Atovaquone and Pibrentasvir Inhibit the SARS-CoV-2 Endoribonuclease and Restrict Infection In Vitro but Not In Vivo
Abstract
:1. Introduction
2. Materials and Methods
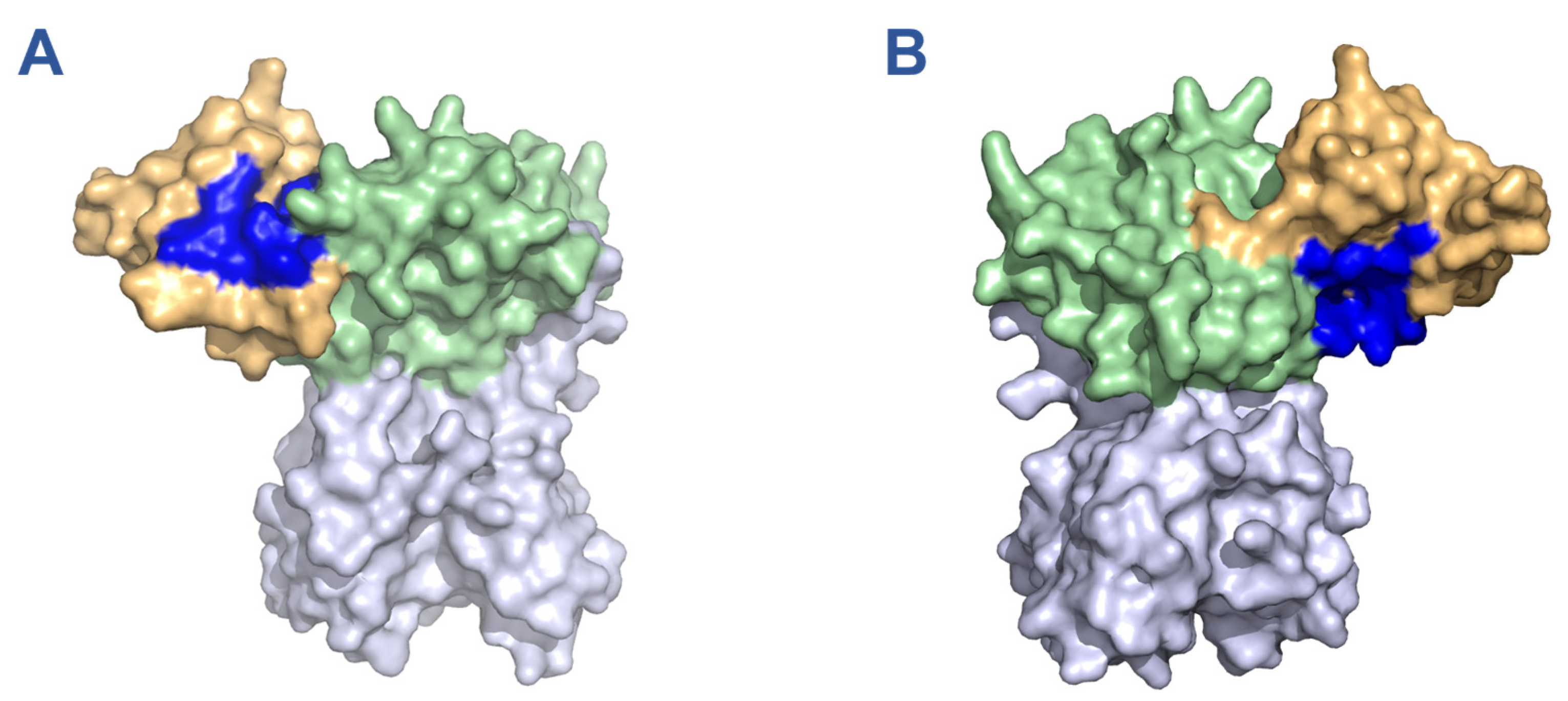
2.1. In Silico Screening of FDA Approved Drugs for Nsp15 Binding Activity
2.2. Growth and Purification of Recombinant SARS-CoV-2 Nsp15
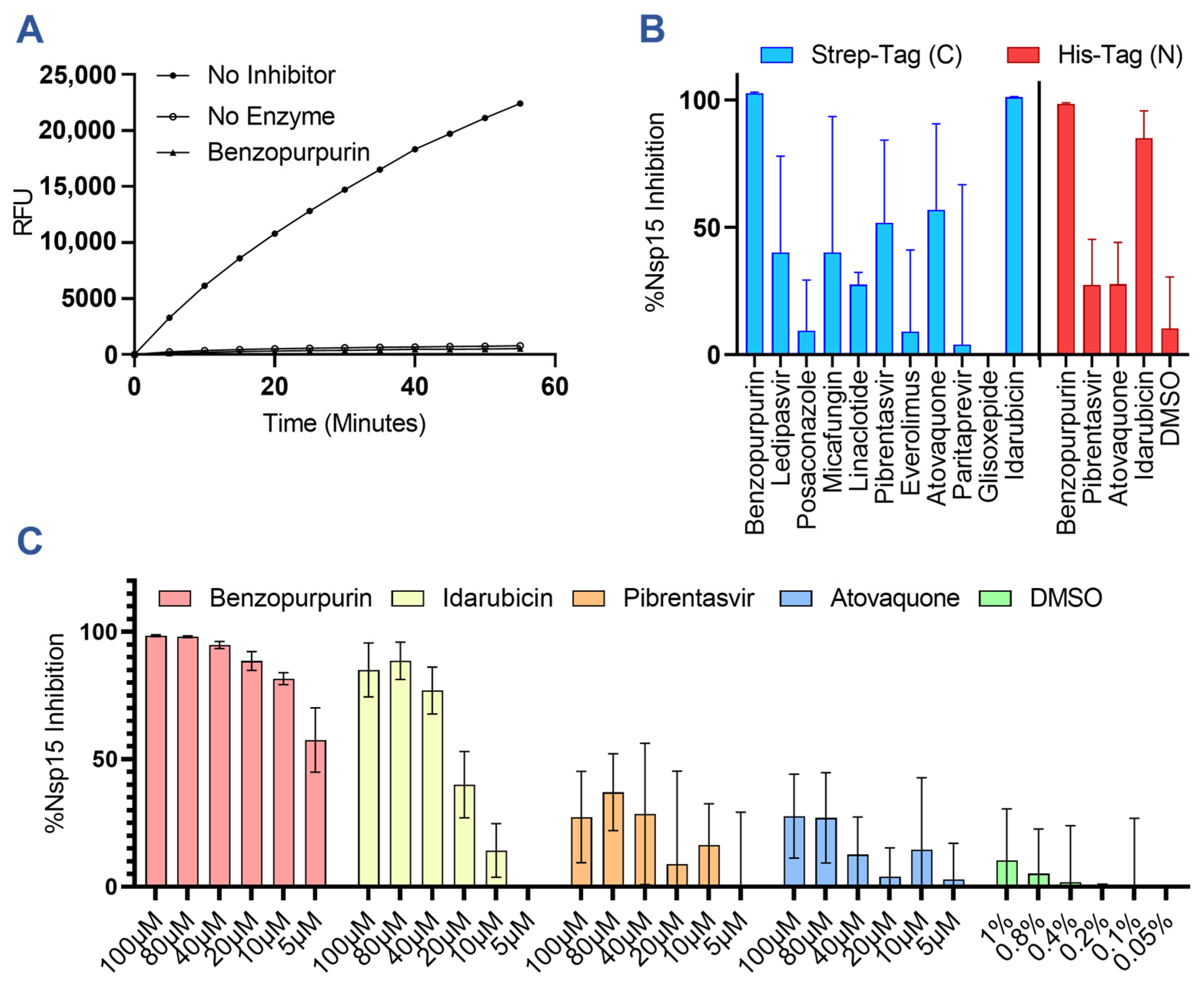
2.3. Library Screening for SARS-CoV-2 Nsp15 Inhibitory Activity
2.4. Evaluation of Drug-Induced Nsp15 Aggregation
2.5. Cell Lines and Viruses
2.6. Coronavirus Infection Inhibition Assay
2.7. Cell Viability Assay
2.8. Flow Cytometry of HCoV-OC43 Infected Cells
2.9. Immunohistochemistry of Viral dsRNA in HCoV-OC43 Infected Cells
2.10. Infection of Mice with SARS-CoV-2 B.1.351
2.11. Quantitative Reverse Transcription-PCR of Lung Tissues
3. Results
3.1. Predicted Nsp15 Binding Drugs Inhibit Nuclease Activity In Vitro
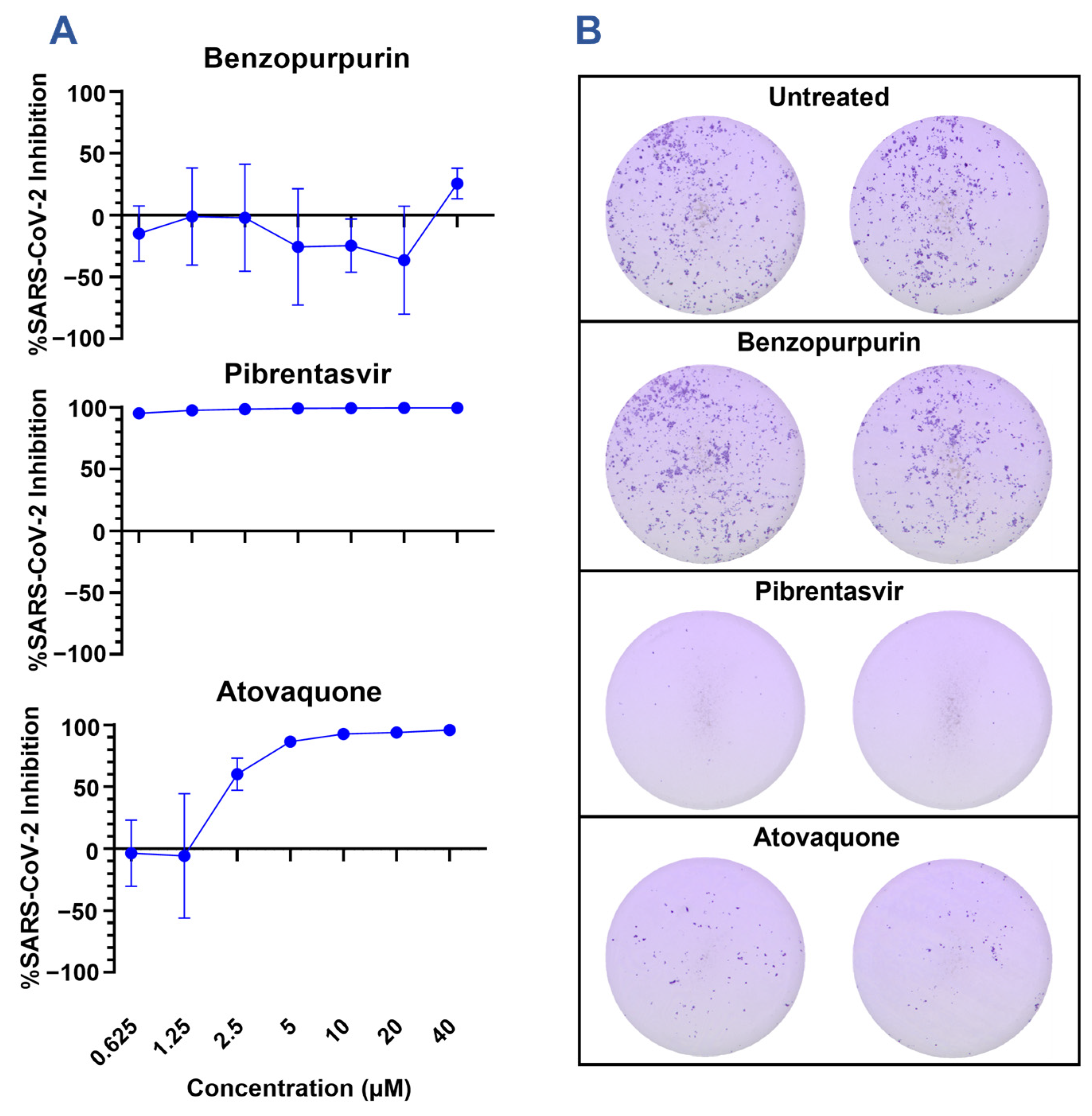
3.2. Nsp15 Inhibitors Restrict SARS-CoV-2 Infection In Vitro
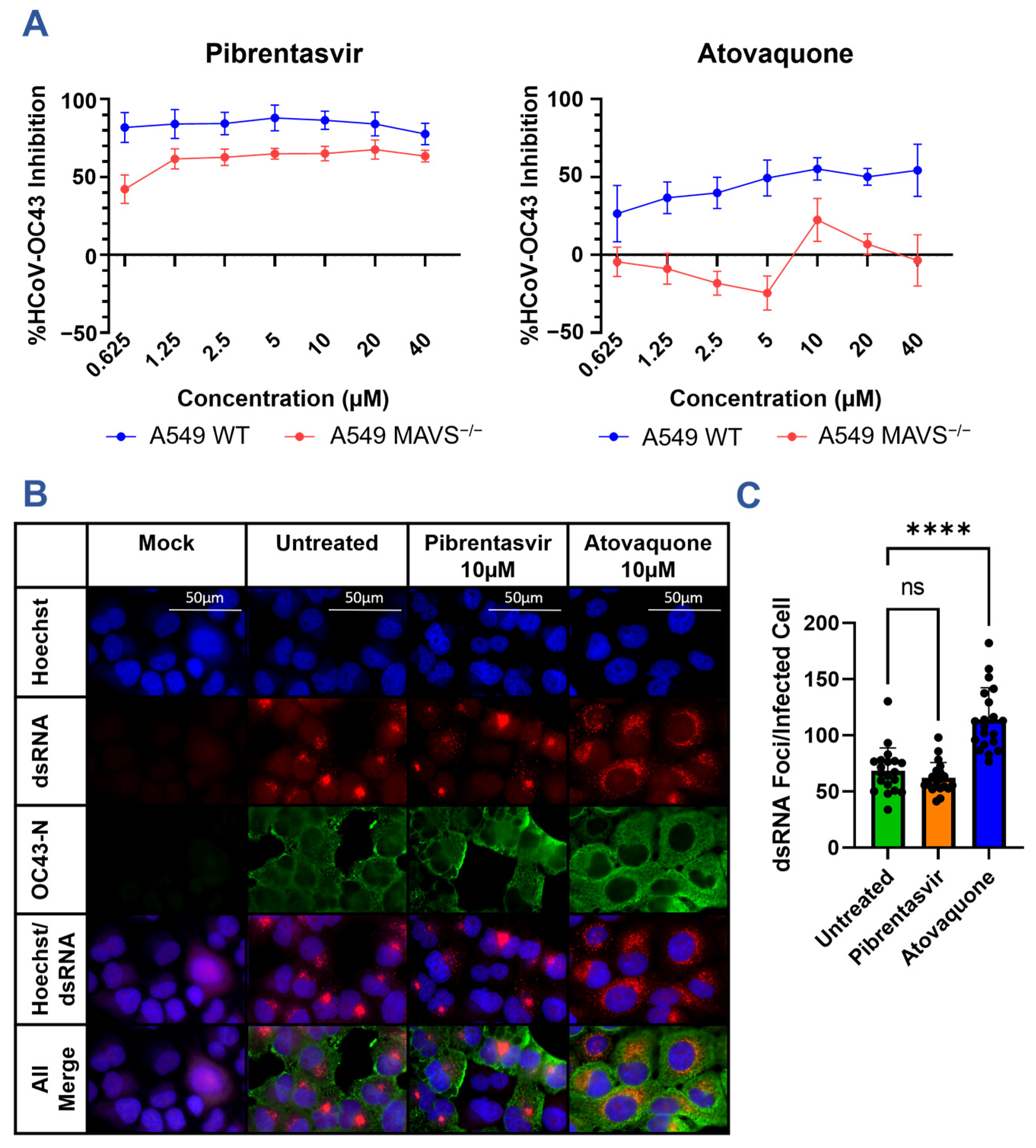
3.3. Nsp15 Inhibitors Restrict HCoV-OC43 Infection In Vitro
3.4. Atovaquone and Pibrentasvir Are Not Protective in a Mouse Model of SARS-CoV-2 Infection
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Antinori, A.; Bausch-Jurken, M. The Burden of COVID-19 in the Immunocompromised Patient: Implications for Vaccination and Needs for the Future. J. Infect. Dis. 2023, 228 (Suppl. S1), S4–S12. [Google Scholar] [CrossRef]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [PubMed]
- Chen, R.E.; Zhang, X.; Case, J.B.; Winkler, E.S.; Liu, Y.; VanBlargan, L.A.; Liu, J.; Errico, J.M.; Xie, X.; Suryadevara, N.; et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021, 27, 717–726. [Google Scholar] [PubMed]
