Influenza A Virus Infection Alters Lipid Packing and Surface Electrostatic Potential of the Host Plasma Membrane
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmids
2.2. Cell Culture, Transfection and Infection
2.3. Alteration of PM Properties to Obtain Control Samples
2.4. Membrane Labelling with Laurdan and Di-4-ANEPPDHQ
2.5. Confocal Spectral Imaging
2.6. FRET Analysis
2.7. GP Index Analysis
2.8. sFCS Analysis
2.9. Statistical Analysis
3. Results
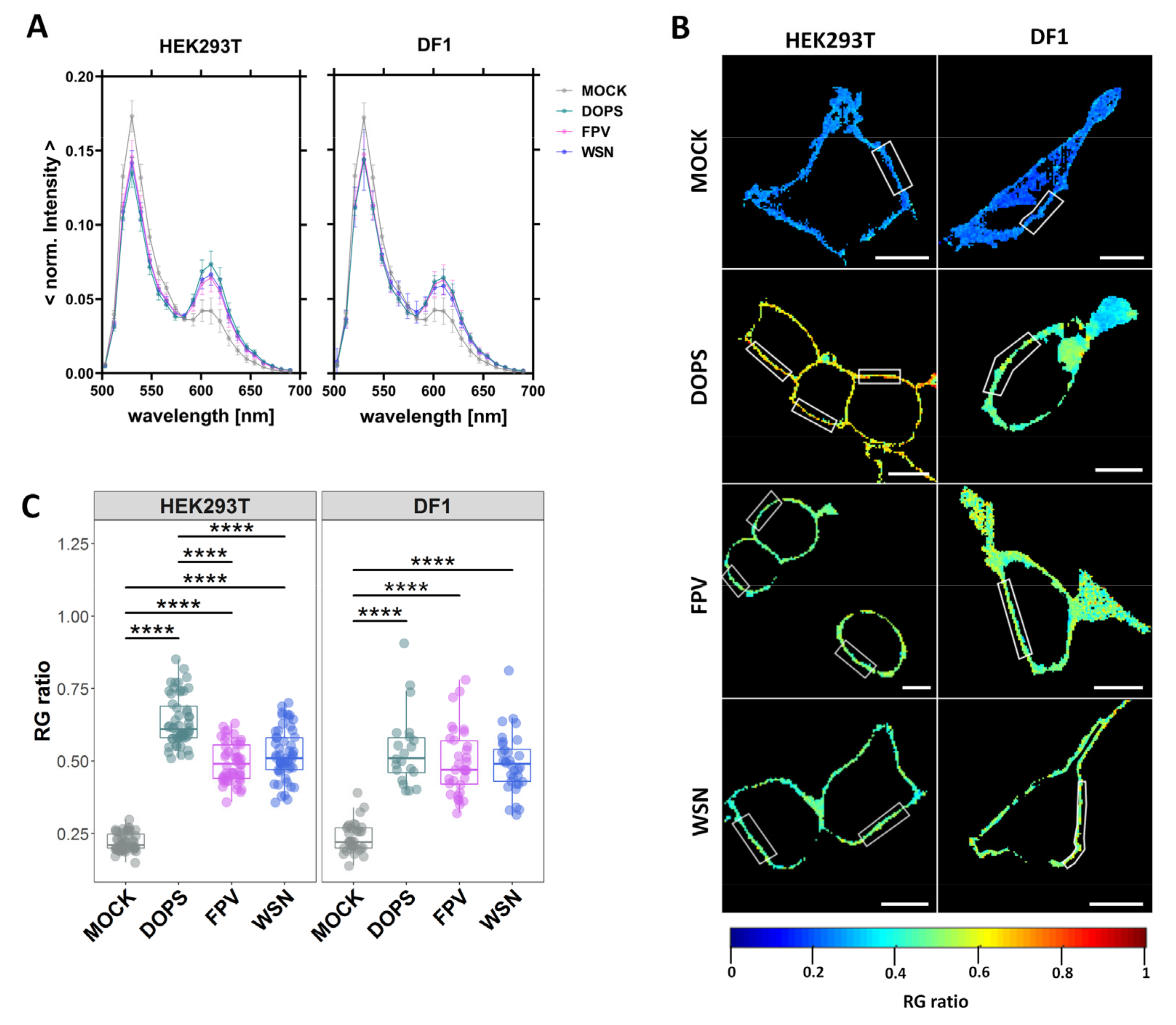
3.1. Infection Increases the Negative Surface Charge of the Inner Leaflet of the PM
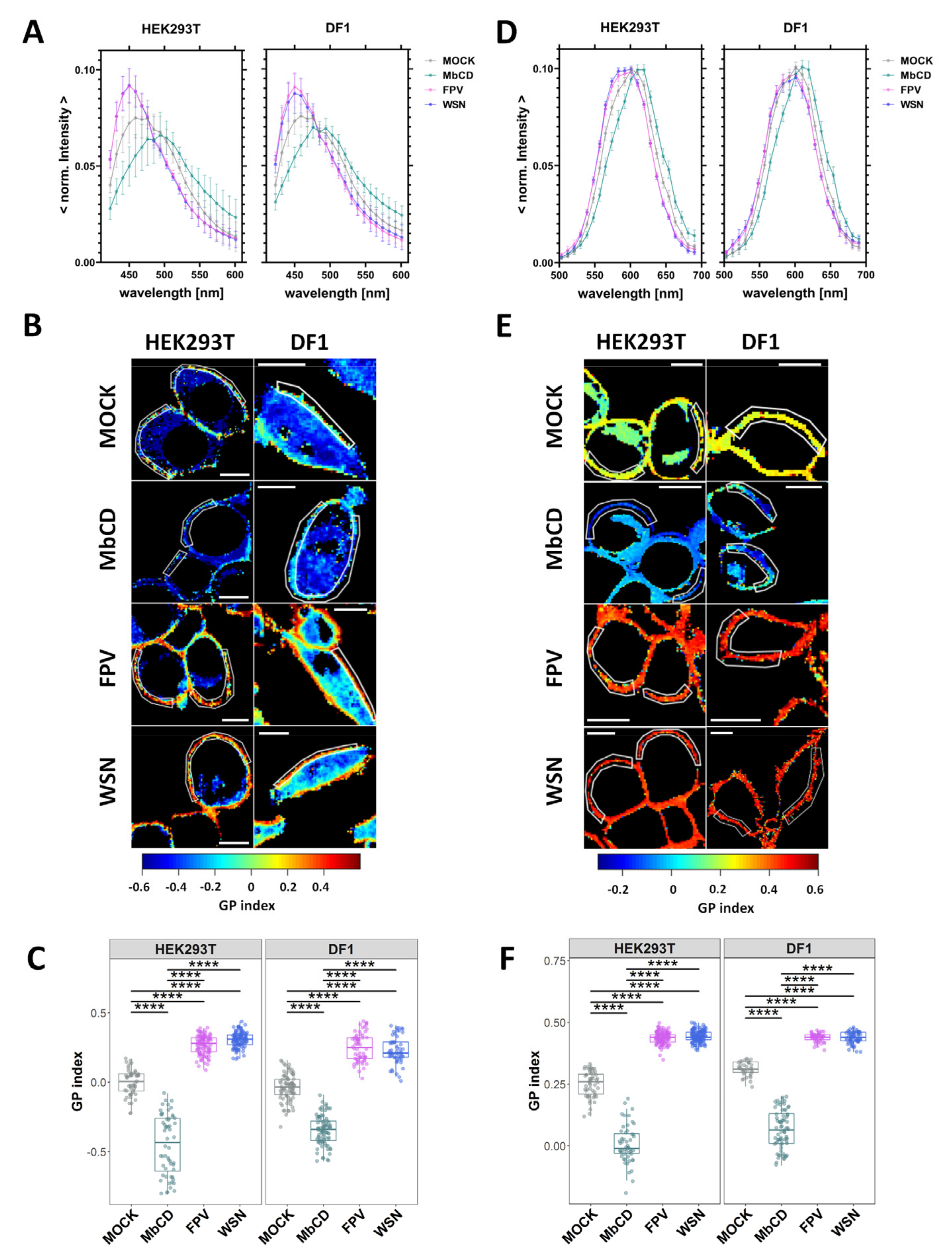
3.2. IAV Infection Increases Lipid Packing in the Plasma Membrane Lipid Bilayer
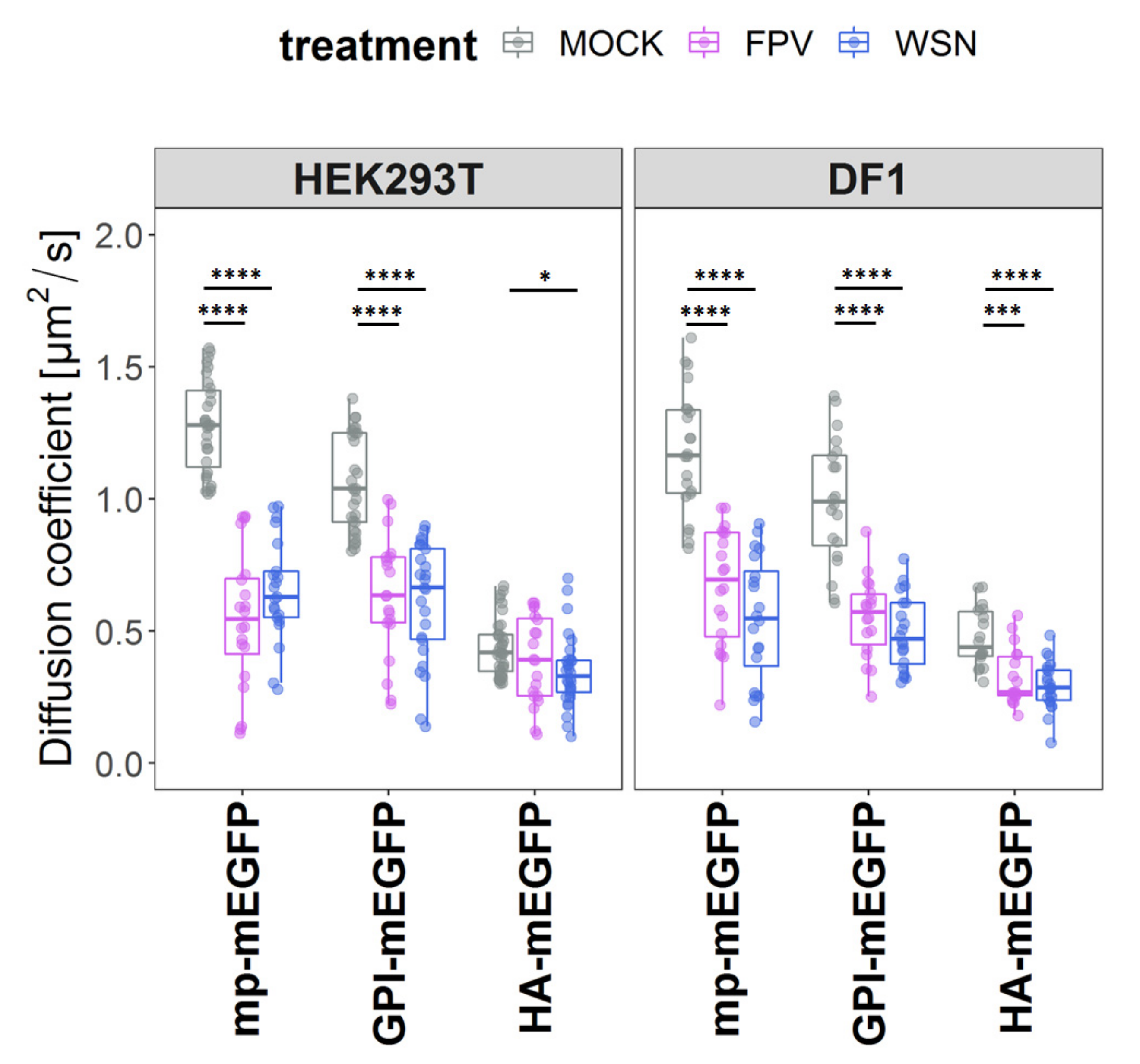
3.3. IAV Infection Reduces Membrane Protein Dynamics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tam, V.C.; Quehenberger, O.; Oshassnsky, C.M.; Suen, R.; Armando, A.M.; Treuting, P.M.; Thomas, P.G.; Dennis, E.A.; Aderem, A. Lipidomic Profiling of Influenza Infection Identifies Mediators that Induce and Resolve Inflammation. Cell 2013, 154, 213–227. [Google Scholar] [CrossRef]
- Taubenberger, J.K.; Kash, J.C. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe 2010, 7, 440–451. [Google Scholar] [CrossRef]
- Schrauwen, E.J.A.; Fouchier, R.A.M. Host adaptation and transmission of influenza A viruses in mammals. Emerg. Microbes Infect. 2014, 3, e9. [Google Scholar] [CrossRef]
- Cui, L.; Zheng, D.; Lee, Y.H.; Chan, T.K.; Kumar, Y.; Ho, W.E.; Chen, J.Z.; Tannenbaum, S.R.; Ong, C.N. Metabolomics Investigation Reveals Metabolite Mediators Associated with Acute Lung Injury and Repair in a Murine Model of Influenza Pneumonia. Sci. Rep. 2016, 6, 26076. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, P.T.; Myers, D.S.; Milne, S.B.; McClaren, J.L.; Thomas, P.G.; Brown, H.A. Lipid composition of viral envelope of three strains of influenza virus—Not all viruses are created equal. ACS Infect. Dis. 2015, 1, 399–452. [Google Scholar] [CrossRef]
- Lin, S.; Liu, N.; Yang, Z.; Song, W.; Wang, P.; Chen, H.; Lucio, M.; Schmitt-Kopplin, P.; Chen, G.; Cai, Z. GC/MS-based metabolomics reveals fatty acid biosynthesis and cholesterol metabolism in cell lines infected with influenza A virus. Talanta 2010, 83, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Tanner, L.B.; Chng, C.; Guan, X.L.; Lei, Z.; Rozen, S.G.; Wenk, M.R. Lipidomics identifies a requirement for peroxisomal function during influenza virus replication. J. Lipid Res. 2014, 55, 1357–1365. [Google Scholar] [CrossRef]
- Tisoncik-Go, J.; Gasper, D.J.; Kyle, J.E.; Eisfeld, A.J.; Selinger, C.; Hatta, M.; Morrison, J.; Korth, M.J.; Zink, E.M.; Kim, Y.M.; et al. Integrated Omics Analysis of Pathogenic Host Responses during Pandemic H1N1 Influenza Virus Infection: The Crucial Role of Lipid Metabolism. Cell Host Microbe 2016, 19, 254–266. [Google Scholar] [CrossRef]
- Woods, P.S.; Doolittle, L.M.; Rosas, L.E.; Joseph, L.M.; Calomeni, E.P.; Davis, I.C. Lethal H1N1 influenza A virus infection alters the murine alveolar type II cell surfactant lipidome. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2016, 311, L1160–L1169. [Google Scholar] [CrossRef]
