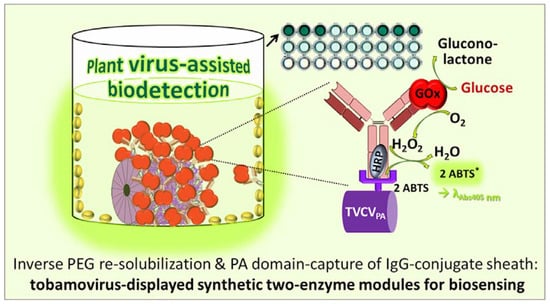
Facile Purification and Use of Tobamoviral Nanocarriers for Antibody-Mediated Display of a Two-Enzyme System
Abstract
:1. Introduction
2. Material and Methods
2.1. Plant Infection
2.1.1. Delivery of Vector-Encoded Viral Constructs via Agroinfiltration
2.1.2. Mechanical Inoculation of Plants with Leaf Extracts or Purified Virus
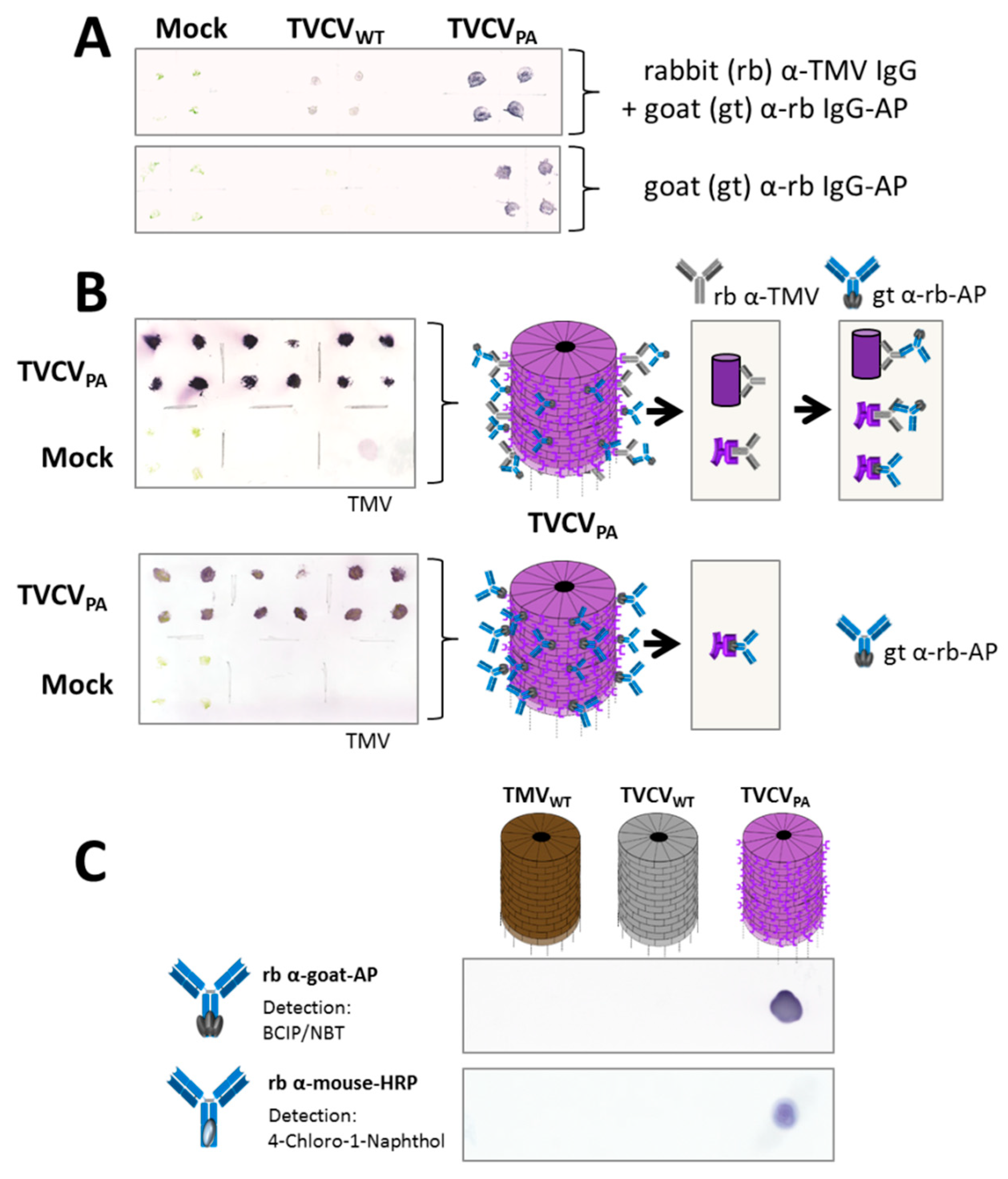
2.1.3. Plant Harvest and Virus Immunodetection on Tissue Blots
2.1.4. Virus Isolation via Stepwise Enrichment: Butanol Extraction, Two-Fold PEG Precipitation, and Ultracentrifugation
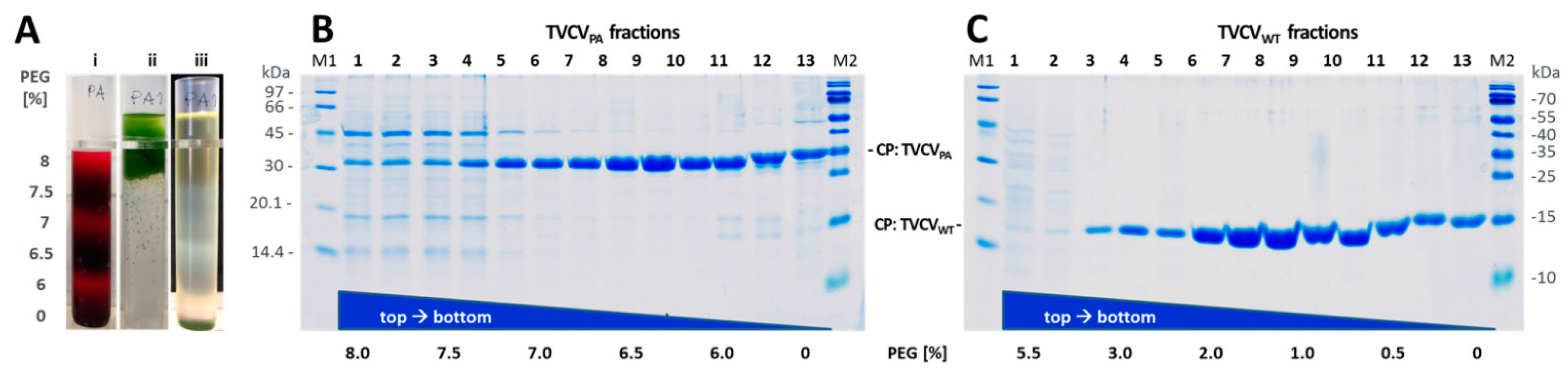
2.1.5. Virus Isolation via Precipitates from Cleared Crude Extracts: Selective Inverse PEG-Sucrose Solubility Gradients
2.2. Characterization of Virus Preparations
2.2.1. Gel Electrophoresis
2.2.2. Transmission Electron Microscopy (TEM)
2.2.3. Gold Labeling of TVCVPA via Antibody Capture
2.2.4. Antibody Immobilization on TVCVPA in Solution and Pull-Down Assay
2.3. Antibody-Mediated Display of Functional Molecules on TVCVPA on Solid Supports
2.3.1. IgG–Enzyme Conjugate-Coupling and Enzyme Activities on Dot-Blots
2.3.2. Microtiter Plate Coating with TVCV Variants and IgG or IgG Conjugate Binding
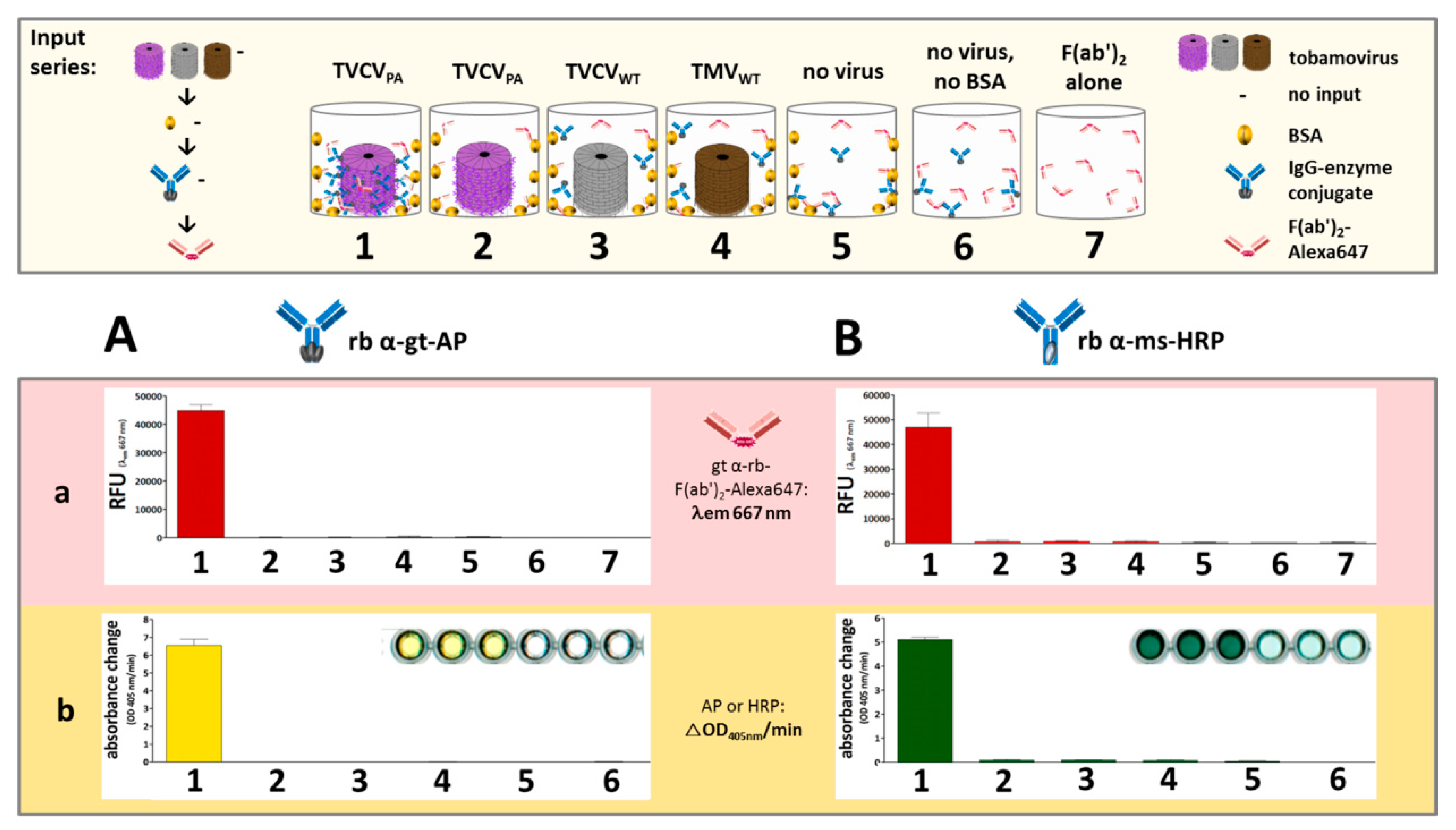
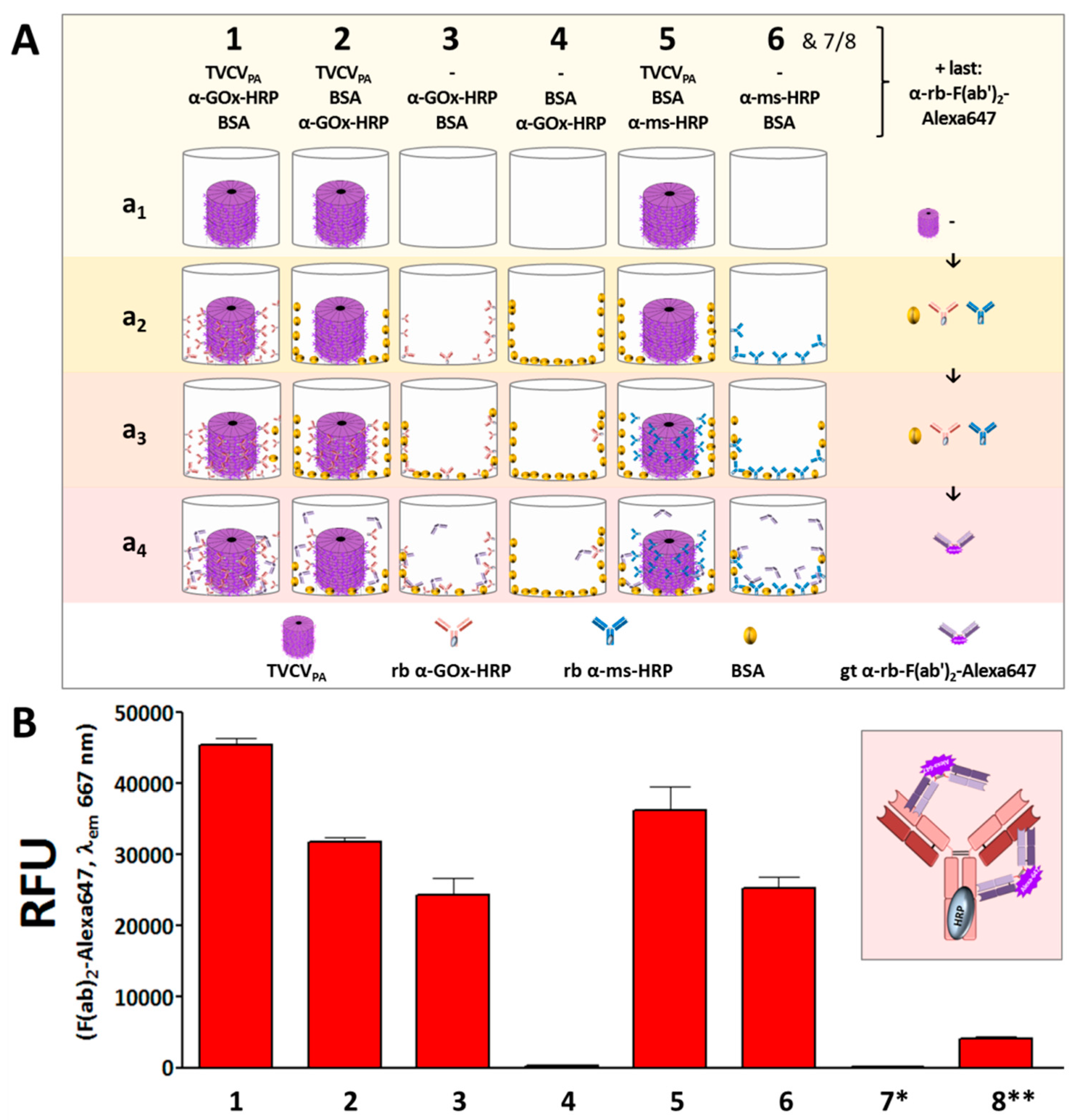
2.3.3. Detection of Bound Antibody Conjugates with Fluorescent F(ab′)2-Fragments
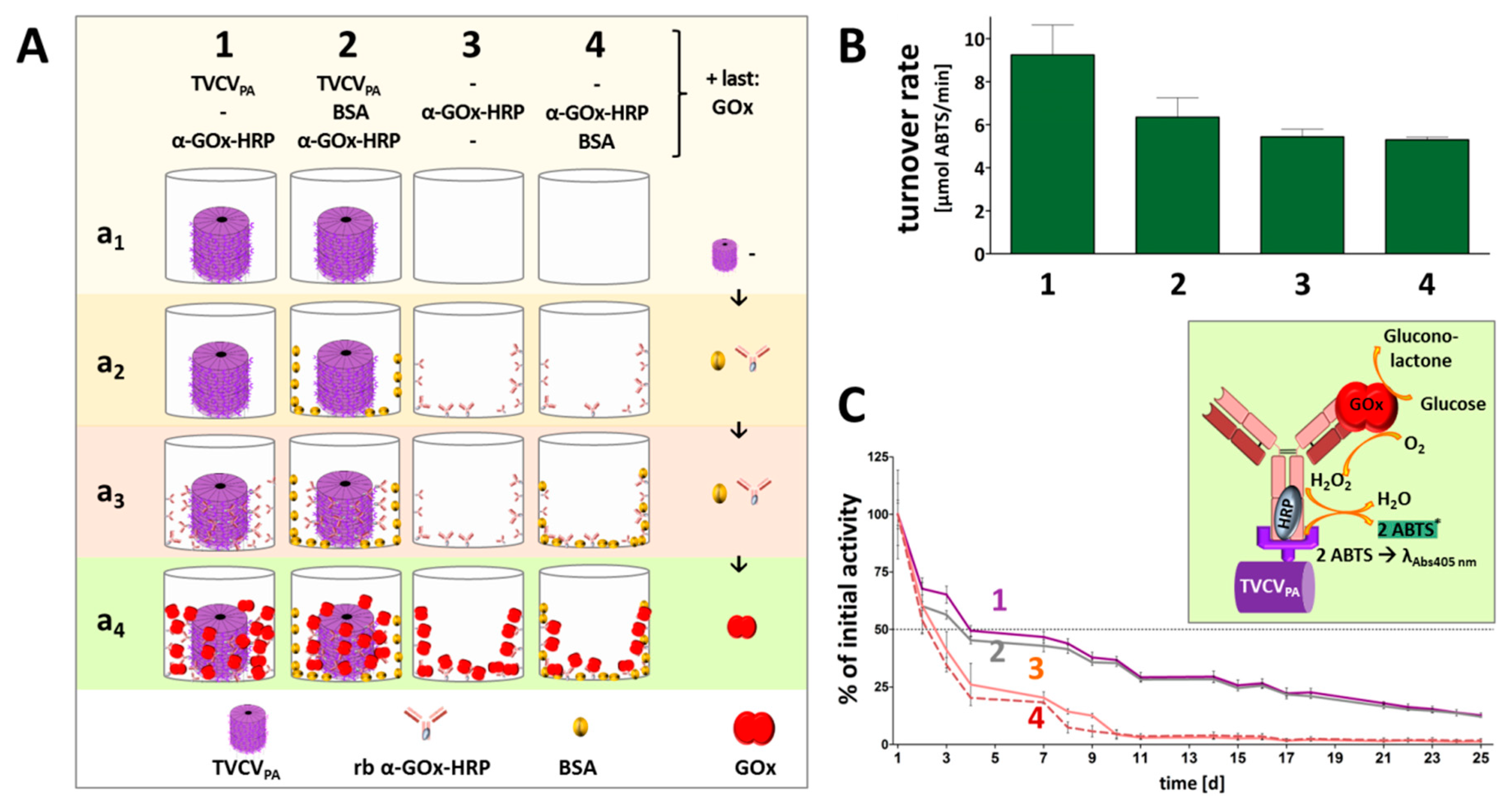
2.4. Antibody-Mediated Immobilization of Enzymes or Control Protein on TVCVPA on Microtiter Plate Surfaces and Activity/Functionality Measurement
2.5. Sensor Chip Preparation and Immobilization of PAH (poly(allylamine hydrochloride), TVCVPA, TVCVWT, and TMV
2.6. Scanning Electron Microscopy (SEM)
2.7. Statistical Evaluation
3. Results
3.1. TVCVWT and TVCVPA Production, Isolation and Characterization
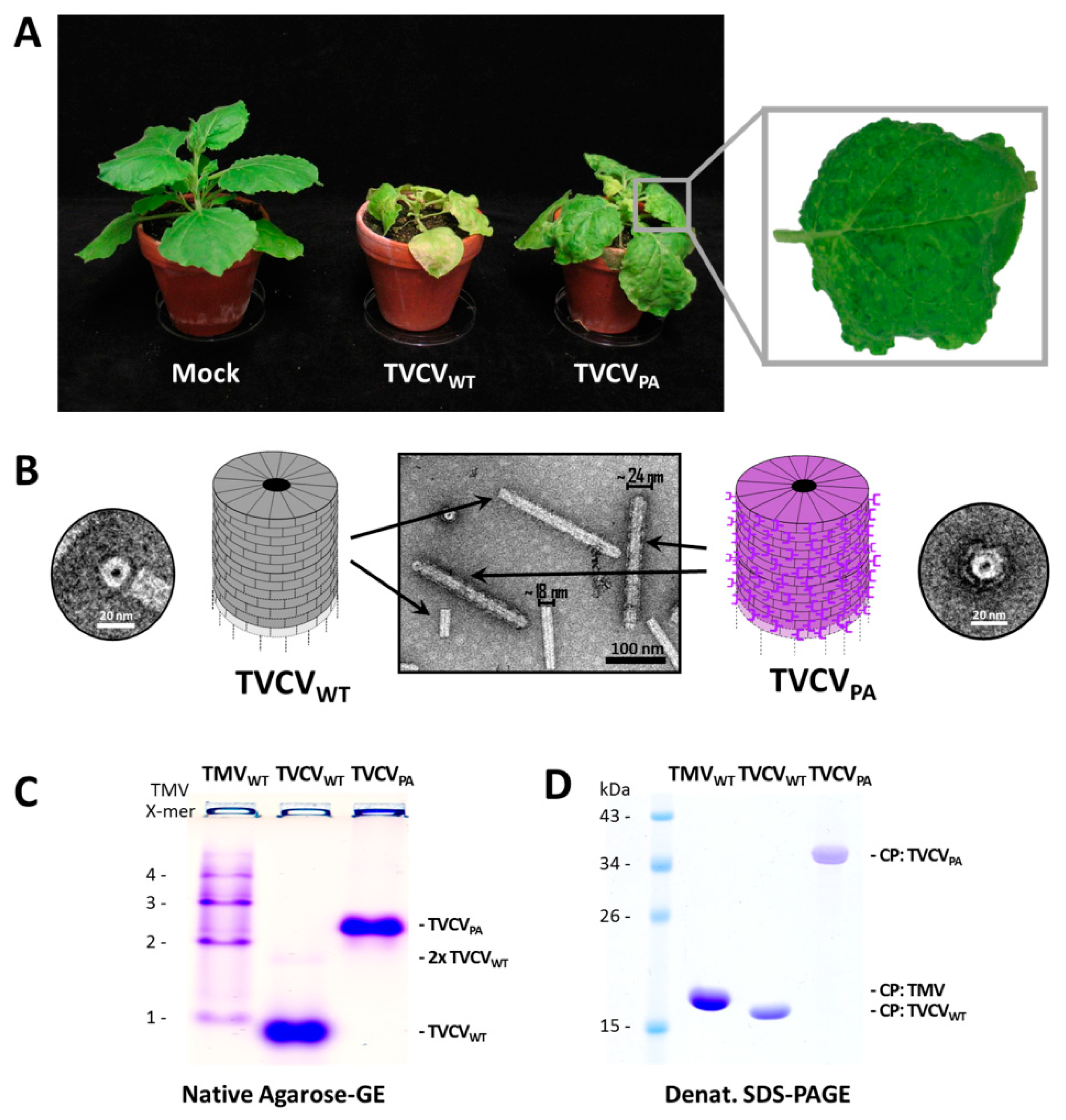
3.1.1. Both TVCVWT and TVCVPA Accumulate Efficiently in N. benthamiana, but Only the Wildtype Virus Infects N. tabacum
3.1.2. A Standard Tobamovirus Purification Procedure via Stepwise Enrichment Needs Adaptation for TVCVPA and Yields the Expected Shortened Particles
3.1.3. Virus Isolation from Crude Precipitates by Selective Inverse PEG-Sucrose Solubility Gradients Reduces Effort and Improves Particle Integrity
3.2. Antibody Display on TVCVPA as Plant Virus-Based Immunosorbent Nanoparticle (VIN)
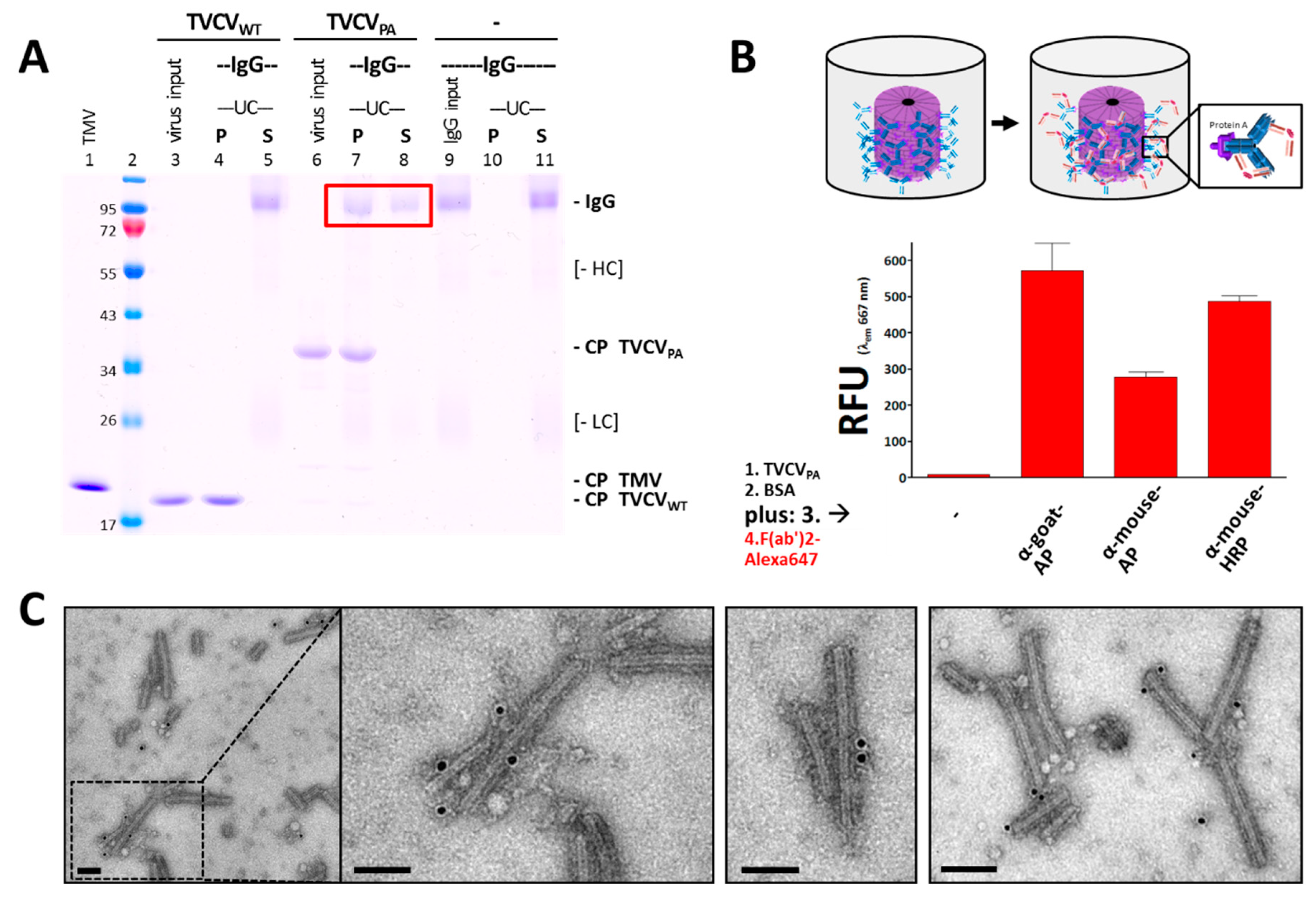
3.2.1. Selective Binding of Rabbit IgGs to Purified TVCVPA
3.2.2. Antibody Fc Conjugates with Enzymes or Gold Nanoparticles Bind to TVCVPA
3.3. Display of Functional Enzymes on TVCVPA Nanocarriers
Rabbit and Goat Antibody-Conjugated Enzymes Remain Active after Coupling to TVCVPA Nanocarriers
3.4. A Two Enzyme-Cascade of GOx and HRP Installed on TVCVPA via the Capture of a Single Antibody-Conjugate
3.4.1. Anti-GOx Antibodies Conjugated with HRP Are Immobilized to Superior Amounts in Microtiter Plates via TVCVPA Adapters
3.4.2. TVCVPA-Displayed HRP-Conjugated AntiGOx–IgGs Tether Both Enzymes into a Durable Cooperating System
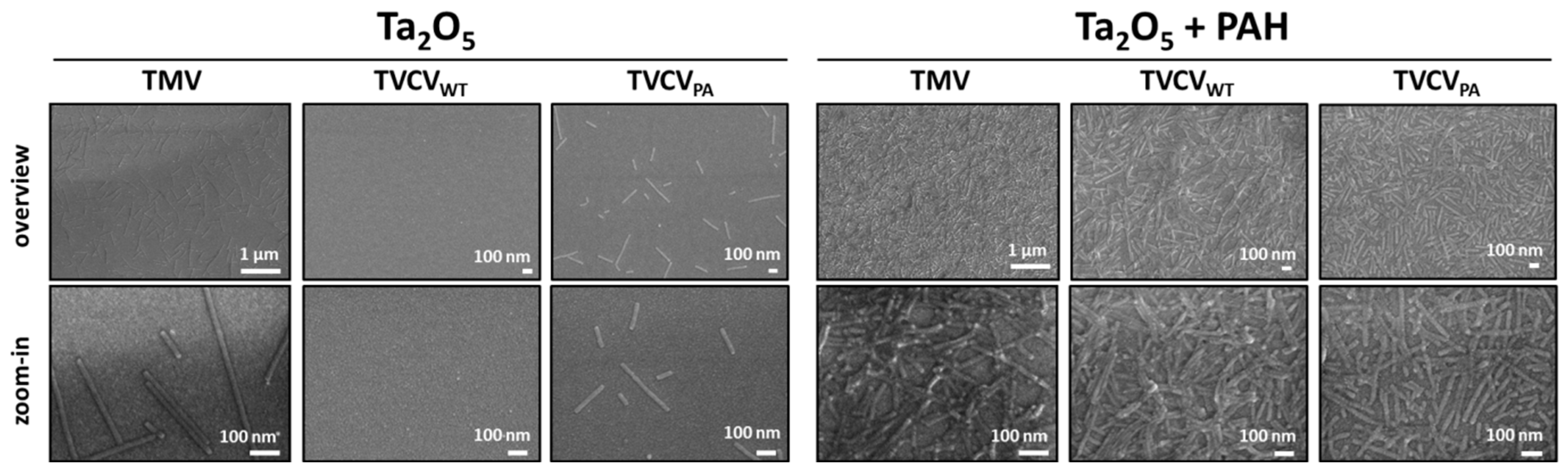
3.5. PAH-Promoted Immobilization of TVCVPA Layers at High Surface Densities: Towards Applications in Biosensor Devices
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alonso, J.M.; Górzny, M.Ł.; Bittner, A.M. The physics of tobacco mosaic virus and virus-based devices in biotechnology. Trends Biotechnol. 2013, 31, 530–538. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.A.; Niu, Z.W.; Wang, Q. Viruses and virus-like protein assemblies—Chemically programmable nanoscale building blocks. Nano Res. 2009, 2, 349–364. [Google Scholar] [CrossRef]
- Wen, A.M.; Steinmetz, N.F. Design of virus-based nanomaterials for medicine, biotechnology, and energy. Chem. Soc. Rev. 2016, 45, 4074–4126. [Google Scholar] [CrossRef] [PubMed]