- Mari, A.; Roloff, T.; Stange, M.; Søgaard, K.K.; Asllanaj, E.; Tauriello, G.; Alexander, L.T.; Schweitzer, M.; Leuzinger, K.; Gensch, A.; et al. Global Genomic Analysis of SARS-CoV-2 RNA Dependent RNA Polymerase Evolution and Antiviral Drug Resistance. Microorganisms 2021, 9, 1094. [Google Scholar] [CrossRef] [PubMed]
- Martinot, M.; Jary, A.; Fafi-Kremer, S.; Leducq, V.; Delagreverie, H.; Garnier, M.; Pacanowski, J.; Mékinian, A.; Pirenne, F.; Tiberghien, P.; et al. Remdesivir failure with SARS-CoV-2 RNA-dependent RNA-polymerase mutation in a B-cell immunodeficient patient with protracted COVID-19. Clin. Infect. Dis. 2020, 28, ciaa1474. [Google Scholar]
- Hackbart, M.; Deng, X.; Baker, S.C. Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors. Proc. Natl. Acad. Sci. USA 2020, 117, 8094–8103. [Google Scholar]
- Frazier, M.N.; Dillard, L.B.; Krahn, J.M.; Perera, L.; Williams, J.G.; Wilson, I.M.; Stewart, Z.D.; Pillon, M.C.; Deterding, L.J.; Borgnia, M.J.; et al. Characterization of SARS2 Nsp15 nuclease activity reveals it’s mad about U. Nucleic Acids Res. 2021, 49, 10136–10149. [Google Scholar]
- Ivanov, K.A.; Hertzig, T.; Rozanov, M.; Bayer, S.; Thiel, V.; Gorbalenya, A.E.; Ziebuhr, J. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc. Natl. Acad. Sci. USA 2004, 101, 12694–12699. [Google Scholar] [CrossRef] [PubMed]
- Kindler, E.; Gil-Cruz, C.; Spanier, J.; Li, Y.; Wilhelm, J.; Rabouw, H.H.; Züst, R.; Hwang, M.; V’kovski, P.; Stalder, H.; et al. Early endonuclease-mediated evasion of RNA sensing ensures efficient coronavirus replication. PLoS Pathog. 2017, 13, e1006195. [Google Scholar]