- Knepper, J.; Schierhorn, K.L.; Becher, A.; Budt, M.; Tönnies, M.; Bauer, T.T.; Schneider, P.; Neudecker, J.; Rückert, J.C.; Gruber, A.D.; et al. The novel human influenza A(H7N9) virus is naturally adapted to efficient growth in human lung tissue. mBio 2013, 4, e00601-13. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-J.; Chen, C.-Y.; Yang, J.-H.; Chiu, Y.-F. Modulating cholesterol-rich lipid rafts to disrupt influenza A virus infection. Front. Immunol. 2022, 13, 982264. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhu, W.; Yang, L.; Shu, Y. The Epidemiology, Virology, and Pathogenicity of Human Infections with Avian Influenza Viruses. Cold Spring Harb. Perspect. Med. 2021, 11, a038620. [Google Scholar] [CrossRef] [PubMed]
- Audi, A.; Soudani, N.; Dbaibo, G.; Zaraket, H. Depletion of Host and Viral Sphingomyelin Impairs Influenza Virus Infection. Front. Microbiol. 2020, 11, 612. [Google Scholar] [CrossRef] [PubMed]
- Hidari, K.I.P.J.; Suzuki, Y.; Suzuki, T. Suppression of the Biosynthesis of Cellular Sphingolipids Results in the Inhibition of the Maturation of Influenza Virus Particles in MDCK Cells. Biol. Pharm. Bull. 2006, 29, 1575–1579. [Google Scholar] [CrossRef][Green Version]
- Ohkura, T.; Momose, F.; Ichikawa, R.; Takeuchi, K.; Morikawa, Y. Influenza A virus hemagglutinin and neuraminidase mutually accelerate their apical targeting through clustering of lipid rafts. J. Virol. 2014, 88, 10039–10055. [Google Scholar] [CrossRef]
- Rossman, J.S.; Lamb, R.A. Influenza virus assembly and budding. Virology 2011, 411, 229–236. [Google Scholar] [CrossRef]
- Tafesse, F.G.; Sanyal, S.; Ashour, J.; Guimaraes, C.P.; Hermansson, M.; Somerharju, P.; Ploegh, H.L. Intact sphingomyelin biosynthetic pathway is essential for intracellular transport of influenza virus glycoproteins. Proc. Natl. Acad. Sci. USA 2013, 110, 6406–6411. [Google Scholar] [CrossRef]
- Sato, R.; Okura, T.; Kawahara, M.; Takizawa, N.; Momose, F.; Morikawa, Y. Apical Trafficking Pathways of Influenza A Virus HA and NA via Rab17- and Rab23-Positive Compartments. Front. Microbiol. 2019, 10, 1857. [Google Scholar] [CrossRef]
- Scheiffele, P.; Rietveld, A.; Wilk, T.; Simons, K. Influenza viruses select ordered lipid domains during budding from the plasma membrane. J. Biol. Chem. 1999, 274, 2038–2044. [Google Scholar] [CrossRef]
- Veit, M.; Thaa, B. Association of influenza virus proteins with membrane rafts. Adv. Virol. 2011, 2011, 370606. [Google Scholar] [CrossRef]
- Zhang, J.; Pekosz, A.; Lamb, R.A. Influenza virus assembly and lipid raft microdomains: A role for the cytoplasmic tails of the spike glycoproteins. J. Virol. 2000, 74, 4634–4644. [Google Scholar] [CrossRef]
- Gerl, M.J.; Sampaio, J.L.; Urban, S.; Kalvodova, L.; Verbavatz, J.M.; Binnington, B.; Lindemann, D.; Lingwood, C.A.; Shevchenko, A.; Schroeder, C.; et al. Quantitative analysis of the lipidomes of the influenza virus envelope and MDCK cell apical membrane. J. Cell Biol. 2012, 196, 213–221. [Google Scholar] [CrossRef]
