Transcriptional Differential Analysis of Nitazoxanide-Mediated Anticanine Parvovirus Effect in F81 Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell and Virus
2.2. Reagents and Antibodies
2.3. Validation of Anti-CPV Effect of NTZ In Vitro
2.4. Western Blot
2.5. Absolute Quantitative PCR
2.6. RNA Extraction and cDNA Library Construction and Sequencing
2.7. Data Processing and Bioinformatics Analysis
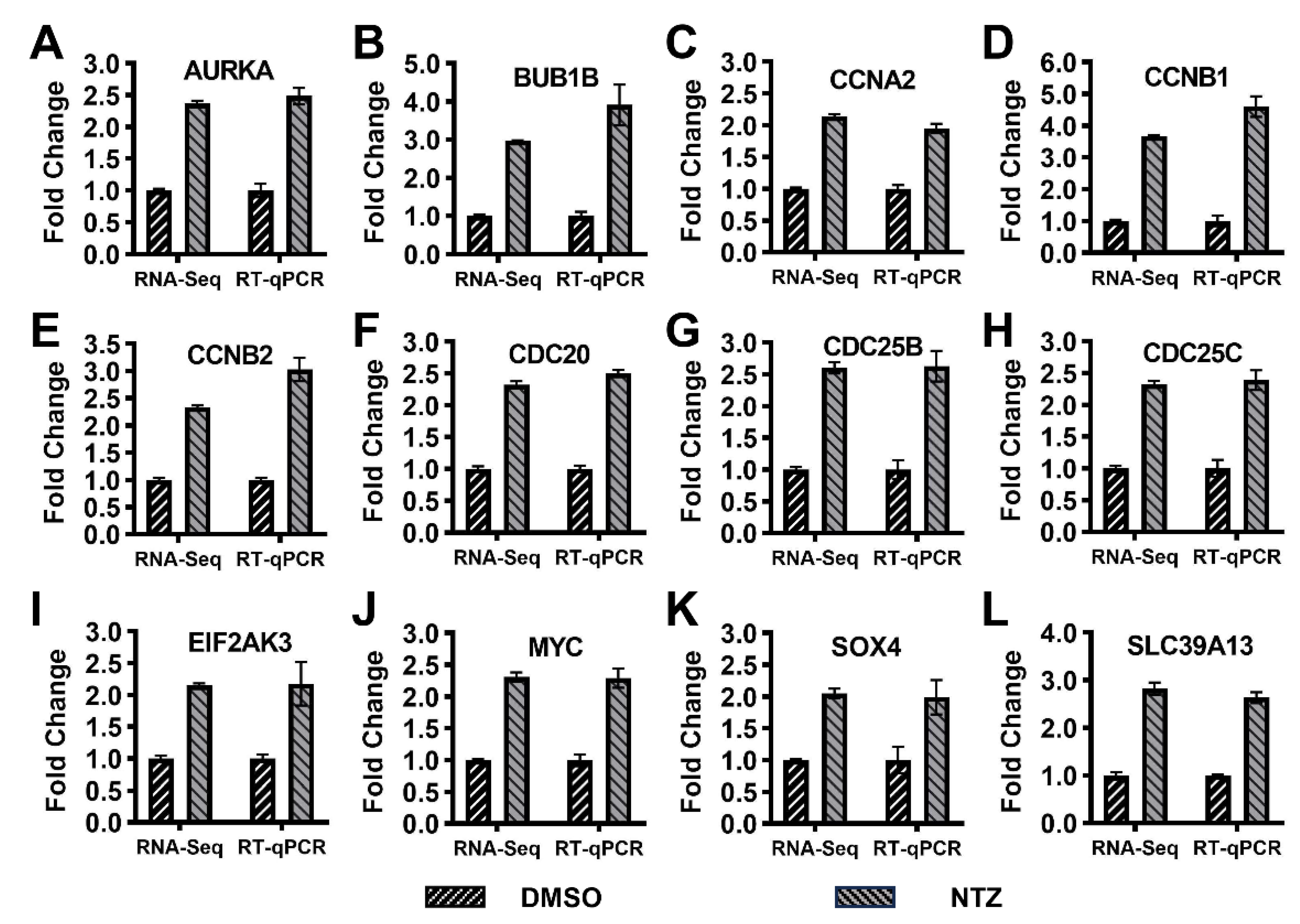
2.8. Verifying the Expression Level of DEGs Using qRT-PCR
2.9. Effect of Cell Cycle Checkpoint Kinase 1 (Chk1) Inhibitor on CPV Replication
2.10. Statistical Analysis
3. Results
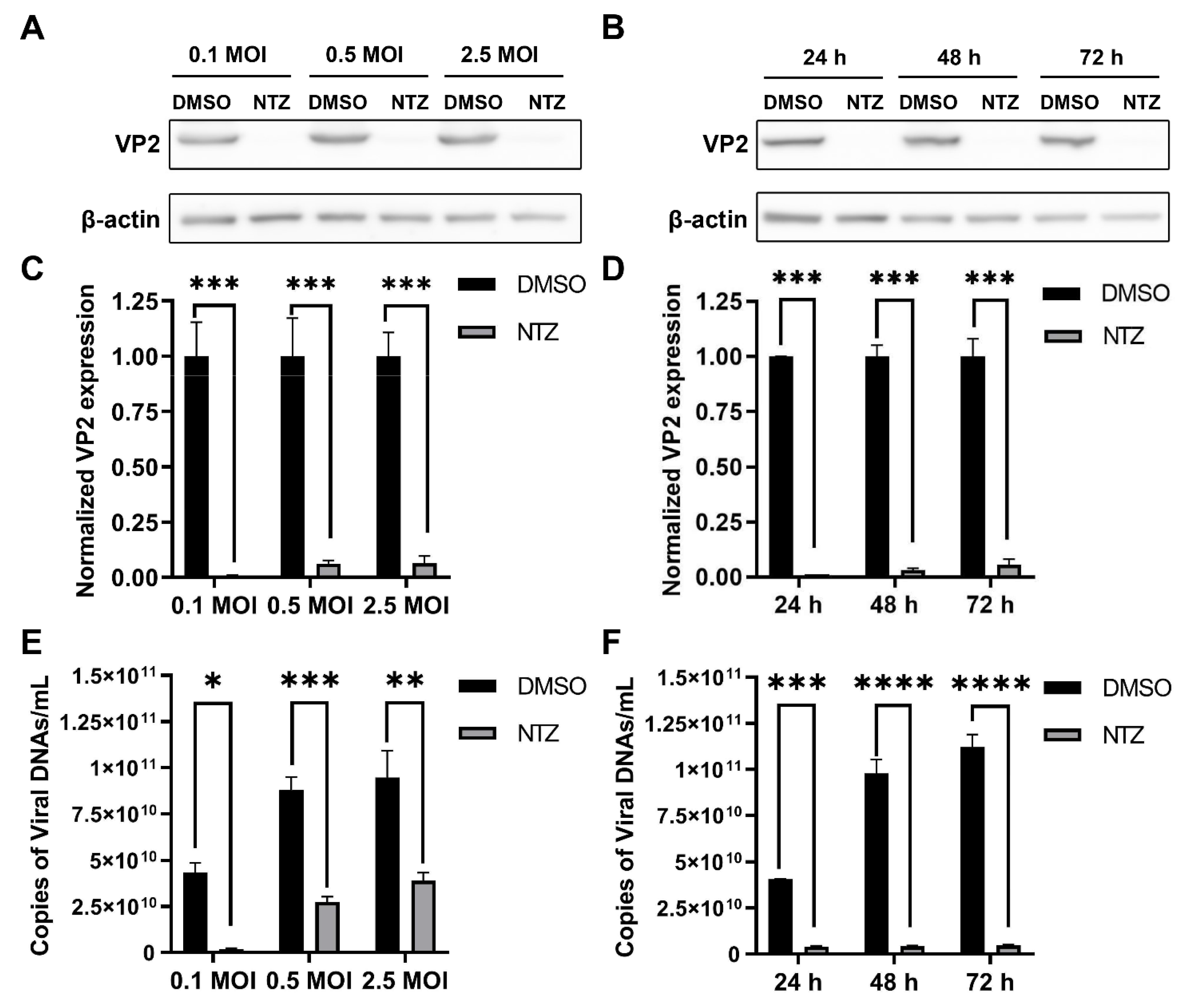
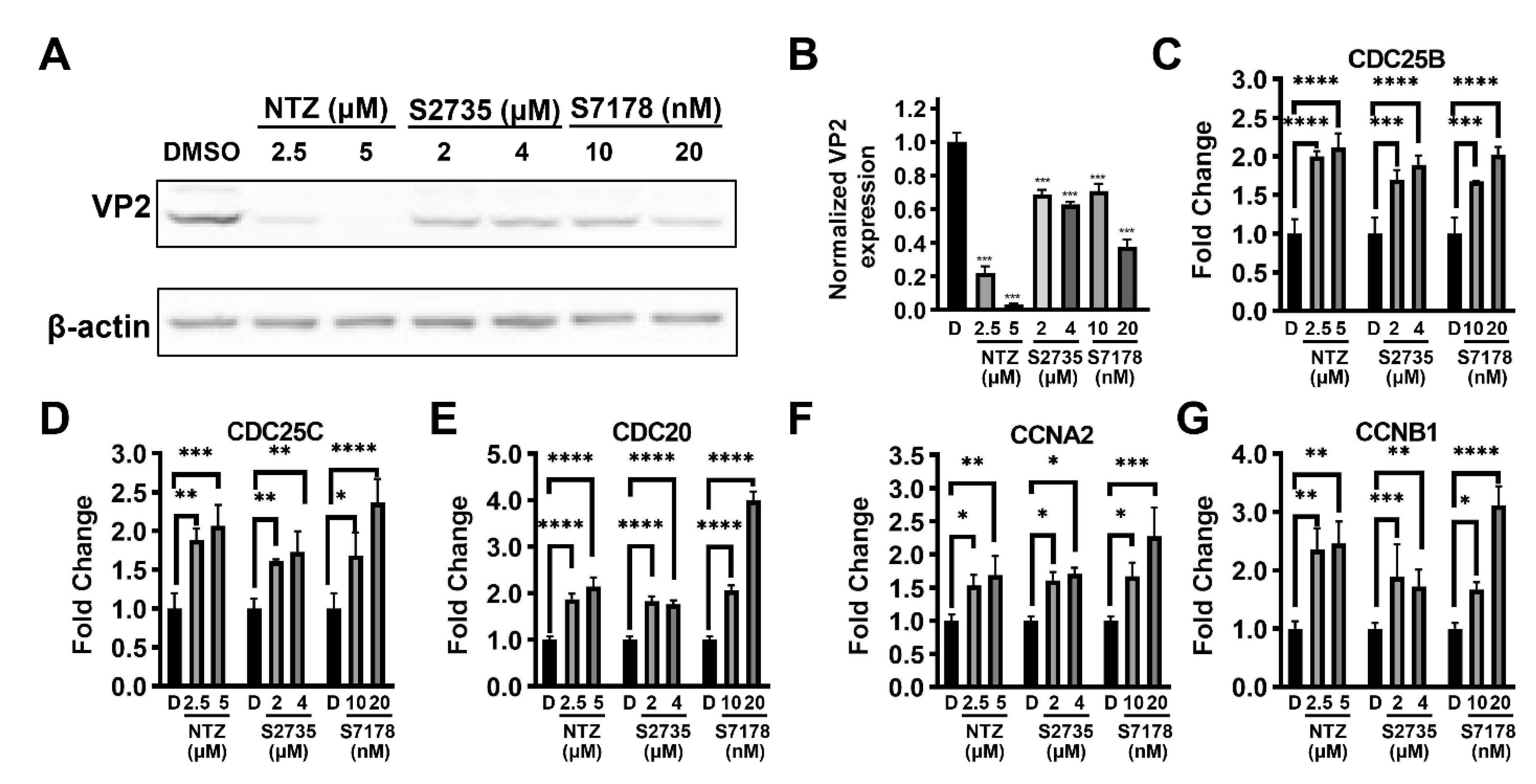
3.1. Effect of NTZ on Virus Replication
3.2. Overview of Transcriptome Sequencing Data
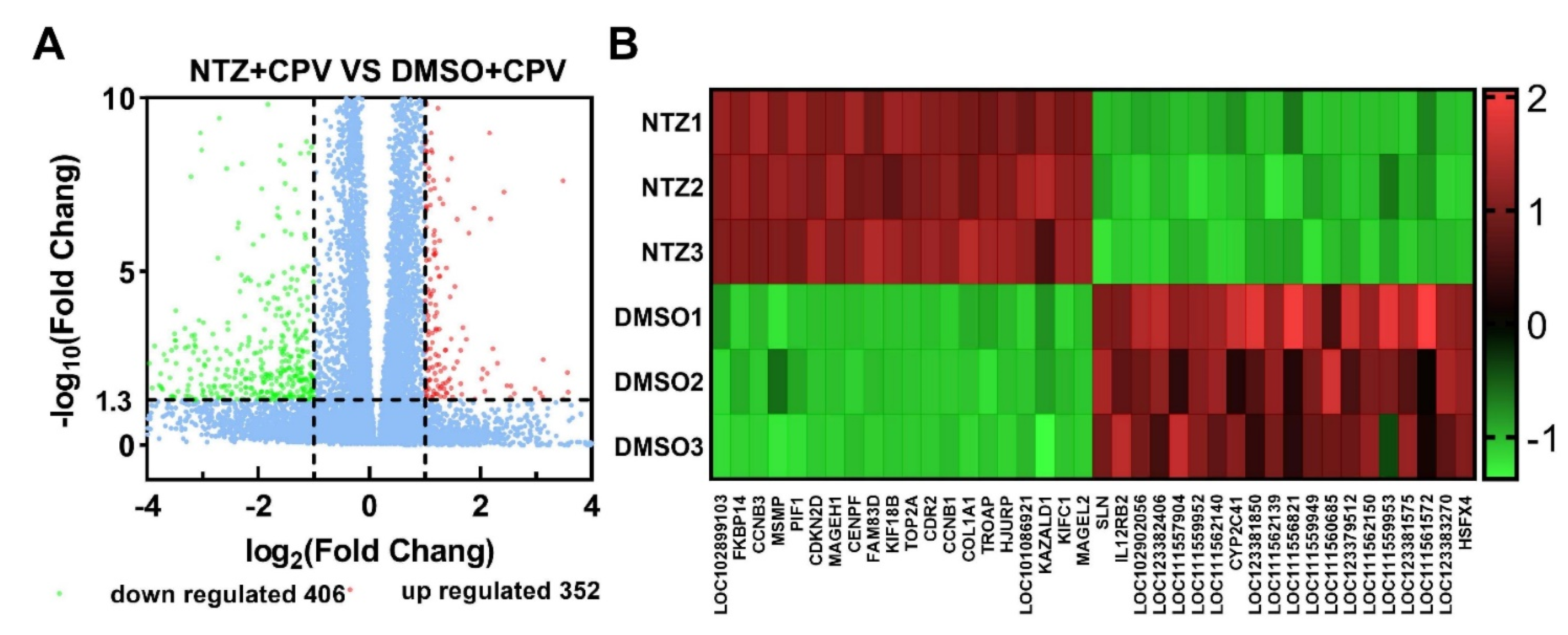
3.3. Analysis of Differentially Expressed Genes
3.4. Enrichment Analysis of GO Terms and KEGG Pathways
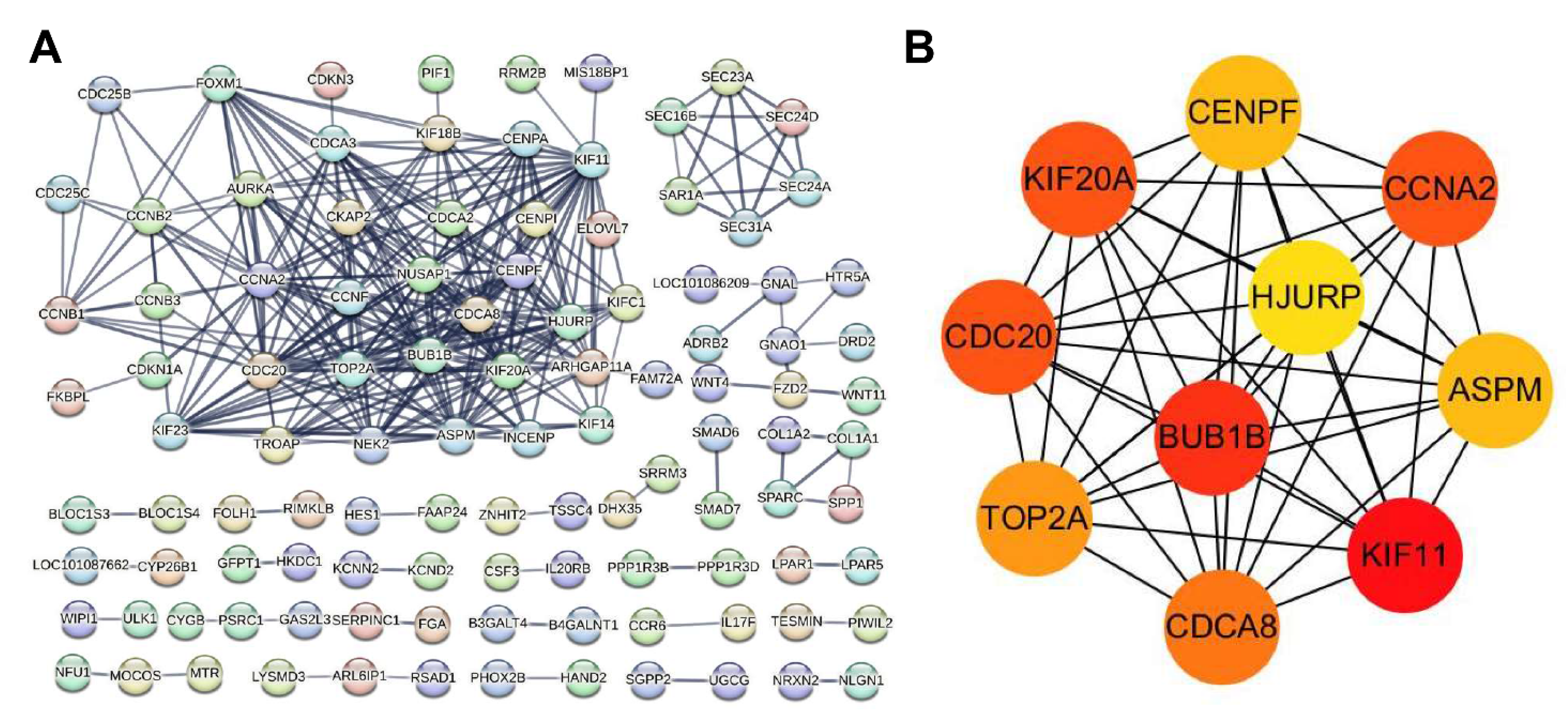
3.5. Protein–Protein Interaction (PPI) Network Analysis of DEGs
3.6. Differential Expression of Enriched or Associated Genes
3.7. Cell Cycle Checkpoint Kinase 1 (Chk1) Inhibitors Inhibit CPV Replication
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Carmichael, L.E. An annotated historical account of canine parvovirus. J. Vet. Med. B Infect. Dis. Vet. Public Health 2005, 52, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.L.; Tu, Y.C.; Lee, M.S.; Wu, L.H.; Chen, T.Y.; Wu, C.H.; Tsao, E.H.; Chin, S.C.; Li, W.T. Fatal canine parvovirus-2 (CPV-2) infection in a rescued free-ranging Taiwanese pangolin (Manis pentadactyla pentadactyla). Transbound. Emerg. Dis. 2020, 67, 1074–1081. [Google Scholar] [CrossRef] [PubMed]