- Deng, X.; Hackbart, M.; Mettelman, R.C.; O’Brien, A.; Mielech, A.M.; Yi, G.; Kao, C.C.; Baker, S.C. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc. Natl. Acad. Sci. USA 2017, 114, E4251–E4260. [Google Scholar] [CrossRef]
- Gao, B.; Gong, X.; Fang, S.; Weng, W.; Wang, H.; Chu, H.; Sun, Y.; Meng, C.; Tan, L.; Song, C.; et al. Inhibition of anti-viral stress granule formation by coronavirus endoribonuclease nsp15 ensures efficient virus replication. PLoS Pathog. 2021, 17, e1008690. [Google Scholar]
- Ulferts, R.; Ziebuhr, J. Nidovirus ribonucleases: Structures and functions in viral replication. RNA Biol. 2011, 8, 295–304. [Google Scholar] [CrossRef]
- Kang, H.; Bhardwaj, K.; Li, Y.; Palaninathan, S.; Sacchettini, J.; Guarino, L.; Leibowitz, J.L.; Kao, C.C. Biochemical and genetic analyses of murine hepatitis virus Nsp15 endoribonuclease. J. Virol. 2007, 81, 13587–13597. [Google Scholar] [CrossRef]
- Deng, X.; van Geelen, A.; Buckley, A.C.; O’Brien, A.; Pillatzki, A.; Lager, K.M.; Faaberg, K.S.; Baker, S.C. Coronavirus Endoribonuclease Activity in Porcine Epidemic Diarrhea Virus Suppresses Type I and Type III Interferon Responses. J. Virol. 2019, 93, e02000-18. [Google Scholar] [CrossRef] [PubMed]
- Ng, Y.L.; Salim, C.K.; Chu, J.J.H. Drug repurposing for COVID-19: Approaches, challenges and promising candidates. Pharmacol. Ther. 2021, 228, 107930. [Google Scholar] [PubMed]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Guedes, I.A.; Barreto, A.M.; Marinho, D.; Krempser, E.; Kuenemann, M.A.; Sperandio, O.; Dardenne, L.E.; Miteva, M.A. New machine learning and physics-based scoring functions for drug discovery. Sci. Rep. 2021, 11, 3198. [Google Scholar]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Cao, H.; Skolnick, J. FRAGSITE: A Fragment-Based Approach for Virtual Ligand Screening. J. Chem. Inf. Model. 2021, 61, 2074–2089. [Google Scholar] [CrossRef]