- Barman, S.; Nayak, D.P. Lipid raft disruption by cholesterol depletion enhances influenza A virus budding from MDCK cells. J. Virol. 2007, 81, 12169–12178. [Google Scholar] [CrossRef]
- Rossman, J.S.; Jing, X.; Leser, G.P.; Balannik, V.; Pinto, L.H.; Lamb, R.A. Influenza virus m2 ion channel protein is necessary for filamentous virion formation. J. Virol. 2010, 84, 5078–5088. [Google Scholar] [CrossRef]
- Barman, S.; Nayak, D.P. Analysis of the transmembrane domain of influenza virus neuraminidase, a type II transmembrane glycoprotein, for apical sorting and raft association. J. Virol. 2000, 74, 6538–6545. [Google Scholar] [CrossRef]
- Engel, S.; de Vries, M.; Herrmann, A.; Veit, M. Mutation of a raft-targeting signal in the transmembrane region retards transport of influenza virus hemagglutinin through the Golgi. FEBS Lett. 2012, 586, 277–282. [Google Scholar] [CrossRef]
- Wilson, R.L.; Frisz, J.F.; Klitzing, H.A.; Zimmerberg, J.; Weber, P.K.; Kraft, M.L. Hemagglutinin Clusters in the Plasma Membrane Are Not Enriched with Cholesterol and Sphingolipids. Biophys. J. 2015, 108, 1652–1659. [Google Scholar] [CrossRef]
- Leser, G.P.; Lamb, R.A. Lateral Organization of Influenza Virus Proteins in the Budozone Region of the Plasma Membrane. J. Virol. 2017, 91, e02104-16. [Google Scholar] [CrossRef]
- Leser, G.P.; Lamb, R.A. Influenza virus assembly and budding in raft-derived microdomains: A quantitative analysis of the surface distribution of HA, NA and M2 proteins. Virology 2005, 342, 215–227. [Google Scholar] [CrossRef]
- Veit, M.; Engel, S.; Thaa, B.; Scolari, S.; Herrmann, A. Lipid domain association of influenza virus proteins detected by dynamic fluorescence microscopy techniques. Cell. Microbiol. 2013, 15, 179–189. [Google Scholar] [CrossRef]
- Curthoys, N.M.; Mlodzianoski, M.J.; Parent, M.; Butler, M.B.; Raut, P.; Wallace, J.; Lilieholm, J.; Mehmood, K.; Maginnis, M.S.; Waters, H.; et al. Influenza Hemagglutinin Modulates Phosphatidylinositol 4,5-Bisphosphate Membrane Clustering. Biophys. J. 2019, 116, 893–909. [Google Scholar] [CrossRef]
- Raut, P.; Obeng, B.; Waters, H.; Zimmerberg, J.; Gosse, J.A.; Hess, S.T. Phosphatidylinositol 4,5-Bisphosphate Mediates the Co-Distribution of Influenza A Hemagglutinin and Matrix Protein M1 at the Plasma Membrane. Viruses 2022, 14, 2509. [Google Scholar] [CrossRef]
- Ekanayake, E.V.; Fu, R.; Cross, T.A. Structural Influences: Cholesterol, Drug, and Proton Binding to Full-Length Influenza A M2 Protein. Biophys. J. 2016, 110, 1391–1399. [Google Scholar] [CrossRef] [PubMed]
- Elkins, M.R.; Williams, J.K.; Gelenter, M.D.; Dai, P.; Kwon, B.; Sergeyev, I.V.; Pentelute, B.L.; Hong, M. Cholesterol-binding site of the influenza M2 protein in lipid bilayers from solid-state NMR. Proc. Natl. Acad. Sci. USA 2017, 114, 12946–12951. [Google Scholar] [CrossRef] [PubMed]
- Georgieva, E.R.; Borbat, P.P.; Norman, H.D.; Freed, J.H. Mechanism of influenza A M2 transmembrane domain assembly in lipid membranes. Sci. Rep. 2015, 5, 11757. [Google Scholar] [CrossRef] [PubMed]