- Qi, S.; Zhao, J.; Guo, D.; Sun, D. A Mini-Review on the Epidemiology of Canine Parvovirus in China. Front. Vet. Sci. 2020, 7, 5. [Google Scholar] [CrossRef] [PubMed]
- Decaro, N.; Buonavoglia, C.; Barrs, V.R. Canine parvovirus vaccination and immunisation failures: Are we far from disease eradication? Vet. Microbiol. 2020, 247, 108760. [Google Scholar] [CrossRef] [PubMed]
- Voorhees, I.E.H.; Lee, H.; Allison, A.B.; Lopez-Astacio, R.; Goodman, L.B.; Oyesola, O.O.; Omobowale, O.; Fagbohun, O.; Dubovi, E.J.; Hafenstein, S.L.; et al. Limited Intrahost Diversity and Background Evolution Accompany 40 Years of Canine Parvovirus Host Adaptation and Spread. J. Virol. 2019, 94, e01162-19. [Google Scholar] [CrossRef]
- Parrish, C.R. Pathogenesis of feline panleukopenia virus and canine parvovirus. Baillieres Clin. Haematol. 1995, 8, 57–71. [Google Scholar] [CrossRef]
- Hao, X.; He, Y.; Wang, C.; Xiao, W.; Liu, R.; Xiao, X.; Zhou, P.; Li, S. The increasing prevalence of CPV-2c in domestic dogs in China. PeerJ 2020, 8, e9869. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhang, H.; Zhang, L.; Zhang, Q.; Zhou, N.; Du, T.; Zhao, Q.; Zhou, E.M.; Du, Y.; Sun, Y. Isolation and Genetic Characterization of Parvoviruses From Dogs, Cats, Minks, and Raccoon Dogs in the Eastern Region of Shandong Province, China. Front. Microbiol. 2022, 13, 862352. [Google Scholar] [CrossRef]
- Decaro, N.; Buonavoglia, C. Canine parvovirus—A review of epidemiological and diagnostic aspects, with emphasis on type 2c. Vet. Microbiol. 2012, 155, 1–12. [Google Scholar] [CrossRef]
