weeks, K.M. Accurate SHAPE-directed RNA secondary structure modeling, including pseudoknots. Proc. Natl. Acad. Sci. USA 2013, 110, 5498–5503. [Google Scholar] [CrossRef]
- Darty, K.; Denise, A.; Ponty, Y. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics 2009, 25, 1974–1975. [Google Scholar] [CrossRef]
- Schmidt, M.D.; Kirkpatrick, A.; Heitsch, C. RNAStructViz: Graphical base pairing analysis. Bioinformatics 2021, 37, 3660–3661. [Google Scholar] [CrossRef]
- Ferhadian, D. Structure de l’ARN Au Sein Des Ribonucléoprotéines de Influenza Virus A. Ph.D. Thesis, Université de Strasbourg, Strasbourg, France, 2018. [Google Scholar]
- Rajkowitsch, L.; Chen, D.; Stampfl, S.; Semrad, K.; Waldsich, C.; Mayer, O.; Jantsch, M.F.; Konrat, R.; Bläsi, U.; Schroeder, R. RNA chaperones, RNA annealers and RNA helicases. RNA Biol. 2007, 4, 118–130. [Google Scholar] [CrossRef]
- Gilmer, O.; Mailler, E.; Paillart, J.-C.; Mouhand, A.; Tisné, C.; Mak, J.; Smyth, R.P.; Marquet, R.; Vivet-Boudou, V. Structural maturation of the HIV-1 RNA 5′ untranslated region by Pr55Gag and its maturation products. RNA Biol. 2022, 19, 191–205. [Google Scholar] [CrossRef] [PubMed]
- Pong, W.-L.; Huang, Z.-S.; Teoh, P.-G.; Wang, C.-C.; Wu, H.-N. RNA binding property and RNA chaperone activity of dengue virus core protein and other viral RNA-interacting proteins. FEBS Lett. 2011, 585, 2575–2581. [Google Scholar] [CrossRef] [PubMed]
- Batisse, J.; Guerrero, S.; Bernacchi, S.; Sleiman, D.; Gabus, C.; Darlix, J.-L.; Marquet, R.; Tisné, C.; Paillart, J.-C. The role of Vif oligomerization and RNA chaperone activity in HIV-1 replication. Virus. Res. 2012, 169, 361–376. [Google Scholar] [CrossRef]
- Mailler, E.; Paillart, J.-C.; Marquet, R.; Smyth, R.P.; Vivet-Boudou, V. The evolution of RNA structural probing methods: From gels to next-generation sequencing. Wiley Interdiscip. Rev. RNA 2019, 10, e1518. [Google Scholar] [CrossRef]
- Gilmer, O.; Quignon, E.; Jousset, A.-C.; Paillart, J.-C.; Marquet, R.; Vivet-Boudou, V. Chemical and Enzymatic Probing of Viral RNAs: From Infancy to Maturity and Beyond. Viruses 2021, 13, 1894. [Google Scholar] [CrossRef]
- McGinnis, J.L.; Duncan, C.D.S.; Weeks, K.M. High-throughput SHAPE and hydroxyl radical analysis of RNA structure and ribonucleoprotein assembly. Meth. Enzymol. 2009, 468, 67–89. [Google Scholar] [CrossRef]
- Deigan, K.E.; Li, T.W.; Mathews, D.H.; Weeks, K.M. Accurate SHAPE-directed RNA structure determination. Proc. Natl. Acad. Sci. USA 2009, 106, 97–102. [Google Scholar] [CrossRef]
- Mathews, D.H.; Moss, W.N.; Turner, D.H. Folding and finding RNA secondary structure. Cold Spring Harb. Perspect. Biol. 2010, 2, a003665. [Google Scholar] [CrossRef]
- Smola, M.J.; Calabrese, J.M.; Weeks, K.M. Detection of RNA-Protein Interactions in Living Cells with SHAPE. Biochemistry 2015, 54, 6867–6875. [Google Scholar] [CrossRef]