- Kuroki, T.; Hatta, T.; Natsume, T.; Sakai, N.; Yagi, A.; Kato, K.; Nagata, K.; Kawaguchi, A. ARHGAP1 Transported with Influenza Viral Genome Ensures Integrity of Viral Particle Surface through Efficient Budozone Formation. mBio 2022, 13, e0072122. [Google Scholar] [CrossRef]
- Manzoor, R.; Igarashi, M.; Takada, A. Influenza A Virus M2 Protein: Roles from Ingress to Egress. Int. J. Mol. Sci. 2017, 18, 2649. [Google Scholar] [CrossRef]
- Martyna, A.; Bahsoun, B.; Madsen, J.J.; Jackson, F.; Badham, M.D.; Voth, G.A.; Rossman, J.S. Cholesterol Alters the Orientation and Activity of the Influenza Virus M2 Amphipathic Helix in the Membrane. J. Phys. Chem. B 2020, 124, 6738–6747. [Google Scholar] [CrossRef] [PubMed]
- Paulino, J.; Pang, X.; Hung, I.; Zhou, H.X.; Cross, T.A. Influenza A M2 Channel Clustering at High Protein/Lipid Ratios: Viral Budding Implications. Biophys. J. 2019, 116, 1075–1084. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, M.; Tran, N.; Hong, M. Clustering of tetrameric influenza M2 peptides in lipid bilayers investigated by (19)F solid-state NMR. Biochim. Biophys. Acta Biomembr. 2022, 1864, 183909. [Google Scholar] [CrossRef]
- Beale, R.; Wise, H.; Stuart, A.; Ravenhill, B.J.; Digard, P.; Randow, F. A LC3-interacting motif in the influenza A virus M2 protein is required to subvert autophagy and maintain virion stability. Cell Host Microbe 2014, 15, 239–247. [Google Scholar] [CrossRef]
- Dunsing, V.; Petrich, A.; Chiantia, S. Multicolor fluorescence fluctuation spectroscopy in living cells via spectral detection. Elife 2021, 10, e69687. [Google Scholar] [CrossRef] [PubMed]
- Durgan, J.; Florey, O. Many roads lead to CASM: Diverse stimuli of noncanonical autophagy share a unifying molecular mechanism. Sci. Adv. 2022, 8, eabo1274. [Google Scholar] [CrossRef] [PubMed]
- Bobone, S.; Hilsch, M.; Storm, J.; Dunsing, V.; Herrmann, A.; Chiantia, S. Phosphatidylserine Lateral Organization Influences the Interaction of Influenza Virus Matrix Protein 1 with Lipid Membranes. J. Virol. 2017, 91, e00267-17. [Google Scholar] [CrossRef] [PubMed]
- Kordyukova, L.V.; Shtykova, E.V.; Baratova, L.A.; Svergun, D.I.; Batishchev, O.V. Matrix proteins of enveloped viruses: A case study of Influenza A virus M1 protein. J. Biomol. Struct. Dyn. 2019, 37, 671–690. [Google Scholar] [CrossRef]
- Petrich, A.; Dunsing, V.; Bobone, S.; Chiantia, S. Influenza A M2 recruits M1 to the plasma membrane: A fluorescence fluctuation microscopy study. Biophys. J. 2021, 120, 5478–5490. [Google Scholar] [CrossRef]
- Zhou, Y.; Pu, J.; Wu, Y. The Role of Lipid Metabolism in Influenza A Virus Infection. Pathogens 2021, 10, 303. [Google Scholar] [CrossRef]
- Kawaguchi, A.; Hirohama, M.; Harada, Y.; Osari, S.; Nagata, K. Influenza Virus Induces Cholesterol-Enriched Endocytic Recycling Compartments for Budozone Formation via Cell Cycle-Independent Centrosome Maturation. PLoS Pathog. 2015, 11, e1005284. [Google Scholar] [CrossRef]
- Ma, Y.; Yamamoto, Y.; Nicovich, P.R.; Goyette, J.; Rossy, J.; Gooding, J.J.; Gaus, K. A FRET sensor enables quantitative measurements of membrane charges in live cells. Nat. Biotechnol. 2017, 35, 363–370. [Google Scholar] [CrossRef]