- Martin, V.; Najbar, W.; Gueguen, S.; Grousson, D.; Eun, H.M.; Lebreux, B.; Aubert, A. Treatment of canine parvoviral enteritis with interferon-omega in a placebo-controlled challenge trial. Vet. Microbiol. 2002, 89, 115–127. [Google Scholar] [CrossRef]
- Zhou, P.; Fu, X.; Yan, Z.; Fang, B.; Huang, S.; Fu, C.; Hong, M.; Li, S. Antiviral effect of lithium chloride on infection of cells by canine parvovirus. Arch. Virol. 2015, 160, 2799–2805. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Fan, J.; Yang, S.; Zhao, X.; Yi, X. Antiviral activity of phosphorylated Radix Cyathulae officinalis polysaccharide against Canine Parvovirus in vitro. Int. J. Biol. Macromol. 2017, 99, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Su, X.; Lin, L.; Zhang, J.; Qi, Q.; Guo, F.; Xu, F.; Yang, B. Inhibitory Effects of Antiviral Drug Candidates on Canine Parvovirus in F81 cells. Viruses 2019, 11, 742. [Google Scholar] [CrossRef]
- Al-Kuraishy, H.M.; Al-Gareeb, A.I.; Elekhnawy, E.; Batiha, G.E. Nitazoxanide and COVID-19: A review. Mol. Biol. Rep. 2022, 49, 11169–11176. [Google Scholar] [CrossRef] [PubMed]
- Shakya, A.; Bhat, H.R.; Ghosh, S.K. Update on Nitazoxanide: A Multifunctional Chemotherapeutic Agent. Curr. Drug Discov. Technol. 2018, 15, 201–213. [Google Scholar] [CrossRef] [PubMed]
- Rossignol, J.F. Nitazoxanide: A first-in-class broad-spectrum antiviral agent. Antivir. Res. 2014, 110, 94–103. [Google Scholar] [CrossRef] [PubMed]
- Belardo, G.; Cenciarelli, O.; La Frazia, S.; Rossignol, J.F.; Santoro, M.G. Synergistic effect of nitazoxanide with neuraminidase inhibitors against influenza A viruses in vitro. Antimicrob. Agents Chemother. 2015, 59, 1061–1069. [Google Scholar] [CrossRef]
- Rossignol, J.F.; Keeffe, E.B. Thiazolides: A new class of drugs for the treatment of chronic hepatitis B and C. Future Microbiol. 2008, 3, 539–545. [Google Scholar] [CrossRef]
