Inactivation of Bacteriophage ɸ6 and SARS-CoV-2 in Antimicrobial Surface Tests
Abstract
:1. Introduction
2. Materials and Methods
2.1. Coating Preparation
2.2. Antiviral Surface Tests
2.2.1. Testing of Antiviral Activity with ɸ6
2.2.2. Testing of Antiviral Activity with SARS-CoV-2
3. Results
3.1. Effect of Cooper Coated Surfaces on Bacteriophage ɸ6
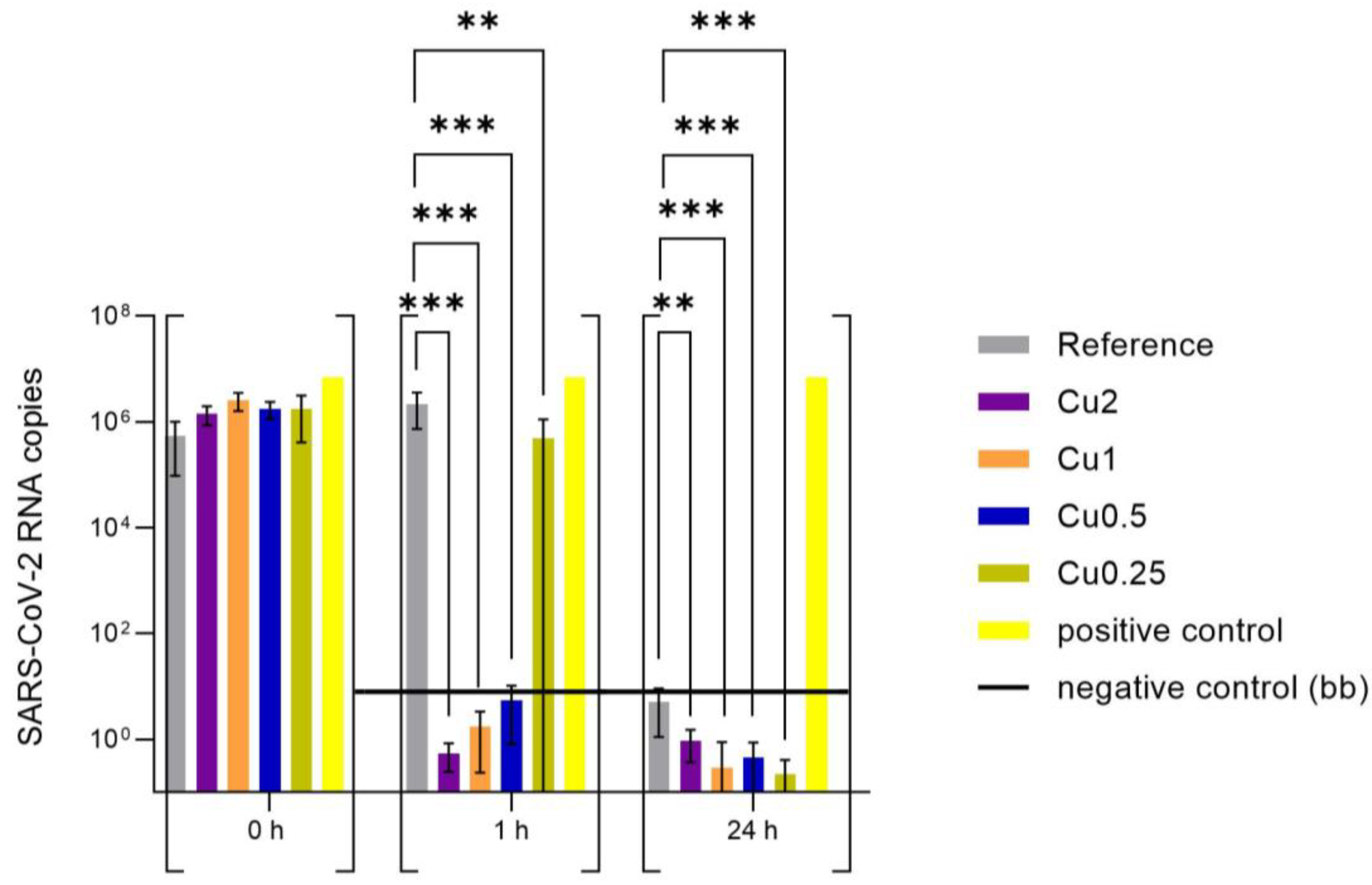
3.2. Effect of Copper-Coated Surfaces on SARS-CoV-2
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chams, N.; Chams, S.; Badran, R.; Shams, A.; Araji, A.; Raad, M.; Mukhopadhyay, S.; Stroberg, E.; Duval, E.J.; Barton, L.M.; et al. COVID-19: A Multidisciplinary Review. Front. Public Health 2020, 8, 383. [Google Scholar] [CrossRef] [PubMed]
- Byun, W.S.; Heo, S.W.; Jo, G.; Kim, J.W.; Kim, S.; Lee, S.; Park, H.E.; Baek, J.-H. Is coronavirus disease (COVID-19) seasonal? A critical analysis of empirical and epidemiological studies at global and local scales. Environ. Res. 2021, 196, 110972. [Google Scholar] [CrossRef] [PubMed]
- Schwarzendahl, F.J.; Grauer, J.; Liebchen, B.; Löwen, H. Mutation induced infection waves in diseases like COVID-19. Sci. Rep. 2022, 12, 9641. [Google Scholar] [CrossRef] [PubMed]
- Van Doremalen, N.; Bushmaker, T.; Morris, D.H.; Holbrook, M.G.; Gamble, A.; Williamson, B.N.; Tamin, A.; Harcourt, J.L.; Thornburg, N.J.; Gerber, S.I.; et al. Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1. N. Engl. J. Med. 2020, 382, 1564–1567. [Google Scholar] [CrossRef] [PubMed]
- Wolfgruber, S.; Loibner, M.; Puff, M.; Melischnig, A.; Zatloukal, K. SARS-CoV2 neutralizing activity of ozone on porous and non-porous materials. New Biotechnol. 2022, 66, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Warnes, S.L.; Little, Z.R.; Keevil, C.W. Human Coronavirus 229E Remains Infectious on Common Touch Surface Materials. mBio 2015, 6, e01697-15. [Google Scholar] [CrossRef] [PubMed]