- Amaro, M.; Reina, F.; Hof, M.; Eggeling, C.; Sezgin, E. Laurdan and Di-4-ANEPPDHQ probe different properties of the membrane. J. Phys. D Appl. Phys. 2017, 50, 134004. [Google Scholar] [CrossRef]
- Owen, D.M.; Rentero, C.; Magenau, A.; Abu-Siniyeh, A.; Gaus, K. Quantitative imaging of membrane lipid order in cells and organisms. Nat. Protoc. 2011, 7, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Chiantia, S.; Ries, J.; Schwille, P. Fluorescence correlation spectroscopy in membrane structure elucidation. Biochim. Biophys. Acta 2009, 1788, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Dunsing, V.; Chiantia, S. A Fluorescence Fluctuation Spectroscopy Assay of Protein-Protein Interactions at Cell-Cell Contacts. JoVE 2018, 142, e58582. [Google Scholar] [CrossRef]
- Tzoneva, R.; Stoyanova, T.; Petrich, A.; Popova, D.; Uzunova, V.; Momchilova, A.; Chiantia, S. Effect of Erufosine on Membrane Lipid Order in Breast Cancer Cell Models. Biomolecules 2020, 10, 802. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.-S.; Wagenknecht-Wiesner, A.; Yin, B.; Suresh, P.; London, E.; Baird, B.; Bag, N. Lipid Driven Inter-leaflet Coupling of Plasma Membrane Order Regulates FcεRI Signaling in Mast Cells. Biophys. J. 2023. Online ahead of print. [Google Scholar] [CrossRef]
- Dunsing, V.; Luckner, M.; Zühlke, B.; Petazzi, R.A.; Herrmann, A.; Chiantia, S. Optimal fluorescent protein tags for quantifying protein oligomerization in living cells. Sci. Rep. 2018, 8, 10634. [Google Scholar] [CrossRef]
- Wagner, R.; Gabriel, G.; Schlesner, M.; Alex, N.; Herwig, A.; Werner, O.; Klenk, H.-D. Protease activation mutants elicit protective immunity against highly pathogenic avian influenza viruses of subtype H7 in chickens and mice. Emerg. Microbes Infect. 2013, 2, e7. [Google Scholar] [CrossRef]
- Sezgin, E.; Waithe, D.; Bernardino de la Serna, J.; Eggeling, C. Spectral imaging to measure heterogeneity in membrane lipid packing. Chemphyschem 2015, 16, 1387–1394. [Google Scholar] [CrossRef]
- Petrich, A.; Koikkarah Aji, A.; Dunsing, V.; Chiantia, S. Benchmarking of novel green fluorescent proteins for the quantification of protein oligomerization in living cells. PLoS ONE 2023, 18, e0285486. [Google Scholar] [CrossRef]
- Petrášek, Z.; Schwille, P. Precise Measurement of Diffusion Coefficients using Scanning Fluorescence Correlation Spectroscopy. Biophys. J. 2008, 94, 1437–1448. [Google Scholar] [CrossRef]
- Ries, J.; Chiantia, S.; Schwille, P. Accurate determination of membrane dynamics with line-scan FCS. Biophys. J. 2009, 96, 1999–2008. [Google Scholar] [CrossRef]
- Ries, J.; Schwille, P. Studying Slow Membrane Dynamics with Continuous Wave Scanning Fluorescence Correlation Spectroscopy. Biophys. J. 2006, 91, 1915–1924. [Google Scholar] [CrossRef]
- Liu, S.; Mok, B.W.-Y.; Deng, S.; Liu, H.; Wang, P.; Song, W.; Chen, P.; Huang, X.; Zheng, M.; Lau, S.-Y.; et al. Mammalian cells use the autophagy process to restrict avian influenza virus replication. Cell Rep. 2021, 35, 109213. [Google Scholar] [CrossRef]