- Trabattoni, D.; Gnudi, F.; Ibba, S.V.; Saulle, I.; Agostini, S.; Masetti, M.; Biasin, M.; Rossignol, J.F.; Clerici, M. Thiazolides Elicit Anti-Viral Innate Immunity and Reduce HIV Replication. Sci. Rep. 2016, 6, 27148. [Google Scholar] [CrossRef]
- Mercorelli, B.; Luganini, A.; Nannetti, G.; Tabarrini, O.; Palu, G.; Gribaudo, G.; Loregian, A. Drug Repurposing Approach Identifies Inhibitors of the Prototypic Viral Transcription Factor IE2 that Block Human Cytomegalovirus Replication. Cell Chem. Biol. 2016, 23, 340–351. [Google Scholar] [CrossRef]
- Korba, B.E.; Montero, A.B.; Farrar, K.; Gaye, K.; Mukerjee, S.; Ayers, M.S.; Rossignol, J.F. Nitazoxanide, tizoxanide and other thiazolides are potent inhibitors of hepatitis B virus and hepatitis C virus replication. Antivir. Res. 2008, 77, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Xu, Y.; Yu, N.; Zhang, M.; Wang, J.; Wan, D.; Tian, Z.; Zhu, H. Catalpol Alleviates Ischemic Stroke Through Promoting Angiogenesis and Facilitating Proliferation and Differentiation of Neural Stem Cells via the VEGF-A/KDR Pathway. Mol. Neurobiol. 2023, 60, 6227–6247. [Google Scholar] [CrossRef] [PubMed]
- Yi, L.; Tong, M.; Cheng, Y.; Song, W.; Cheng, S. Phylogenetic Analysis of Canine Parvovirus VP2 Gene in China. Transbound. Emerg. Dis. 2016, 63, e262–e269. [Google Scholar] [CrossRef]
- Zhou, L.; Tang, Q.; Shi, L.; Kong, M.; Liang, L.; Mao, Q.; Bu, B.; Yao, L.; Zhao, K.; Cui, S.; et al. Full-length genomic characterization and molecular evolution of canine parvovirus in China. Virus Genes. 2016, 52, 411–416. [Google Scholar] [CrossRef]