- Mizielińska, M.; Nawrotek, P.; Stachurska, X.; Ordon, M.; Bartkowiak, A. Packaging Covered with Antiviral and Antibacterial Coatings Based on ZnO Nanoparticles Supplemented with Geraniol and Carvacrol. Int. J. Mol. Sci. 2021, 22, 1717. [Google Scholar] [CrossRef] [PubMed]
- Turgeon, N.; Toulouse, M.-J.; Martel, B.; Moineau, S.; Duchaine, C. Comparison of Five Bacteriophages as Models for Viral Aerosol Studies. Appl. Environ. Microbiol. 2014, 80, 4242–4250. [Google Scholar] [CrossRef] [PubMed]
- String, G.M.; White, M.R.; Gute, D.M.; Mühlberger, E.; Lantagne, D.S. Selection of a SARS-CoV-2 Surrogate for Use in Surface Disinfection Efficacy Studies with Chlorine and Antimicrobial Surfaces. Environ. Sci. Technol. Lett. 2021, 8, 995–1001. [Google Scholar] [CrossRef] [PubMed]
- Tuñón-Molina, A.; Martí, M.; Muramoto, Y.; Noda, T.; Takayama, K.; Serrano-Aroca, Á. Antimicrobial Face Shield: Next Generation of Facial Protective Equipment against SARS-CoV-2 and Multidrug-Resistant Bacteria. Int. J. Mol. Sci. 2021, 22, 9518. [Google Scholar] [CrossRef] [PubMed]
- Prussin, A.J., II; Schwake, D.O.; Lin, K.; Gallagher, D.L.; Buttling, L.; Marr, L.C. Survival of the Enveloped Virus Phi6 in Droplets as a Function of Relative Humidity, Absolute Humidity, and Temperature. Appl. Environ. Microbiol. 2018, 84, e00551-18. [Google Scholar] [CrossRef] [PubMed]
- Laurinavičius, S.; Käkelä, R.; Bamford, D.H.; Somerharju, P. The origin of phospholipids of the enveloped bacteriophage phi6. Virology 2004, 326, 182–190. [Google Scholar] [CrossRef] [PubMed]
- ISO 21702:2019; Measurement of Antiviral Activity on Plastics and Other Non-Porous Surfaces. ISO: Geneva, Switzerland, 2019.