- Soto, C.M.; Ratna, B.R. Virus hybrids as nanomaterials for biotechnology. Curr. Opin. Biotechnol. 2010, 21, 426–438. [Google Scholar] [CrossRef] [PubMed]
- Glasgow, J.; Tullman-Ercek, D. Production and applications of engineered viral capsids. Appl. Microbiol. Biotechnol. 2014, 98, 5847–5858. [Google Scholar] [CrossRef]
- Li, F.; Wang, Q. Fabrication of nanoarchitectures templated by virus-based nanoparticles: Strategies and applications. Small 2014, 10, 230–245. [Google Scholar] [CrossRef]
- Manchester, M.; Singh, P. Virus-based nanoparticles (VNPs): Platform technologies for diagnostic imaging. Adv. Drug Deliv. Rev. 2006, 58, 1505–1522. [Google Scholar] [CrossRef]
- Hull, R. Plant Virology, 5th ed.; Academic Press: Boston, MA, USA, 2014; p. 1118. [Google Scholar]
- Mao, C.; Liu, A.; Cao, B. Virus-based chemical and biological sensing. Angew. Chem. Int. Ed. 2009, 48, 6790–6810. [Google Scholar] [CrossRef]
- Khudyakov, Y.; Pumpens, P. Viral Nanotechnology; CRC Press, Taylor & Francis: Boca Raton, FL, USA; London, UK; New York, NY, USA, 2016. [Google Scholar]
- Chen, M.Y.; Butler, S.S.; Chen, W.; Suh, J. Physical, chemical, and synthetic virology: Reprogramming viruses as controllable nanodevices. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2019, 11, e1545. [Google Scholar] [CrossRef]
- Chung, Y.H.; Church, D.; Koellhoffer, E.C.; Osota, E.; Shukla, S.; Rybicki, E.P.; Pokorski, J.K.; Steinmetz, N.F. Integrating plant molecular farming and materials research for next-generation vaccines. Nat. Rev. Mater. 2021, 7, 372–388. [Google Scholar] [CrossRef]
- Shukla, S.; Hu, H.; Cai, H.; Chan, S.-K.; Boone, C.E.; Beiss, V.; Chariou, P.L.; Steinmetz, N.F. Plant viruses and bacteriophage-based reagents for diagnosis and therapy. Annu. Rev. Virol. 2020, 7, 559–587. [Google Scholar] [CrossRef] [PubMed]
- Wen, A.M.; Lee, K.L.; Steinmetz, N.F. Plant Virus-Based Nanotechnologies. In Women in Nanotechnology: Contributions from the Atomic Level and Up; Norris, P.M., Friedersdorf, L.E., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 57–69. [Google Scholar]
- Eiben, S.; Koch, C.; Altintoprak, K.; Southan, A.; Tovar, G.; Laschat, S.; Weiss, I.; Wege, C. Plant virus-based materials for biomedical applications: Trends and prospects. Adv. Drug Deliv. Rev. 2019, 145, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Cuenca, S.; Mansilla, C.; Aguado, M.; Yuste-Calvo, C.; Sánchez, F.; Sánchez-Montero, J.M.; Ponz, F. Nanonets derived from turnip mosaic virus as scaffolds for increased enzymatic activity of immobilized Candida antarctica lipase B. Front. Plant Sci. 2016, 7, 464. [Google Scholar] [CrossRef] [PubMed]
- Yuste-Calvo, C.; González-Gamboa, I.; Pacios, L.F.; Sánchez, F.; Ponz, F. Structure-based multifunctionalization of flexuous elongated viral nanoparticles. ACS Omega 2019, 4, 5019–5028. [Google Scholar] [CrossRef]
- Dickmeis, C.; Kauth, L.; Commandeur, U. From infection to healing: The use of plant viruses in bioactive hydrogels. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2021, 13, e1662. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Qiao, J.; Niu, Z.; Wang, Q. Natural supramolecular building blocks: From virus coat proteins to viral nanoparticles. Chem. Soc. Rev. 2012, 41, 6178–6194. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.Z.; Naves, L.; Siwak, N.P.; Brown, A.; Culver, J.; Ghodssi, R. Integration of genetically modified virus-like-particles with an optical resonator for selective bio-detection. Nanotechnology 2015, 26, 205501. [Google Scholar] [CrossRef]
- Wu, F.; Zhang, H.; Zhou, Q.; Wu, M.; Ballard, Z.; Tian, Y.; Wang, J.; Niu, Z.; Huang, Y. Expanding the genetic code for site-specific labelling of tobacco mosaic virus coat protein and building biotin-functionalized virus-like particles. Chem. Commun. 2014, 50, 4007–4009. [Google Scholar] [CrossRef]