- Pollock, R.V.; Coyne, M.J. Canine parvovirus. Vet. Clin. North Am. Small Anim. Pract. 1993, 23, 555–568. [Google Scholar] [CrossRef]
- Bordicchia, M.; Fumian, T.M.; Van Brussel, K.; Russo, A.G.; Carrai, M.; Le, S.J.; Pesavento, P.A.; Holmes, E.C.; Martella, V.; White, P.; et al. Feline Calicivirus Virulent Systemic Disease: Clinical Epidemiology, Analysis of Viral Isolates and In Vitro Efficacy of Novel Antivirals in Australian Outbreaks. Viruses 2021, 13, 2040. [Google Scholar] [CrossRef]
- Fan, X.X.; Gao, Y.; Shu, L.; Wei, Y.Q.; Yao, X.P.; Cao, S.Z.; Peng, G.N.; Liu, X.T.; Sun, S.Q. Transcriptome profiling indicating canine parvovirus type 2a as a potential immune activator. Virus Genes 2016, 52, 768–779. [Google Scholar] [CrossRef] [PubMed]
- Vannamahaxay, S.; Sornpet, B.; Pringproa, K.; Patchanee, P.; Chuammitri, P. Transcriptome analysis of infected Crandell Rees Feline Kidney (CRFK) cells by canine parvovirus type 2c Laotian isolates. Gene 2022, 822, 146324. [Google Scholar] [CrossRef]
- Fenizia, C.; Ibba, S.V.; Vanetti, C.; Strizzi, S.; Rossignol, J.F.; Biasin, M.; Trabattoni, D.; Clerici, M. The Modulation of Cholesterol Metabolism Is Involved in the Antiviral Effect of Nitazoxanide. Infect. Dis. Rep. 2021, 13, 636–644. [Google Scholar] [CrossRef]
- Jasenosky, L.D.; Cadena, C.; Mire, C.E.; Borisevich, V.; Haridas, V.; Ranjbar, S.; Nambu, A.; Bavari, S.; Soloveva, V.; Sadukhan, S.; et al. The FDA-Approved Oral Drug Nitazoxanide Amplifies Host Antiviral Responses and Inhibits Ebola Virus. iScience 2019, 19, 1279–1290. [Google Scholar] [CrossRef]
- Riccio, A.; Santopolo, S.; Rossi, A.; Piacentini, S.; Rossignol, J.F.; Santoro, M.G. Impairment of SARS-CoV-2 spike glycoprotein maturation and fusion activity by nitazoxanide: An effect independent of spike variants emergence. Cell Mol. Life Sci. 2022, 79, 227. [Google Scholar] [CrossRef] [PubMed]