- Chaurio, R.A.; Janko, C.; Muñoz, L.E.; Frey, B.; Herrmann, M.; Gaipl, U.S. Phospholipids: Key Players in Apoptosis and Immune Regulation. Molecules 2009, 14, 4892–4914. [Google Scholar] [CrossRef]
- Gui, R.; Chen, Q. Molecular Events Involved in Influenza A Virus-Induced Cell Death. Front. Microbiol. 2021, 12, 797789. [Google Scholar] [CrossRef]
- Bailey, R.W.; Nguyen, T.; Robertson, L.; Gibbons, E.; Nelson, J.; Christensen, R.E.; Bell, J.P.; Judd, A.M.; Bell, J.D. Sequence of Physical Changes to the Cell Membrane During Glucocorticoid-Induced Apoptosis in S49 Lymphoma Cells. Biophys. J. 2009, 96, 2709–2718. [Google Scholar] [CrossRef]
- Darwich, Z.; Klymchenko, A.S.; Kucherak, O.A.; Richert, L.; Mély, Y. Detection of apoptosis through the lipid order of the outer plasma membrane leaflet. Biochim. Biophys. Acta 2012, 1818, 3048–3054. [Google Scholar] [CrossRef]
- Tochigi, M.; Inoue, T.; Suzuki-Karasaki, M.; Ochiai, T.; Ra, C.; Suzuki-Karasaki, Y. Hydrogen peroxide induces cell death in human TRAIL-resistant melanoma through intracellular superoxide generation. Int. J. Oncol. 2013, 42, 863–872. [Google Scholar] [CrossRef]
- Wu, C.A.; Yang, Y.W. Induction of cell death by saponin and antigen delivery. Pharm. Res. 2004, 21, 271–277. [Google Scholar] [CrossRef]
- Leavesley, S.J.; Britain, A.L.; Cichon, L.K.; Nikolaev, V.O.; Rich, T.C. Assessing FRET using spectral techniques. Cytom. Part A 2013, 83, 898–912. [Google Scholar] [CrossRef]
- Pokorna, S.; Ventura, A.E.; Santos, T.C.B.; Hof, M.; Prieto, M.; Futerman, A.H.; Silva, L.C. Laurdan in live cell imaging: Effect of acquisition settings, cell culture conditions and data analysis on generalized polarization measurements. J. Photochem. Photobiol. B 2022, 228, 112404. [Google Scholar] [CrossRef]
- Fellmann, P.; Hervé, P.; Pomorski, T.; Müller, P.; Geldwerth, D.; Herrmann, A.; Devaux, P.F. Transmembrane movement of diether phospholipids in human erythrocytes and human fibroblasts. Biochemistry 2000, 39, 4994–5003. [Google Scholar] [CrossRef]
- Herrera, S.A.; Grifell-Junyent, M.; Pomorski, T.G. NBD-lipid Uptake Assay for Mammalian Cell Lines. Bio-Protocol 2022, 12, e4330. [Google Scholar] [CrossRef]
- Klenk, H.D.; Rott, R.; Becht, H. On the structure of the influenza virus envelope. Virology 1972, 47, 579–591. [Google Scholar] [CrossRef]
- Landsberger, F.R.; Lenard, J.; Paxton, J.; Compans, R.W. Spin-labeled electron spin resonance study of the lipid-containing membrane of influenza virus. Proc. Natl. Acad. Sci. USA 1971, 68, 2579–2583. [Google Scholar] [CrossRef]
- Golfetto, O.; Hinde, E.; Gratton, E. The Laurdan spectral phasor method to explore membrane micro-heterogeneity and lipid domains in live cells. Methods Mol. Biol. 2015, 1232, 273–290. [Google Scholar] [CrossRef]
- Jin, L.; Millard, A.C.; Wuskell, J.P.; Dong, X.; Wu, D.; Clark, H.A.; Loew, L.M. Characterization and application of a new optical probe for membrane lipid domains. Biophys. J. 2006, 90, 2563–2575. [Google Scholar] [CrossRef]
- Sezgin, E.; Sadowski, T.; Simons, K. Measuring lipid packing of model and cellular membranes with environment sensitive probes. Langmuir 2014, 30, 8160–8166. [Google Scholar] [CrossRef] [PubMed]