- Culver, J.N.; Brown, A.D.; Zang, F.; Gnerlich, M.; Gerasopoulos, K.; Ghodssi, R. Plant virus directed fabrication of nanoscale materials and devices. Virology 2015, 479–480, 200–212. [Google Scholar] [CrossRef]
- Lico, C.; Benvenuto, E.; Baschieri, S. The two-faced Potato Virus X: From plant pathogen to smart nanoparticle. Front. Plant Sci. 2015, 6, 1009. [Google Scholar] [CrossRef]
- Koch, C.; Eber, F.J.; Azucena, C.; Foerste, A.; Walheim, S.; Schimmel, T.; Bittner, A.; Jeske, H.; Gliemann, H.; Eiben, S.; et al. Novel roles for well-known players: From tobacco mosaic virus pests to enzymatically active assemblies. Beilstein J. Nanotechnol. 2016, 7, 613–629. [Google Scholar] [CrossRef] [PubMed]
- Schuphan, J.; Commandeur, U. Analysis of engineered tobacco mosaic virus and potato virus X nanoparticles as carriers for biocatalysts. Front. Plant Sci. 2021, 12, 710869. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.Z.; Pomerantseva, E.; Gnerlich, M.; Brown, A.; Gerasopoulos, K.; McCarthy, M.; Culver, J.; Ghodssi, R. Tobacco mosaic virus: A biological building block for micro/nano/biosystems. J. Vac. Sci. Technol. A 2013, 31, 050815. [Google Scholar] [CrossRef]
- Lee, K.Z.; Basnayake Pussepitiyalage, V.; Lee, Y.-H.; Loesch-Fries, L.S.; Harris, M.T.; Hemmati, S.; Solomon, K.V. Engineering tobacco mosaic virus and its virus-like-particles for synthesis of biotemplated nanomaterials. Biotechnol. J. 2021, 16, 2000311. [Google Scholar] [CrossRef] [PubMed]
- Röder, J.; Dickmeis, C.; Commandeur, U. Small, smaller, nano: New applications for potato virus X in nanotechnology. Front. Plant Sci. 2019, 10, 158. [Google Scholar] [CrossRef]
- Vaidya, A.J.; Solomon, K.V. Surface functionalization of rod-shaped viral particles for biomedical applications. ACS Appl. Bio Mater. 2022, 5, 1980–1989. [Google Scholar] [CrossRef]
- Venkataraman, S.; Hefferon, K. Application of plant viruses in biotechnology, medicine, and human health. Viruses 2021, 13, 1697. [Google Scholar] [CrossRef]
- Wege, C.; Koch, C. From stars to stripes: RNA-directed shaping of plant viral protein templates—Structural synthetic virology for smart biohybrid nanostructures. WIREs Nanomed. Nanobiotechnol. 2020, 12, e1591. [Google Scholar] [CrossRef]
- Hou, C.; Xu, H.; Jiang, X.; Li, Y.; Deng, S.; Zang, M.; Xu, J.; Liu, J. Virus-based supramolecular structure and materials: Concept and prospects. ACS Appl. Bio Mater. 2021, 4, 5961–5974. [Google Scholar] [CrossRef]
- Nellist, C.F.; Ohshima, K.; Ponz, F.; Walsh, J.A. Turnip mosaic virus, a virus for all seasons. Ann. Appl. Biol. 2022, 180, 312–327. [Google Scholar] [CrossRef]
- Zahmanova, G.; Aljabali, A.A.; Takova, K.; Toneva, V.; Tambuwala, M.M.; Andonov, A.P.; Lukov, G.L.; Minkov, I. The plant viruses and molecular farming: How beneficial they might be for human and animal health? Int. J. Mol. Sci. 2023, 24, 1533. [Google Scholar] [CrossRef]
- Lomonossoff, G.P.; Wege, C. Chapter Six—TMV Particles: The Journey From Fundamental Studies to Bionanotechnology Applications. In Advances in Virus Research; Palukaitis, P., Roossinck, M.J., Eds.; Academic Press: Cambridge, MA, USA, 2018; Volume 102, pp. 149–176. [Google Scholar]
- Jablonski, M.; Poghossian, A.; Severins, R.; Keusgen, M.; Wege, C.; Schöning, M.J. Capacitive field-effect biosensor studying adsorption of tobacco mosaic virus particles. Micromachines 2021, 12, 57. [Google Scholar] [CrossRef] [PubMed]
- Hou, C.; Luo, Q.; Liu, J.; Miao, L.; Zhang, C.; Gao, Y.; Zhang, X.; Xu, J.; Dong, Z.; Liu, J. Construction of GPx active centers on natural protein nanodisk/nanotube: A new way to develop artificial nanoenzyme. ACS Nano 2012, 6, 8692–8701. [Google Scholar] [CrossRef] [PubMed]
- Zang, F.; Gerasopoulos, K.; Brown, A.D.; Culver, J.N.; Ghodssi, R. Capillary microfluidics-assembled virus-like particle bionanoreceptor interfaces for label-free biosensing. ACS Appl. Mater. Interfaces 2017, 9, 8471–8479. [Google Scholar] [CrossRef]