- Stachulski, A.V.; Taujanskas, J.; Pate, S.L.; Rajoli, R.K.R.; Aljayyoussi, G.; Pennington, S.H.; Ward, S.A.; Hong, W.D.; Biagini, G.A.; Owen, A.; et al. Therapeutic Potential of Nitazoxanide: An Appropriate Choice for Repurposing versus SARS-CoV-2? ACS Infect. Dis. 2021, 7, 1317–1331. [Google Scholar] [CrossRef] [PubMed]
- Adeyemi, R.O.; Pintel, D.J. Replication of minute virus of mice in murine cells is facilitated by virally induced depletion of p21. J. Virol. 2012, 86, 8328–8332. [Google Scholar] [CrossRef] [PubMed]
- Hristov, G.; Kramer, M.; Li, J.; El-Andaloussi, N.; Mora, R.; Daeffler, L.; Zentgraf, H.; Rommelaere, J.; Marchini, A. Through its nonstructural protein NS1, parvovirus H-1 induces apoptosis via accumulation of reactive oxygen species. J. Virol. 2010, 84, 5909–5922. [Google Scholar] [CrossRef]
- Op De Beeck, A.; Anouja, F.; Mousset, S.; Rommelaere, J.; Caillet-Fauquet, P. The nonstructural proteins of the autonomous parvovirus minute virus of mice interfere with the cell cycle, inducing accumulation in G2. Cell Growth Differ. 1995, 6, 781–787. [Google Scholar]
- Op De Beeck, A.; Sobczak-Thepot, J.; Sirma, H.; Bourgain, F.; Brechot, C.; Caillet-Fauquet, P. NS1- and minute virus of mice-induced cell cycle arrest: Involvement of p53 and p21(cip1). J. Virol. 2001, 75, 11071–11078. [Google Scholar] [CrossRef]
- Chen, A.Y.; Qiu, J. Parvovirus infection-induced cell death and cell cycle arrest. Future Virol. 2010, 5, 731–743. [Google Scholar] [CrossRef]