- Zhou, H.; Skolnick, J. Template-based protein structure modeling using TASSER(VMT.). Proteins 2012, 80, 352–361. [Google Scholar] [CrossRef]
- Bernstein, F.C.; Koetzle, T.F.; Williams, G.J.; Meyer Jr, E.F.; Brice, M.D.; Rodgers, J.R.; Kennard, O.; Shimanouchi, T.; Tasumi, M. The Protein Data Bank: A computer-based archival file for macromolecular structures. J. Mol. Biol. 1977, 112, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Mysinger, M.M.; Carchia, M.; Irwin, J.J.; Shoichet, B.K. Directory of useful decoys, enhanced (DUD-E): Better ligands and decoys for better benchmarking. J. Med. Chem. 2012, 55, 6582–6594. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Shrivastava, S.; Hassanali, M.; Stothard, P.; Chang, Z.; Woolsey, J. DrugBank: A comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006, 34, D668–D672. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Wower, J.; Maltseva, N.; Chang, C.; Jedrzejczak, R.; Wilamowski, M.; Kang, S.; Nicolaescu, V.; Randall, G.; Michalska, K.; et al. Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2. Commun. Biol. 2021, 4, 193. [Google Scholar] [PubMed]
- Corbett, K.S.; Werner, A.P.; Connell, S.O.; Gagne, M.; Lai, L.; Moliva, J.I.; Flynn, B.; Choi, A.; Koch, M.; Foulds, K.E.; et al. mRNA-1273 protects against SARS-CoV-2 beta infection in nonhuman primates. Nat. Immunol. 2021, 22, 1306–1315. [Google Scholar] [CrossRef] [PubMed]
- Vanderheiden, A.; Thomas, J.; Soung, A.L.; Davis-Gardner, M.E.; Floyd, K.; Jin, F.; Cowan, D.A.; Pellegrini, K.; Creanga, A.; Pegu, A.; et al. CCR2 Signaling Restricts SARS-CoV-2 Infection. mBio 2021, 12, e0274921. [Google Scholar] [CrossRef]