- ISO 18071:2016; Fine Ceramics—Determination of Antiviral Activity of Semiconducting Photocatalytic Materials under Indoor Lighting Environment—Test Method Using Bacteriophage Q-Beta. ISO: Geneva, Switzerland, 2016.
- Hardt, M.; Föderl-Höbenreich, E.; Freydl, S.; Kouros, A.; Loibner, M.; Zatloukal, K. Pre-analytical sample stabilization by different sampling devices for PCR-based COVID-19 diagnostics. New Biotechnol. 2022, 70, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Loibner, M.; Langner, C.; Regitnig, P.; Gorkiewicz, G.; Zatloukal, K. Biosafety Requirements for Autopsies of Patients with COVID-19: Example of a BSL-3 Autopsy Facility Designed for Highly Pathogenic Agents. Pathobiology 2021, 88, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Kicker, E.; Tittel, G.; Schaller, T.; Pferschy-Wenzig, E.-M.; Zatloukal, K.; Bauer, R. SARS-CoV-2 neutralizing activity of polyphenols in a special green tea extract preparation. Phytomedicine 2022, 98, 153970. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention Division of Viral Diseases. CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel. CDC EUA, 2020; Volume 3, pp. 1–42. Available online: https://www.fda.gov/media/134922/download (accessed on 3 July 2023).
- Adcock, N.J.; Rice, E.W.; Sivaganesan, M.; Brown, J.D.; Stallknecht, D.E.; Swayne, D.E. The use of bacteriophages of the family Cystoviridae as surrogates for H5N1 highly pathogenic avian influenza viruses in persistence and inactivation studies. J. Environ. Sci. Health Part A Toxic/Hazard. Subst. Environ. Eng. 2009, 44, 1362–1366. [Google Scholar] [CrossRef]




| 2019-nCoV_N2-F 2019-nCoV_N2 Forward Primer | 5′-TTA CAA ACA TTG GCC GCA AA-3′ |
| 2019-nCoV_N2-R 2019-nCoV_N2 Reverse Primer | 5′-GCG CGA CAT TCC GAA GAA-3′ |
| 2019-nCoV_N2-P 2019-nCoV_N2 Probe | 5′-FAM-ACA ATT TGC/ZEN/CCC CAG CGC TTC AG-3IABkFQ-3′ FAM, BHQ-1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Poelzl, S.; Rieger, J.; Zatloukal, K.; Augl, S.; Stummer, M.; Hinterer, A.; Kittinger, C. Inactivation of Bacteriophage ɸ6 and SARS-CoV-2 in Antimicrobial Surface Tests. Viruses 2023, 15, 1833. https://doi.org/10.3390/v15091833
Poelzl S, Rieger J, Zatloukal K, Augl S, Stummer M, Hinterer A, Kittinger C. Inactivation of Bacteriophage ɸ6 and SARS-CoV-2 in Antimicrobial Surface Tests. Viruses. 2023; 15(9):1833. https://doi.org/10.3390/v15091833
Chicago/Turabian StylePoelzl, Sabine, Julia Rieger, Kurt Zatloukal, Stefan Augl, Maximilian Stummer, Andreas Hinterer, and Clemens Kittinger. 2023. "Inactivation of Bacteriophage ɸ6 and SARS-CoV-2 in Antimicrobial Surface Tests" Viruses 15, no. 9: 1833. https://doi.org/10.3390/v15091833
APA StylePoelzl, S., Rieger, J., Zatloukal, K., Augl, S., Stummer, M., Hinterer, A., & Kittinger, C. (2023). Inactivation of Bacteriophage ɸ6 and SARS-CoV-2 in Antimicrobial Surface Tests. Viruses, 15(9), 1833. https://doi.org/10.3390/v15091833