- Nykky, J.; Tuusa, J.E.; Kirjavainen, S.; Vuento, M.; Gilbert, L. Mechanisms of cell death in canine parvovirus-infected cells provide intuitive insights to developing nanotools for medicine. Int. J. Nanomed. 2010, 5, 417–428. [Google Scholar] [CrossRef]

| Gene ID | Primers | Sequence 5′-3′ | Size (bp) |
|---|---|---|---|
| 101091184 | AURKA-F | TCGTCTCCAGCCATAAACCG | 86 |
| AURKA-R | CAGCGGCCTAGAGACAGAAC | ||
| 101087670 | BUB1B-F | AAATGATCCTCTGGATGTTTGG | 184 |
| BUB1B-R | GCATAAATGCCCCAATTTGAGCC | ||
| 101088573 | CCNA2-F | GGATGGTAGTTTTGAGTCACCAC | 202 |
| CCNA2-R | CACAAGGATAGCCCTCATACTGT | ||
| 101086284 | CCNB1-F | TCGGAGACATTGGTAACAAAGTC | 208 |
| CCNB1-R | ATAGGCTCCGGAGAAAGCTTTT | ||
| 101080972 | CCNB2-F | CCGACGGTGTCCACTGATTT | 180 |
| CCNB2-R | ATTTGTTTTGGCGGGTTGAACA | ||
| 101097259 | CDC20-F | GCTTTGAATCTGAACGGCTTTG | 77 |
| CDC20-R | TCTGGGGCATTTTGTGGTTTT | ||
| 101092757 | CDC25B-F | GGCCGAGGAACCTAAAGCCCG | 139 |
| CDC25B-R | CTTCCCATCCACAGTCTGCAGAA | ||
| 101086942 | CDC25C-F | CCGCTATCCATATGAGTACCAG | 147 |
| CDC25C-R | AATTCACAGTGGAACACGA | ||
| 101095973 | EIF2AK3-F | GGAAACTAGAGCCGGATTTATT | 111 |
| EIF2AK3-R | ACTGTGTCCATCATGGCAGCTTC | ||
| 100379628 | MYC-F | CCCCTCCACTAGGAAGGAC | 96 |
| MYC-R | CTGATGCATTTGCGGTTGTTG | ||
| 111560412 | SOX4-F | GACCTGCTCGACCTGAACC | 107 |
| SOX4-R | CCGGGCTCGAAGTTAAAATCC | ||
| 101095193 | SLC39A13-F | TCAGTGGCTATCTCAACCT | 100 |
| SLC39A13-R | AGGAGCCCAATCTTCTTGCT | ||
| 101098507 | ACTB-F | CATGTACGTGGCCATCCAGGC | 250 |
| ACTB-R | CTCCTTGATGTCACGCACAAT |
| Sample | Raw Reads | Clean Reads | Valid Ratio (%) | Q30 (%) | GC Content (%) | Mapped Reads | Unique Mapped Reads |
|---|---|---|---|---|---|---|---|
| DMSO 1 | 38,970,132 | 37,459,332 | 96.12 | 97.77 | 45 | 19,169,896 (51.18%) | 18,422,453 (49.18%) |