- Moeller, A.; Kirchdoerfer, R.N.; Potter, C.S.; Carragher, B.; Wilson, I.A. Organization of the influenza virus replication machinery. Science 2012, 338, 1631–1634. [Google Scholar] [CrossRef]
- Coloma, R.; Arranz, R.; de la Rosa-Trevín, J.M.; Sorzano, C.O.S.; Munier, S.; Carlero, D.; Naffakh, N.; Ortín, J.; Martín-Benito, J. Structural insights into influenza A virus ribonucleoproteins reveal a processive helical track as transcription mechanism. Nat. Microbiol. 2020, 5, 727–734. [Google Scholar] [CrossRef]
- Gerlach, W.; Giegerich, R. GUUGle: A utility for fast exact matching under RNA complementary rules including G-U base pairing. Bioinformatics 2006, 22, 762–764. [Google Scholar] [CrossRef]
- Doetsch, M.; Schroeder, R.; Fürtig, B. Transient RNA-protein interactions in RNA folding. FEBS J. 2011, 278, 1634–1642. [Google Scholar] [CrossRef] [PubMed]
- Wandzik, J.M.; Kouba, T.; Karuppasamy, M.; Pflug, A.; Drncova, P.; Provaznik, J.; Azevedo, N.; Cusack, S. A Structure-Based Model for the Complete Transcription Cycle of Influenza Polymerase. Cell 2020, 181, 877–893.e21. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Fodor, E.; Keown, J.R. A structural understanding of influenza virus genome replication. Trends Microbiol. 2023, 31, 308–319. [Google Scholar] [CrossRef] [PubMed]
- Hagey, R.J.; Elazar, M.; Pham, E.A.; Tian, S.; Ben-Avi, L.; Bernardin-Souibgui, C.; Yee, M.F.; Moreira, F.R.; Rabinovitch, M.V.; Meganck, R.M.; et al. Programmable antivirals targeting critical conserved viral RNA secondary structures from influenza A virus and SARS-CoV-2. Nat. Med. 2022, 28, 1944–1955. [Google Scholar] [CrossRef] [PubMed]
- French, H.; Pitré, E.; Oade, M.S.; Elshina, E.; Bisht, K.; King, A.; Bauer, D.L.V.; Te Velthuis, A.J.W. Transient RNA structures cause aberrant influenza virus replication and innate immune activation. Sci. Adv. 2022, 8, eabp8655. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.-S.; Xu, S.; Chen, Y.-W.; Wang, J.-H.; Shaw, P.-C. Crystal structures of influenza nucleoprotein complexed with nucleic acid provide insights into the mechanism of RNA interaction. Nucleic Acids Res. 2021, 49, 4144–4154. [Google Scholar] [CrossRef] [PubMed]
- Mirska, B.; Woźniak, T.; Lorent, D.; Ruszkowska, A.; Peterson, J.M.; Moss, W.N.; Mathews, D.H.; Kierzek, R.; Kierzek, E. In vivo secondary structural analysis of Influenza A virus genomic RNA. Cell Mol. Life Sci. 2023, 80, 136. [Google Scholar] [CrossRef]
- Takizawa, N.; Kawaguchi, R.K. Comprehensive in virio structure probing analysis of the influenza A virus identifies functional RNA structures involved in viral genome replication. Comput. Struct. Biotechnol. J. 2023, 21, 5259–5272. [Google Scholar] [CrossRef]
- Nakano, M.; Sugita, Y.; Kodera, N.; Miyamoto, S.; Muramoto, Y.; Wolf, M.; Noda, T. Ultrastructure of influenza virus ribonucleoprotein complexes during viral RNA synthesis. Commun. Biol. 2021, 4, 858. [Google Scholar] [CrossRef] [PubMed]


| Primer Name | 5′ Fluorophore | Sequence (5′→3′) |