- Sixto-Lopez, Y.; Martinez-Archundia, M. Drug repositioning to target NSP15 protein on SARS-CoV-2 as possible COVID-19 treatment. J. Comput. Chem. 2021, 42, 897–907. [Google Scholar] [CrossRef]
- Chandra, A.; Gurjar, V.; Qamar, I.; Singh, N. Identification of potential inhibitors of SARS-CoV-2 endoribonuclease (EndoU) from FDA approved drugs: A drug repurposing approach to find therapeutics for COVID-19. J. Biomol. Struct. Dyn. 2021, 39, 4201–4211. [Google Scholar] [CrossRef]
- Ortiz-Alacantare, J.; Bhardwaj, K.; Palaninathan, S.; Frieman, M.; Baric, R.S.; Kao, C.C. Small molecule inhibitors of the SARS-CoV Nsp15 endoribonuclease. Virus Adapt. Treat. 2010, 2010, 125–133. [Google Scholar]
- Wang, X.; Sacramento, C.Q.; Jockusch, S.; Chaves, O.A.; Tao, C.; Fintelman-Rodrigues, N.; Chien, M.; Temerozo, J.R.; Li, X.; Kumar, S.; et al. Combination of antiviral drugs inhibits SARS-CoV-2 polymerase and exonuclease and demonstrates COVID-19 therapeutic potential in viral cell culture. Commun. Biol. 2022, 5, 154. [Google Scholar] [CrossRef]
- Ahmed, M.S.; Farag, A.B.; Boys, I.N.; Wang, P.; Menendez-Montes, I.; Nguyen, N.U.N.; Eitson, J.L.; Ohlson, M.B.; Fan, W.; McDougal, M.B.; et al. FDA approved drugs with antiviral activity against SARS-CoV-2: From structure-based repurposing to host-specific mechanisms. Biomed. Pharmacother. 2023, 162, 114614. [Google Scholar] [CrossRef]
- Carter-Timofte, M.E.; Arulanandam, R.; Kurmasheva, N.; Fu, K.; Laroche, G.; Taha, Z.; van Der Horst, D.; Cassin, L.; van der Sluis, R.M.; Palermo, E.; et al. Antiviral Potential of the Antimicrobial Drug Atovaquone against SARS-CoV-2 and Emerging Variants of Concern. ACS Infect. Dis. 2021, 7, 3034–3051. [Google Scholar] [CrossRef]