| DMSO 2 | 39,582,814 | 38,127,200 | 96.32 | 97.52 | 45.50 | 20,344,108 (53.36%) | 19,557,030 (51.29%) |
| DMSO 3 | 39,418,474 | 38,071,304 | 96.58 | 97.58 | 45 | 20,064,280 (52.70%) | 19,315,191 (50.73%) |
| NTZ 1 | 39,292,554 | 37,896,286 | 96.45 | 97.83 | 50 | 33,196,056 (87.60%) | 31,342,275 (82.71%) |
| NTZ 2 | 39,735,696 | 38,298,006 | 96.38 | 98.00 | 49.50 | 33,471,408 (87.40%) | 31,946,335 (83.42%) |
| NTZ 3 | 41,529,962 | 40,036,868 | 96.40 | 97.91 | 50 | 35,142,284 (87.77%) | 33,508,501 (83.69%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, X.; Zhou, H.; Han, Z.; Xu, F.; Xiao, B.; Zhang, J.; Qi, Q.; Lin, L.; Zhang, H.; Li, S.; et al. Transcriptional Differential Analysis of Nitazoxanide-Mediated Anticanine Parvovirus Effect in F81 Cells. Viruses 2024, 16, 282. https://doi.org/10.3390/v16020282
Su X, Zhou H, Han Z, Xu F, Xiao B, Zhang J, Qi Q, Lin L, Zhang H, Li S, et al. Transcriptional Differential Analysis of Nitazoxanide-Mediated Anticanine Parvovirus Effect in F81 Cells. Viruses. 2024; 16(2):282. https://doi.org/10.3390/v16020282
Chicago/Turabian StyleSu, Xia, Hongzhuan Zhou, Ziwei Han, Fuzhou Xu, Bing Xiao, Jin Zhang, Qi Qi, Lulu Lin, Huanhuan Zhang, Songping Li, and et al. 2024. "Transcriptional Differential Analysis of Nitazoxanide-Mediated Anticanine Parvovirus Effect in F81 Cells" Viruses 16, no. 2: 282. https://doi.org/10.3390/v16020282
APA StyleSu, X., Zhou, H., Han, Z., Xu, F., Xiao, B., Zhang, J., Qi, Q., Lin, L., Zhang, H., Li, S., & Yang, B. (2024). Transcriptional Differential Analysis of Nitazoxanide-Mediated Anticanine Parvovirus Effect in F81 Cells. Viruses, 16(2), 282. https://doi.org/10.3390/v16020282