- Zang, F.; Gerasopoulos, K.; Fan, X.Z.; Brown, A.D.; Culver, J.N.; Ghodssi, R. An electrochemical sensor for selective TNT sensing based on Tobacco mosaic virus-like particle binding agents. Chem. Commun. 2014, 50, 12977–12980. [Google Scholar] [CrossRef] [PubMed]
- Zang, F.; Gerasopoulos, K.; Fan, X.Z.; Brown, A.D.; Culver, J.N.; Ghodssi, R. Real-time monitoring of macromolecular biosensing probe self-assembly and on-chip ELISA using impedimetric microsensors. Biosens. Bioelectron. 2016, 81, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Welden, M.; Poghossian, A.; Vahidpour, F.; Wendlandt, T.; Keusgen, M.; Christina, W.; Schöning, M.J. Capacitive field-effect biosensor modified with a stacked bilayer of weak polyelectrolyte and plant virus particles as enzyme nanocarriers. Bioelectrochemistry 2023, 151, 108397. [Google Scholar] [CrossRef] [PubMed]
- Welden, M.; Poghossian, A.; Vahidpour, F.; Wendlandt, T.; Keusgen, M.; Wege, C.; Schöning, M.J. Towards multi-analyte detection with field-effect capacitors modified with tobacco mosaic virus bioparticles as enzyme nanocarriers. Biosensors 2022, 12, 43. [Google Scholar] [CrossRef]
- Paiva, T.O.; Schneider, A.; Bataille, L.; Chovin, A.; Anne, A.; Michon, T.; Wege, C.; Demaille, C. Enzymatic activity of individual bioelectrocatalytic viral nanoparticles: Dependence of catalysis on the viral scaffold and its length. Nanoscale 2022, 14, 875–889. [Google Scholar] [CrossRef]
- Welden, M.; Severins, R.; Poghossian, A.; Wege, C.; Bongaerts, J.; Siegert, P.; Keusgen, M.; Schöning, M.J. Detection of acetoin and diacetyl by a tobacco mosaic virus-assisted field-effect biosensor. Chemosensors 2022, 10, 218. [Google Scholar] [CrossRef]
- Welden, R.; Jablonski, M.; Wege, C.; Keusgen, M.; Wagner, P.H.; Wagner, T.; Schöning, M.J. Light-addressable actuator-sensor platform for monitoring and manipulation of pH gradients in microfluidics: A case study with the enzyme penicillinase. Biosensors 2021, 11, 171. [Google Scholar] [CrossRef] [PubMed]
- Koch, C.; Poghossian, A.; Schöning, M.J.; Wege, C. Penicillin detection by tobacco mosaic virus-assisted colorimetric biosensors. Nanotheranostics 2018, 2, 184–196. [Google Scholar] [CrossRef] [PubMed]
- Koch, C.; Poghossian, A.; Wege, C.; Schöning, M.J. TMV-Based Adapter Templates for Enhanced Enzyme Loading in Biosensor Applications. In Virus-Derived Nanoparticles for Advanced Technologies; Methods in Molecular Biology; Wege, C., Lomonossoff, G.P., Walker, J.M., Eds.; Humana Press, Springer Science+Business Media: Heidelberg, Germany; London, UK; New York, NY, USA, 2018; Volume 1776, pp. 553–568. [Google Scholar]
- Poghossian, A.; Jablonski, M.; Koch, C.; Bronder, T.S.; Rolka, D.; Wege, C.; Schöning, M.J. Field-effect biosensor using virus particles as scaffolds for enzyme immobilization. Biosens. Bioelectron. 2018, 110, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Bäcker, M.; Koch, C.; Eiben, S.; Geiger, F.; Eber, F.J.; Gliemann, H.; Poghossian, A.; Wege, C.; Schöning, M.J. Tobacco mosaic virus as enzyme nanocarrier for electrochemical biosensors. Sens. Actuators B 2017, 238, 716–722. [Google Scholar] [CrossRef]
- Koch, C.; Wabbel, K.; Eber, F.J.; Krolla-Sidenstein, P.; Azucena, C.; Gliemann, H.; Eiben, S.; Geiger, F.; Wege, C. Modified TMV particles as beneficial scaffolds to present sensor enzymes. Front. Plant Sci. 2015, 6, 1137. [Google Scholar] [CrossRef]
- Geiger, F.; Wendlandt, T.; Berking, T.; Spatz, J.P.; Wege, C. Convenient site-selective protein coupling from bacterial raw lysates to coenzyme A-modified tobacco mosaic virus (TMV) by Bacillus subtilis Sfp phosphopantetheinyl transferase. Virology 2023, 578, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Chapman, S.; Faulkner, C.; Kaiserli, E.; Garcia-Mata, C.; Savenkov, E.I.; Roberts, A.G.; Oparka, K.J.; Christie, J.M. The photoreversible fluorescent protein iLOV outperforms GFP as a reporter of plant virus infection. Proc. Natl. Acad. Sci. USA 2008, 105, 20038–20043. [Google Scholar] [CrossRef] [PubMed]