- Bakshi, R.P.; Tatham, L.M.; Savage, A.C.; Tripathi, A.K.; Mlambo, G.; Ippolito, M.M.; Nenortas, E.; Rannard, S.P.; Owen, A.; Shapiro, T.A. Long-acting injectable atovaquone nanomedicines for malaria prophylaxis. Nat. Commun. 2018, 9, 315. [Google Scholar] [CrossRef]
- Osawa, M.; Uchida, T.; Imamura, M.; Teraoka, Y.; Fujino, H.; Nakahara, T.; Ono, A.; Murakami, E.; Kawaoka, T.; Miki, D.; et al. Efficacy of glecaprevir and pibrentasvir treatment for genotype 1b hepatitis C virus drug resistance-associated variants in humanized mice. J. Gen. Virol. 2019, 100, 1123–1131. [Google Scholar] [CrossRef]
- Ma, C.D.; Imamura, M.; Talley, D.C.; Rolt, A.; Xu, X.; Wang, A.Q.; Le, D.; Uchida, T.; Osawa, M.; Teraoka, Y.; et al. Fluoxazolevir inhibits hepatitis C virus infection in humanized chimeric mice by blocking viral membrane fusion. Nat. Microbiol. 2020, 5, 1532–1541. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Cao, H.; Skolnick, J. FINDSITE(comb2.0): A New Approach for Virtual Ligand Screening of Proteins and Virtual Target Screening of Biomolecules. J. Chem. Inf. Model. 2018, 58, 2343–2354. [Google Scholar] [CrossRef] [PubMed]
- Gaulton, A.; Bellis, L.J.; Bento, A.P.; Chambers, J.; Davies, M.; Hersey, A.; Light, Y.; McGlinchey, S.; Michalovich, D.; Al-Lazikani, B.; et al. ChEMBL: A large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012, 40, D1100–D1107. [Google Scholar] [CrossRef]
- Canal, B.; Fujisawa, R.; McClure, A.W.; Deegan, T.; Wu, M.; Ulferts, R.; Weissmann, F.; Drury, L.S.; Bertolin, A.P.; Zeng, J.; et al. Identifying SARS-CoV-2 antiviral compounds by screening for small molecule inhibitors of nsp15 endoribonuclease. Biochem. J. 2021, 478, 2465–2479. [Google Scholar] [CrossRef]