- Dunsing, V.; Mayer, M.; Liebsch, F.; Multhaup, G.; Chiantia, S. Direct evidence of amyloid precursor-like protein 1 trans interactions in cell-cell adhesion platforms investigated via fluorescence fluctuation spectroscopy. Mol. Biol. Cell 2017, 28, 3609–3620. [Google Scholar] [CrossRef]
- Goyette, J.; Gaus, K. Mechanisms of protein nanoscale clustering. Curr. Opin. Cell Biol. 2017, 44, 86–92. [Google Scholar] [CrossRef]
- Kakisaka, M.; Yamada, K.; Yamaji-Hasegawa, A.; Kobayashi, T.; Aida, Y. Intrinsically disordered region of influenza A NP regulates viral genome packaging via interactions with viral RNA and host PI(4,5)P2. Virology 2016, 496, 116–126. [Google Scholar] [CrossRef]
- Motsa, B.B.; Stahelin, R.V. Lipid-protein interactions in virus assembly and budding from the host cell plasma membrane. Biochem. Soc. Trans. 2021, 49, 1633–1641. [Google Scholar] [CrossRef]
- Orlikowska-Rzeznik, H.; Krok, E.; Chattopadhyay, M.; Lester, A.; Piatkowski, L. Laurdan Discerns Lipid Membrane Hydration and Cholesterol Content. J. Phys. Chem. B 2023, 127, 3382–3391. [Google Scholar] [CrossRef]
- Doole, F.T.; Gupta, S.; Kumarage, T.; Ashkar, R.; Brown, M.F. Biophysics of Membrane Stiffening by Cholesterol and Phosphatidylinositol 4,5-bisphosphate (PIP2). Adv. Exp. Med. Biol. 2023, 1422, 61–85. [Google Scholar] [CrossRef]
- Frewein, M.P.K.; Piller, P.; Semeraro, E.F.; Czakkel, O.; Gerelli, Y.; Porcar, L.; Pabst, G. Distributing aminophospholipids asymmetrically across leaflets causes anomalous membrane stiffening. Biophys. J. 2023, 122, 2445–2455. [Google Scholar] [CrossRef]
- Maynard, S.A.; Triller, A. Inhibitory Receptor Diffusion Dynamics. Front. Mol. Neurosci. 2019, 12, 313. [Google Scholar] [CrossRef]
- Alenghat, F.J.; Golan, D.E. Membrane protein dynamics and functional implications in mammalian cells. Curr. Top. Membr. 2013, 72, 89–120. [Google Scholar] [CrossRef]
- Sarmento, M.J.; Hof, M.; Šachl, R. Interleaflet Coupling of Lipid Nanodomains—Insights From in vitro Systems. Front. Cell Dev. Biol. 2020, 8, 284. [Google Scholar] [CrossRef]
- Bag, N.; Huang, S.; Wohland, T. Plasma Membrane Organization of Epidermal Growth Factor Receptor in Resting and Ligand-Bound States. Biophys. J. 2015, 109, 1925–1936. [Google Scholar] [CrossRef] [PubMed]
- Gudheti, M.V.; Curthoys, N.M.; Gould, T.J.; Kim, D.; Gunewardene, M.S.; Gabor, K.A.; Gosse, J.A.; Kim, C.H.; Zimmerberg, J.; Hess, S.T. Actin mediates the nanoscale membrane organization of the clustered membrane protein influenza hemagglutinin. Biophys. J. 2013, 104, 2182–2192. [Google Scholar] [CrossRef]
- Huang, S.; Lim, S.Y.; Gupta, A.; Bag, N.; Wohland, T. Plasma membrane organization and dynamics is probe and cell line dependent. Biochim. Biophys. Acta Biomembr. 2017, 1859, 1483–1492. [Google Scholar] [CrossRef]
- Frisz, J.F.; Lou, K.; Klitzing, H.A.; Hanafin, W.P.; Lizunov, V.; Wilson, R.L.; Carpenter, K.J.; Kim, R.; Hutcheon, I.D.; Zimmerberg, J.; et al. Direct chemical evidence for sphingolipid domains in the plasma membranes of fibroblasts. Proc. Natl. Acad. Sci. USA 2013, 110, E613–E622. [Google Scholar] [CrossRef]