|---|---|---|
| NS30V | Vic | GAT CCA AAC ACT GTG TCA AGC |
| NS30N | Ned | GAT CCA AAC ACT GTG TCA AGC |
| NS341V | Vic | CAT ACC CAA GCA GAA AGT GGC |
| NS341N | Ned | CAT ACC CAA GCA GAA AGT GGC |
| NS621V | Vic | CAG AGA TTC GCT TGG TGT TGC |
| NS621N | Ned | CAG AGA TTC GCT TGG TGT TGC |
| M20V | Vic | TGA AAG ATG AGT CTT CTA ACC |
| M20N | Ned | TGA AAG ATG AGT CTT CTA ACC |
| M312V | Vic | CAG TTA AAC TGT ATA GGA AGC |
| M312N | Ned | CAG TTA AAC TGT ATA GGA AGC |
| M622V | Vic | AGC AGA GGC CAT GGA TAT TGC |
| M633N | Ned | AGC AGA GGC CAT GGA TAT TGC |
| M843V | Vic | GAT CGT CTT TTT TTC AAA TGC |
| M843N | Ned | GAT CGT CTT TTT TTC AAA TGC |
| NS vRNA (890 nts) | NoNP | |||
|---|---|---|---|---|
| Low | Intermediate | High | ||
| ProtK | Low | 383 | 27 | 2 |
| Intermediate | 59 | 45 | 7 | |
| High | 13 | 24 | 90 | |
| M vRNA (1027 nts) | NoNP | |||
| Low | Intermediate | High | ||
| ProtK | Low | 413 | 54 | 2 |
| Intermediate | 53 | 63 | 29 | |
| High | 12 | 38 | 100 | |
| NS vRNA (890 nts) | NoNP | |||
|---|---|---|---|---|
| Low | Intermediate | High | ||
| Comp | Low | 296 | 44 | 33 |
| Intermediate | 84 | 25 | 34 | |
| High | 39 | 19 | 32 | |
| M vRNA (1027 nts) | NoNP | |||
| Low | Intermediate | High | ||
| Comp | Low | 297 | 72 | 32 |
| Intermediate | 106 | 45 | 45 | |
| High | 41 | 23 | 46 | |
| NS vRNA (890 nts) | ProtK | |||
|---|---|---|---|---|
| Low | Intermediate | High | ||
| Comp | Low | 278 | 53 | 45 |
| Intermediate | 69 | 29 | 40 | |
| High | 36 | 16 | 38 | |
| M vRNA (1027 nts) | ProtK | |||
| Low | Intermediate | High | ||
| Comp | Low | 277 | 62 | 45 |
| Intermediate | 114 | 42 | 46 | |
| High | 43 | 24 | 44 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quignon, E.; Ferhadian, D.; Hache, A.; Vivet-Boudou, V.; Isel, C.; Printz-Schweigert, A.; Donchet, A.; Crépin, T.; Marquet, R. Structural Impact of the Interaction of the Influenza A Virus Nucleoprotein with Genomic RNA Segments. Viruses 2024, 16, 421. https://doi.org/10.3390/v16030421
Quignon E, Ferhadian D, Hache A, Vivet-Boudou V, Isel C, Printz-Schweigert A, Donchet A, Crépin T, Marquet R. Structural Impact of the Interaction of the Influenza A Virus Nucleoprotein with Genomic RNA Segments. Viruses. 2024; 16(3):421. https://doi.org/10.3390/v16030421
Chicago/Turabian StyleQuignon, Erwan, Damien Ferhadian, Antoine Hache, Valérie Vivet-Boudou, Catherine Isel, Anne Printz-Schweigert, Amélie Donchet, Thibaut Crépin, and Roland Marquet. 2024. "Structural Impact of the Interaction of the Influenza A Virus Nucleoprotein with Genomic RNA Segments" Viruses 16, no. 3: 421. https://doi.org/10.3390/v16030421
APA StyleQuignon, E., Ferhadian, D., Hache, A., Vivet-Boudou, V., Isel, C., Printz-Schweigert, A., Donchet, A., Crépin, T., & Marquet, R. (2024). Structural Impact of the Interaction of the Influenza A Virus Nucleoprotein with Genomic RNA Segments. Viruses, 16(3), 421. https://doi.org/10.3390/v16030421