- Choi, R.; Zhou, M.; Shek, R.; Wilson, J.W.; Tillery, L.; Craig, J.K.; Salukhe, I.A.; Hickson, S.E.; Kumar, N.; James, R.M.; et al. High-throughput screening of the ReFRAME, Pandemic Box, and COVID Box drug repurposing libraries against SARS-CoV-2 nsp15 endoribonuclease to identify small-molecule inhibitors of viral activity. PLoS ONE 2021, 16, e0250019. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Seo, S.H.; Woo, S.J.; Kwon, Y.; Song, M.; Ha, N.C. Epigallocatechin Gallate Inhibits the Uridylate-Specific Endoribonuclease Nsp15 and Efficiently Neutralizes the SARS-CoV-2 Strain. J. Agric. Food Chem. 2021, 69, 5948–5954. [Google Scholar] [CrossRef]
- Stevaert, A.; Krasniqi, B.; Van Loy, B.; Nguyen, T.; Thomas, J.; Vandeput, J.; Jochmans, D.; Thiel, V.; Dijkman, R.; Dehaen, W.; et al. Betulonic Acid Derivatives Interfering with Human Coronavirus 229E Replication via the nsp15 Endoribonuclease. J. Med. Chem. 2021, 64, 5632–5644. [Google Scholar] [CrossRef] [PubMed]
- Nixon, G.L.; Moss, D.M.; Shone, A.E.; Lalloo, D.G.; Fisher, N.; O’Neill, P.M.; Ward, S.A.; Biagini, G.A. Antimalarial pharmacology and therapeutics of atovaquone. J. Antimicrob. Chemother. 2013, 68, 977–985. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.K.; De Lemos, J.A.; McGuire, D.K.; Ayers, C.; Eitson, J.L.; Sanchez, C.L.; Kamel, D.; Meisner, J.A.; Thomas, E.V.; Hegde, A.A.; et al. Atovaquone for treatment of COVID-19: A prospective randomized, double-blind, placebo-controlled clinical trial. Front. Pharmacol. 2022, 13, 1020123. [Google Scholar] [CrossRef] [PubMed]

| Drug Name | Score | Precision | Binding Site |
|---|---|---|---|