- Röder, J.; Fischer, R.; Commandeur, U. Adoption of the 2A ribosomal skip principle to tobacco mosaic virus for peptide display. Front. Plant Sci. 2017, 8, 1125. [Google Scholar] [CrossRef]
- Lartey, R.T.; Voss, T.C.; Melcher, U. Completion of a cDNA sequence from a tobamovirus pathogenic to crucifers. Gene 1995, 166, 331–332. [Google Scholar] [CrossRef]
- Lartey, R.T.; Lane, L.C.; Melcher, U. Electron microscopic and molecular characterization of turnip vein-clearing virus. Arch. Virol. 1994, 138, 287–298. [Google Scholar] [CrossRef]
- Lartey, R.T.; Voss, T.C.; Melcher, U. Erratum. Gene 1997, 188, 295. [Google Scholar] [CrossRef]
- Werner, S.; Marillonnet, S.; Hause, G.; Klimyuk, V.; Gleba, Y. Immunoabsorbent nanoparticles based on a tobamovirus displaying protein A. Proc. Natl. Acad. Sci. USA 2006, 103, 17678–17683. [Google Scholar] [CrossRef] [PubMed]
- McNulty, M.J.; Schwartz, A.; Delzio, J.; Karuppanan, K.; Jacobson, A.; Hart, O.; Dandekar, A.; Giritch, A.; Nandi, S.; Gleba, Y.; et al. Affinity sedimentation and magnetic separation with plant-made immunosorbent nanoparticles for therapeutic protein purification. Front. Bioeng. Biotechnol. 2022, 10, 865481. [Google Scholar] [CrossRef] [PubMed]
- McNulty, M.J.; Hamada, N.; Delzio, J.; McKee, L.; Nandi, S.; Longo, M.L.; McDonald, K.A. Functionalizing silica sol–gel with entrapped plant virus-based immunosorbent nanoparticles. J. Nanobiotechnol. 2022, 20, 105. [Google Scholar] [CrossRef]
- Poghossian, A.; Jablonski, M.; Molinnus, D.; Wege, C.; Schöning, M.J. Field-effect sensors for virus detection: From Ebola to SARS-CoV-2 and plant viral enhancers. Front. Plant Sci. 2020, 11, 598103. [Google Scholar] [CrossRef]
- Adams, M.J.; Adkins, S.; Bragard, C.; Gilmer, D.; Li, D.; MacFarlane, S.A.; Wong, S.-M.; Melcher, U.; Ratti, C.; Ryu, K.H.; et al. ICTV Virus Taxonomy Profile: Virgaviridae. J. Gen. Virol. 2017, 98, 1999–2000. [Google Scholar] [CrossRef]
- Gibbs, A.J.; Wood, J.; Garcia-Arenal, F.; Ohshima, K.; Armstrong, J.S. Tobamoviruses have probably co-diverged with their eudicotyledonous hosts for at least 110 million years. Virus Evol. 2015, 1, vev019. [Google Scholar] [CrossRef]
- Adams, M.J.; Heinze, C.; Jackson, A.O.; Kreuze, J.F.; MacFarlane, S.A.; Torrance, L. Family Virgaviridae. In Virus Taxonomy. Classification and Nomenclature of Viruses. Ninth Report of the International Committee on Taxonomy of Viruses; King, A.M.Q., Adams, M.J., Carstens, E.B., Lefkowitz, E.J., Eds.; Elsevier/Academic Press: Amsterdam, The Netherlands; Boston, MA, USA; Heidelberg, Germany; London, UK; New York, NY, USA; Oxford, UK; Paris, France; San Diego, CA, USA; San Francisco, CA, USA; Singapore; Sydney, Australia; Tokyo, Japan, 2012; pp. 1139–1162. [Google Scholar]
- Min, B.E.; Chung, B.N.; Kim, M.J.; Ha, J.H.; Lee, B.Y.; Ryu, K.H. Cactus mild mottle virus is a new cactus-infecting tobamovirus. Arch. Virol. 2006, 151, 13–21. [Google Scholar] [CrossRef]
- Ilyas, R.; Rohde, M.J.; Richert-Pöggeler, K.R.; Ziebell, H. To be seen or not to be seen: Latent infection by tobamoviruses. Plants 2022, 11, 2166. [Google Scholar] [CrossRef]
- Lartey, R.T.; Voss, T.C.; Melcher, U. Tobamovirus evolution: Gene overlaps, recombination, and taxonomic implications. Mol. Biol. Evol. 1996, 13, 1327–1338. [Google Scholar] [CrossRef]
- Levy, A.; Zheng, J.Y.; Lazarowitz, S.G. The tobamovirus Turnip Vein Clearing Virus 30-kilodalton movement protein localizes to novel nuclear filaments to enhance virus infection. J. Virol. 2013, 87, 6428–6440. [Google Scholar] [CrossRef]