- Chua, S.; Cui, J.; Engelberg, D.; Lim, L.H.K. A Review and Meta-Analysis of Influenza Interactome Studies. Front. Microbiol. 2022, 13, 869406. [Google Scholar] [CrossRef]
- Bajimaya, S.; Frankl, T.; Hayashi, T.; Takimoto, T. Cholesterol is required for stability and infectivity of influenza A and respiratory syncytial viruses. Virology 2017, 510, 234–241. [Google Scholar] [CrossRef]
- Zeng, L.; Li, J.; Lv, M.; Li, Z.; Yao, L.; Gao, J.; Wu, Q.; Wang, Z.; Yang, X.; Tang, G.; et al. Environmental Stability and Transmissibility of Enveloped Viruses at Varied Animate and Inanimate Interfaces. Environ. Health 2023, 1, 15–31. [Google Scholar] [CrossRef] [PubMed]
- Angelova, M.I.; Dimitrov, D.S. Liposome electroformation. Faraday Discuss. Chem. Soc. 1986, 81, 303–311. [Google Scholar] [CrossRef]
- Stein, H.; Spindler, S.; Bonakdar, N.; Wang, C.; Sandoghdar, V. Production of Isolated Giant Unilamellar Vesicles under High Salt Concentrations. Front. Physiol. 2017, 8, 63. [Google Scholar] [CrossRef]
- Malacrida, L.; Astrada, S.; Briva, A.; Bollati-Fogolín, M.; Gratton, E.; Bagatolli, L.A. Spectral phasor analysis of LAURDAN fluorescence in live A549 lung cells to study the hydration and time evolution of intracellular lamellar body-like structures. Biochim. Biophys. Acta 2016, 1858, 2625–2635. [Google Scholar] [CrossRef]
- Klymchenko, A.S.; Kreder, R. Fluorescent probes for lipid rafts: From model membranes to living cells. Chem. Biol. 2014, 21, 97–113. [Google Scholar] [CrossRef]
- Bondelli, G.; Sardar, S.; Chiaravalli, G.; Vurro, V.; Paternò, G.M.; Lanzani, G.; D’Andrea, C. Shedding Light on Thermally Induced Optocapacitance at the Organic Biointerface. J. Phys. Chem. B 2021, 125, 10748–10758. [Google Scholar] [CrossRef]
- Gunther, G.; Malacrida, L.; Jameson, D.M.; Gratton, E.; Sánchez, S.A. LAURDAN since Weber: The Quest for Visualizing Membrane Heterogeneity. Acc. Chem. Res. 2021, 54, 976–987. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Lei, Y.; Ma, Y.; Liu, M.; Zheng, J.; Dan, D.; Gao, P. A Comprehensive Review of Fluorescence Correlation Spectroscopy. Front. Phys. 2021, 9, 644450. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petrich, A.; Chiantia, S. Influenza A Virus Infection Alters Lipid Packing and Surface Electrostatic Potential of the Host Plasma Membrane. Viruses 2023, 15, 1830. https://doi.org/10.3390/v15091830
Petrich A, Chiantia S. Influenza A Virus Infection Alters Lipid Packing and Surface Electrostatic Potential of the Host Plasma Membrane. Viruses. 2023; 15(9):1830. https://doi.org/10.3390/v15091830
Chicago/Turabian StylePetrich, Annett, and Salvatore Chiantia. 2023. "Influenza A Virus Infection Alters Lipid Packing and Surface Electrostatic Potential of the Host Plasma Membrane" Viruses 15, no. 9: 1830. https://doi.org/10.3390/v15091830
APA StylePetrich, A., & Chiantia, S. (2023). Influenza A Virus Infection Alters Lipid Packing and Surface Electrostatic Potential of the Host Plasma Membrane. Viruses, 15(9), 1830. https://doi.org/10.3390/v15091830