| Oritavancin | 1.29 | 0.9 | 2LEU 26ILE 31VAL 50PRO 51VAL 52ASN 53VAL 55PHE 56GLU |
| Ledipasvir | 1.10 | 0.9 | |
| Posaconazole | 1.06 | 0.9 | |
| Micafungin | 1.06 | 0.9 | |
| Linaclotide | 1.02 | 0.9 | |
| Pibrentasvir | 0.97 | 0.87 | |
| Desmopressin | 0.97 | 0.86 | |
| Cyanocobalamin | 0.93 | 0.83 | |
| Rifamixin | 0.99 | 0.88 | 31VAL 42LEU 43PHE 44GLU 46LYS 54ALA 55PHE 58TRP 59ALA 61ARG 86TRP 91ASP |
| Rifapentine | 0.80 | 0.68 | 42LEU 43PHE 44GLU 46LYS 54ALA 55PHE 58TRP 59ALA 61ARG 86TRP 91ASP |
| Everolimus | 0.77 | 0.65 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
von Beck, T.; Mena Hernandez, L.; Zhou, H.; Floyd, K.; Suthar, M.S.; Skolnick, J.; Jacob, J. Atovaquone and Pibrentasvir Inhibit the SARS-CoV-2 Endoribonuclease and Restrict Infection In Vitro but Not In Vivo. Viruses 2023, 15, 1841. https://doi.org/10.3390/v15091841
von Beck T, Mena Hernandez L, Zhou H, Floyd K, Suthar MS, Skolnick J, Jacob J. Atovaquone and Pibrentasvir Inhibit the SARS-CoV-2 Endoribonuclease and Restrict Infection In Vitro but Not In Vivo. Viruses. 2023; 15(9):1841. https://doi.org/10.3390/v15091841
Chicago/Turabian Stylevon Beck, Troy, Luis Mena Hernandez, Hongyi Zhou, Katharine Floyd, Mehul S. Suthar, Jeffrey Skolnick, and Joshy Jacob. 2023. "Atovaquone and Pibrentasvir Inhibit the SARS-CoV-2 Endoribonuclease and Restrict Infection In Vitro but Not In Vivo" Viruses 15, no. 9: 1841. https://doi.org/10.3390/v15091841
APA Stylevon Beck, T., Mena Hernandez, L., Zhou, H., Floyd, K., Suthar, M. S., Skolnick, J., & Jacob, J. (2023). Atovaquone and Pibrentasvir Inhibit the SARS-CoV-2 Endoribonuclease and Restrict Infection In Vitro but Not In Vivo. Viruses, 15(9), 1841. https://doi.org/10.3390/v15091841