- Zhang, Y.; Lartey, R.; Hartson, S.; Voss, T.; Melcher, U. Limitations to tobacco mosaic virus infection of turnip. Arch. Virol. 1999, 144, 957–971. [Google Scholar] [CrossRef] [PubMed]
- Melcher, U. Turnip vein-clearing virus, from pathogen to host expression profile. Mol. Plant Pathol. 2003, 4, 133–140. [Google Scholar] [CrossRef]
- Goelet, P.; Lomonossoff, G.P.; Butler, P.J.; Akam, M.E.; Gait, M.J.; Karn, J. Nucleotide sequence of tobacco mosaic virus RNA. Proc. Natl. Acad. Sci. USA 1982, 79, 5818–5822. [Google Scholar] [CrossRef]
- Kozlowski, L.P. IPC 2.0: Prediction of isoelectric point and pKa dissociation constants. Nucleic Acids Res. 2021, 49, W285–W292. [Google Scholar] [CrossRef]
- Mickoleit, F.; Altintoprak, K.; Wenz, N.L.; Richter, R.; Wege, C.; Schueler, D. Precise assembly of genetically functionalized magnetosomes and tobacco mosaic virus particles generates a magnetic biocomposite. ACS Appl. Mater. Interfaces 2018, 10, 37898–37910. [Google Scholar] [CrossRef]
- Ge, P.; Zhou, Z.H. Hydrogen-bonding networks and RNA bases revealed by cryo electron microscopy suggest a triggering mechanism for calcium switches. Proc. Natl. Acad. Sci. USA 2011, 108, 9637–9642. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Marillonnet, S.; Thoeringer, C.; Kandzia, R.; Klimyuk, V.; Gleba, Y. Systemic Agrobacterium tumefaciens–mediated transfection of viral replicons for efficient transient expression in plants. Nat. Biotechnol. 2005, 23, 718–723. [Google Scholar] [CrossRef] [PubMed]
- Marillonnet, S.; Giritch, A.; Gils, M.; Kandzia, R.; Klimyuk, V.; Gleba, Y. In planta engineering of viral RNA replicons: Efficient assembly by recombination of DNA modules delivered by Agrobacterium. Proc. Natl. Acad. Sci. USA 2004, 101, 6852–6857. [Google Scholar] [CrossRef]
- Kadri, A.; Maiß, E.; Amsharov, N.; Bittner, A.M.; Balci, S.; Kern, K.; Jeske, H.; Wege, C. Engineered Tobacco mosaic virus mutants with distinct physical characteristics in planta and enhanced metallization properties. Virus Res. 2011, 157, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Wege, C.; Geiger, F. Dual Functionalization of Rod-Shaped Viruses on Single Coat Protein Subunits. In Virus-Derived Nanoparticles for Advanced Technologies; Methods in Molecular Biology; Wege, C., Lomonossoff, G.P., Walker, J.M., Eds.; Humana Press, Springer Science+Business Media: Heidelberg, Germany; London, UK; New York, NY, USA, 2018; Volume 1776, pp. 405–424. [Google Scholar]
- Cassab, G.I.; Varner, J.E. Immunocytolocalization of extensin in developing soybean seed coats by immunogold-silver staining and by tissue printing on nitrocellulose paper. J. Cell Biol. 1987, 105, 2581–2588. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J. Molecular Cloning: A Laboratory Manual, 4th ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2012. [Google Scholar]
- Chapman, S.N. Tobamovirus Isolation and RNA Extraction. In Methods in Molecular Biology: Plant Virology Protocols: From Virus Isolation to Transgenic Resistance; Foster, G.D., Taylor, S.C., Eds.; Humana Press Inc.: Totowa, NJ, USA, 1998; Volume 81, pp. 123–129. [Google Scholar]
- Gooding, G.V.; Hebert, T.T. A simple technique for purification of tobacco mosaic virus in large quantities. Phytopathology 1967, 57, 1285. [Google Scholar] [PubMed]
- Dickmeis, C.; Altintoprak, K.; van Rijn, P.; Wege, C.; Commandeur, U. Bioinspired Silica Mineralization on Viral Templates. In Virus-Derived Nanoparticles for Advanced Technologies; Methods in Molecular Biology; Wege, C., Lomonossoff, G.P., Walker, J.M., Eds.; Humana Press, Springer Science+Business Media: Heidelberg, Germany; London, UK; New York, NY, USA, 2018; Volume 1776, pp. 337–362. [Google Scholar]
- Zaitlin, M.; Israel, H.W. Tobacco Mosaic Virus (Type Strain). AAB Descr. Plant Viruses 1975, DPV No. 151. Available online: https://www.dpvweb.net/dpv/showdpv/?dpvno=151 (accessed on 12 September 2023).
- Clark, M.F.; Lister, R.M. The application of polyethylene glycol solubility-concentration gradients in plant virus research. Virology 1971, 43, 338–351. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Wege, C.; Eber, F. Bottom-Up Assembly of TMV-Based Nucleoprotein Architectures on Solid Supports. In Virus-Derived Nanoparticles for Advanced Technologies; Methods in Molecular Biology; Wege, C., Lomonossoff, G.P., Walker, J.M., Eds.; Humana Press, Springer Science+Business Media: Heidelberg, Germany; London, UK; New York, NY, USA, 2018; pp. 169–186. [Google Scholar]
- Rasband, W.S. ImageJ. U. S. National Institutes of Health, Bethesda, Maryland, USA 1997–2011. Available online: https://imagej.net/ij/ (accessed on 12 September 2023).
- Poghossian, A.; Schöning, M.J. Capacitive field-sffect EIS chemical sensors and biosensors: A status report. Sensors 2020, 20, 5639. [Google Scholar] [CrossRef]
- Poghossian, A.; Weil, M.; Cherstvy, A.G.; Schöning, M.J. Electrical monitoring of polyelectrolyte multilayer formation by means of capacitive field-effect devices. Anal. Bioanal. Chem. 2013, 405, 6425–6436. [Google Scholar] [CrossRef]
- Bronder, T.S.; Poghossian, A.; Jessing, M.P.; Keusgen, M.; Schöning, M.J. Surface regeneration and reusability of label-free DNA biosensors based on weak polyelectrolyte-modified capacitive field-effect structures. Biosens. Bioelectron. 2019, 126, 510–517. [Google Scholar] [CrossRef]
- Guss, B.; Eliasson, M.; Olsson, A.; Uhlen, M.; Frej, A.; Jörnvall, H.; Flock, J.; Lindberg, M. Structure of the IgG-binding regions of streptococcal protein G. EMBO J. 1986, 5, 1567–1575. [Google Scholar] [CrossRef]
- Goudswaard, J.; Donk, J.v.d.; Noordzij, A.; Dam, R.v.; Vaerman, J.P. Protein A reactivity of various mammalian immunoglobulins. Scand. J. Immunol. 1978, 8, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Langone, J.J. [125I] protein A: A tracer for general use in immunoassay. J. Immunol. Methods 1978, 24, 269–285. [Google Scholar] [CrossRef] [PubMed]
- Brown, N.L.; Bottomley, S.P.; Scawen, M.D.; Gore, M.G. A study of the interactions between an IgG-binding domain based on the B domain of staphylococcal protein A and rabbit IgG. Mol. Biotechnol. 1998, 10, 9–16. [Google Scholar] [CrossRef]
- Starovasnik, M.A.; O’Connell, M.P.; Fairbrother, W.J.; Kelley, R.F. Antibody variable region binding by Staphylococcal protein A: Thermodynamic analysis and location of the Fv binding site on E-domain. Protein Sci. 1999, 8, 1423–1431. [Google Scholar] [CrossRef] [PubMed]
- Mateo, C.; Palomo, J.M.; Fernandez-Lorente, G.; Guisan, J.M.; Fernandez-Lafuente, R. Improvement of enzyme activity, stability and selectivity via immobilization techniques. Enzym. Microb. Technol. 2007, 40, 1451–1463. [Google Scholar] [CrossRef]
- Singh, R.K.; Tiwari, M.K.; Singh, R.; Lee, J.K. From protein engineering to immobilization: Promising strategies for the upgrade of industrial enzymes. Int. J. Mol. Sci. 2013, 14, 1232–1277. [Google Scholar] [CrossRef]
- Tischer, W.; Kasche, V. Immobilized enzymes: Crystals or carriers? Trends Biotechnol. 1999, 17, 326–335. [Google Scholar] [CrossRef]
- Homaei, A.A.; Sariri, R.; Vianello, F.; Stevanato, R. Enzyme immobilization: An update. J. Chem. Biol. 2013, 6, 185–205. [Google Scholar] [CrossRef]
- Pille, J.; Cardinale, D.; Carette, N.; Di Primo, C.; Besong-Ndika, J.; Walter, J.; Lecoq, H.; van Eldijk, M.B.; Smits, F.C.; Schoffelen, S.; et al. General strategy for ordered noncovalent protein assembly on well-defined nanoscaffolds. Biomacromolecules 2013, 14, 4351–4359. [Google Scholar] [CrossRef]
- Carette, N.; Engelkamp, H.; Akpa, E.; Pierre, S.J.; Cameron, N.R.; Christianen, P.C.M.; Maan, J.C.; Thies, J.C.; Weberskirch, R.; Rowan, A.E.; et al. A virus-based biocatalyst. Nat. Nanotechnol. 2007, 2, 226–229. [Google Scholar] [CrossRef]
- Aljabali, A.A.; Barclay, J.E.; Steinmetz, N.F.; Lomonossoff, G.P.; Evans, D.J. Controlled immobilisation of active enzymes on the cowpea mosaic virus capsid. Nanoscale 2012, 4, 5640–5645. [Google Scholar] [CrossRef] [PubMed]
- Besong-Ndika, J.; Wahlsten, M.; Cardinale, D.; Pille, J.; Walter, J.; Michon, T.; Mäkinen, K. Towards the reconstitution of a two-enzyme cascade for resveratrol synthesis on potyvirus particles. Front. Plant Sci. 2016, 7, 89. [Google Scholar] [CrossRef]
- Bally, J.; Jung, H.; Mortimer, C.; Naim, F.; Philips, J.G.; Hellens, R.; Bombarely, A.; Goodin, M.M.; Waterhouse, P.M. The rise and rise of Nicotiana benthamiana: A plant for all reasons. Annu. Rev. Phytopathol. 2018, 56, 405–426. [Google Scholar] [CrossRef]
- Yang, S.-J.; Carter, S.A.; Cole, A.B.; Cheng, N.-H.; Nelson, R.S. A natural variant of a host RNA-dependent RNA polymerase is associated with increased susceptibility to viruses by Nicotiana benthamiana. Proc. Natl. Acad. Sci. USA 2004, 101, 6297–6302. [Google Scholar] [CrossRef]
- Yamamoto, K.R.; Alberts, B.M.; Benzinger, R.; Lawhorne, L.; Treiber, G. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology 1970, 40, 734–744. [Google Scholar] [CrossRef]
- Juckles, I.R.M. Fractionation of proteins and viruses with polyethylene glycol. Biochim. Biophys. Acta Protein Struct. 1971, 229, 535–546. [Google Scholar] [CrossRef]
- Hebert, T.T. Precipitation of plant viruses by polyethylene glycol. Phytopathology 1963, 53, 362. [Google Scholar]
- Jungblut, S.; Tuinier, R.; Binder, K.; Schilling, T. Depletion induced isotropic-isotropic phase separation in suspensions of rod-like colloids. J. Chem. Phys. 2007, 127, 244909. [Google Scholar] [CrossRef]
- Clark, M.F. Polyethylene glycol solubility gradients, a new and rapid technique for investigations of plant viruses. Virology 1970, 42, 246–247. [Google Scholar] [CrossRef]
- Francki, R.I.B. Some factors affecting particle length distribution in tobacco mosaic virus preparations. Virology 1966, 30, 388–396. [Google Scholar] [CrossRef]
- Williams, R.C.; Steere, R.L. Electron microscopic observations on the unit of length of the particles of tobacco mosaic virus. J. Am. Chem. Soc. 1951, 73, 2057–2061. [Google Scholar] [CrossRef]
- Beachy, R.N.; Zaitlin, M. Characterization and in vitro translation of the RNAs from less-than-full-length, virus-related, nucleoprotein rods present in tobacco mosaic virus preparations. Virology 1977, 81, 160–169. [Google Scholar] [CrossRef]
- Hulett, H.R.; Loring, H.S. Effect of particle length distribution on infectivity of tobacco mosaic virus. Virology 1965, 25, 418–430. [Google Scholar] [CrossRef]
- Ogawa, T.; Watanabe, Y.; Meshi, T.; Okada, Y. Trans complementation of virus-encoded replicase components of tobacco mosaic virus. Virology 1991, 185, 580–584. [Google Scholar] [CrossRef]
- Taliansky, M.E.; Atabekova, T.I.; Kaplan, I.B.; Morozov, S.Y.; Malyshenko, S.I.; Atabekov, J.G. A study of TMV ts mutant Ni2519 I. Complementation experiments. Virology 1982, 118, 301–308. [Google Scholar] [CrossRef]
- Gaddipati, J.P.; Siegel, A. Study of TMV assembly with heterologous RNA containing the origin-of-assembly sequence. Virology 1990, 174, 337–344. [Google Scholar] [CrossRef]
- Rochon, D.; Siegel, A. Chloroplast DNA transcripts are encapsidated by tobacco mosaic virus coat protein. Proc. Natl. Acad. Sci. USA 1984, 81, 1719–1723. [Google Scholar] [CrossRef]
- Siegel, A. Pseudovirions of tobacco mosaic virus. Virology 1971, 46, 50–59. [Google Scholar] [CrossRef]
- Schneider, A.; Eber, F.J.; Wenz, N.; Altintoprak, K.; Jeske, H.; Eiben, S.; Wege, C. Dynamic DNA-controlled “stop-and-go” assembly of well-defined protein domains on RNA-scaffolded TMV-like nanotubes. Nanoscale 2016, 8, 19853–19866. [Google Scholar] [CrossRef] [PubMed]
- Saunders, K.; Thuenemann, E.C.; Peyret, H.; Lomonossoff, G.P. The tobacco mosaic virus origin of assembly sequence is dispensable for specific viral RNA encapsidation but necessary for initiating assembly at a single site. J. Mol. Biol. 2022, 434, 167873. [Google Scholar] [CrossRef] [PubMed]
- Pilz, I.; Kratky, O.; Karush, F. Changes of the conformation of rabbit IgG antibody caused by the specific binding of a hapten. Eur. J. Biochem. FEBS 1974, 41, 91–96. [Google Scholar] [CrossRef]
- Jay, J.W.; Bray, B.; Qi, Y.; Igbinigie, E.; Wu, H.; Li, J.; Ren, G. IgG antibody 3D structures and dynamics. Antibodies 2018, 7, 18. [Google Scholar] [CrossRef] [PubMed]
- Poghossian, A.; Bäcker, M.; Mayer, D.; Schöning, M.J. Gating capacitive field-effect sensors by the charge of nanoparticle/molecule hybrids. Nanoscale 2015, 7, 1023–1031. [Google Scholar] [CrossRef]
- Poghossian, A.; Schöning, M.J. Recent progress in silicon-based biologically sensitive field-effect devices. Curr. Opin. Electrochem. 2021, 29, 100811. [Google Scholar] [CrossRef]
- Wang, Y.; Douglas, T. Tuning properties of biocatalysis using protein cage architectures. J. Mater. Chem. B 2023, 11, 3567–3578. [Google Scholar] [CrossRef]
- Uchida, M.; Manzo, E.; Echeveria, D.; Jiménez, S.; Lovell, L. Harnessing physicochemical properties of virus capsids for designing enzyme confined nanocompartments. Curr. Opin. Virol. 2022, 52, 250–257. [Google Scholar] [CrossRef] [PubMed]
- Wi, S.; Hwang, I.S.; Jo, B.H. Engineering a plant viral coat protein for in vitro hybrid self-assembly of CO2-capturing catalytic nanofilaments. Biomacromolecules 2020, 21, 3847–3856. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.S.; Wang, L.; Jiang, R.R. Specific enzyme immobilization approaches and their application with nanomaterials. Top. Catal. 2012, 55, 1146–1156. [Google Scholar] [CrossRef]
- Liu, M.; Wang, Y.; Jiang, H.; Han, Y.; Xia, J. Synthetic multienzyme assemblies for natural product biosynthesis. ChemBioChem 2023, 24, e202200518. [Google Scholar] [CrossRef]
- Foote, J.; Eisen, H.N. Kinetic and affinity limits on antibodies produced during immune responses. Proc. Natl. Acad. Sci. USA 1995, 92, 1254–1256. [Google Scholar] [CrossRef]
- Moser, A.C.; Hage, D.S. Immunoaffinity chromatography: An introduction to applications and recent developments. Bioanalysis 2010, 2, 769–790. [Google Scholar] [CrossRef]
- Kruljec, N.; Bratkovič, T. Alternative affinity ligands for immunoglobulins. Bioconjugate Chem. 2017, 28, 2009–2030. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Chen, K.; Chu, L.; Kinderman, F.; Apostol, I.; Huang, G. Methionine oxidation in human IgG2 Fc decreases binding affinities to protein A and FcRn. Protein Sci. 2009, 18, 424–433. [Google Scholar] [CrossRef] [PubMed]
- Dias, R.L.; Fasan, R.; Moehle, K.; Renard, A.; Obrecht, D.; Robinson, J.A. Protein ligand design: From phage display to synthetic protein epitope mimetics in human antibody Fc-binding peptidomimetics. J. Am. Chem. Soc. 2006, 128, 2726–2732. [Google Scholar] [CrossRef] [PubMed]
- Shenton, W.; Douglas, T.; Young, M.; Stubbs, G.; Mann, S. Inorganic-organic nanotube composites from template mineralization of tobacco mosaic virus. Adv. Mater. 1999, 11, 253–256. [Google Scholar] [CrossRef]
- Khan, A.; Fox, E.; Górzny, M.; Nikulina, E.; Brougham, D.; Wege, C.; Bittner, A.M. pH control of the electrostatic binding of gold and iron oxide nanoparticles to tobacco mosaic virus. Langmuir 2013, 29, 2094–2098. [Google Scholar] [CrossRef]
- Jablonski, M.; Poghossian, A.; Keusgen, M.; Wege, C.; Schöning, M.J. Detection of plant virus particles with a capacitive field-effect sensor. Anal. Bioanal. Chem. 2021, 413, 5669–5678. [Google Scholar] [CrossRef]
- Röder, J.; Fischer, R.; Commandeur, U. Engineering potato virus X particles for a covalent protein based attachment of enzymes. Small 2017, 13, 1702151. [Google Scholar] [CrossRef]
- Braisted, A.C.; Wells, J.A. Minimizing a binding domain from protein A. Proc. Natl. Acad. Sci. USA 1996, 93, 5688–5692. [Google Scholar] [CrossRef]
- Tashiro, M.; Tejero, R.; Zimmerman, D.E.; Celda, B.; Nilsson, B.; Montelione, G.T. High-resolution solution NMR structure of the Z domain of staphylococcal protein A. J. Mol. Biol. 1997, 272, 573–590. [Google Scholar] [CrossRef]
- Kalnciema, I.; Balke, I.; Skrastina, D.; Ose, V.; Zeltins, A. Potato virus M-like nanoparticles: Construction and characterization. Mol. Biotechnol. 2015, 57, 982–992. [Google Scholar] [CrossRef]
- Sabharwal, P.; Sushmitha, C.; Amritha, C.K.; Natraj, U.; Murthy, M.R.N.; Savithri, H.S. Development of pepper vein banding virus chimeric virus-like particles for potential diagnostic and therapeutic applications. Arch. Virol. 2020, 165, 1163–1176. [Google Scholar] [CrossRef] [PubMed]
- Gulati, A.; Murthy, A.; Abraham, A.; Mohan, K.; Natraj, U.; Savithri, H.S.; Murthy, M.R.N. Structural studies on chimeric Sesbania mosaic virus coat protein: Revisiting SeMV assembly. Virology 2016, 489, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Uhde-Holzem, K.; McBurney, M.; Tiu, B.D.B.; Advincula, R.C.; Fischer, R.; Commandeur, U.; Steinmetz, N.F. Production of immunoabsorbent nanoparticles by displaying single-domain protein A on potato virus X. Macromol. Biosci. 2016, 16, 231–241. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wendlandt, T.; Koch, C.; Britz, B.; Liedek, A.; Schmidt, N.; Werner, S.; Gleba, Y.; Vahidpour, F.; Welden, M.; Poghossian, A.; et al. Facile Purification and Use of Tobamoviral Nanocarriers for Antibody-Mediated Display of a Two-Enzyme System. Viruses 2023, 15, 1951. https://doi.org/10.3390/v15091951
Wendlandt T, Koch C, Britz B, Liedek A, Schmidt N, Werner S, Gleba Y, Vahidpour F, Welden M, Poghossian A, et al. Facile Purification and Use of Tobamoviral Nanocarriers for Antibody-Mediated Display of a Two-Enzyme System. Viruses. 2023; 15(9):1951. https://doi.org/10.3390/v15091951
Chicago/Turabian StyleWendlandt, Tim, Claudia Koch, Beate Britz, Anke Liedek, Nora Schmidt, Stefan Werner, Yuri Gleba, Farnoosh Vahidpour, Melanie Welden, Arshak Poghossian, and et al. 2023. "Facile Purification and Use of Tobamoviral Nanocarriers for Antibody-Mediated Display of a Two-Enzyme System" Viruses 15, no. 9: 1951. https://doi.org/10.3390/v15091951
APA StyleWendlandt, T., Koch, C., Britz, B., Liedek, A., Schmidt, N., Werner, S., Gleba, Y., Vahidpour, F., Welden, M., Poghossian, A., Schöning, M. J., Eber, F. J., Jeske, H., & Wege, C. (2023). Facile Purification and Use of Tobamoviral Nanocarriers for Antibody-Mediated Display of a Two-Enzyme System. Viruses, 15(9), 1951. https://doi.org/10.3390/v15091951