Different Patterns of Codon Usage and Amino Acid Composition across Primate Lentiviruses
Abstract
1. Introduction
2. Materials and Methods
3. Results
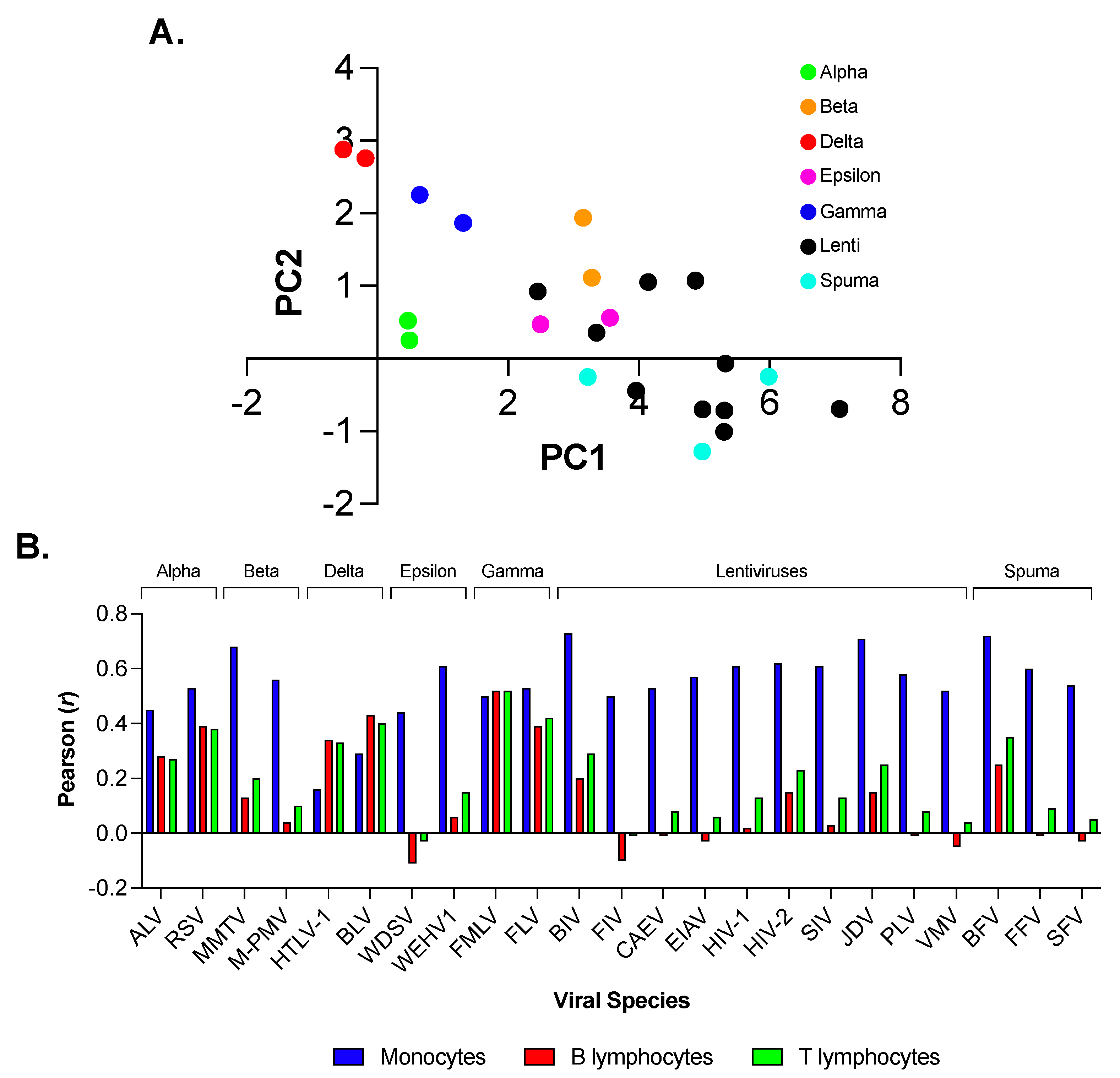
3.1. Principal Component Analysis (PCA) of Codon Usage in the Family of Retroviridae Revealed a Considerable Degree of Variation
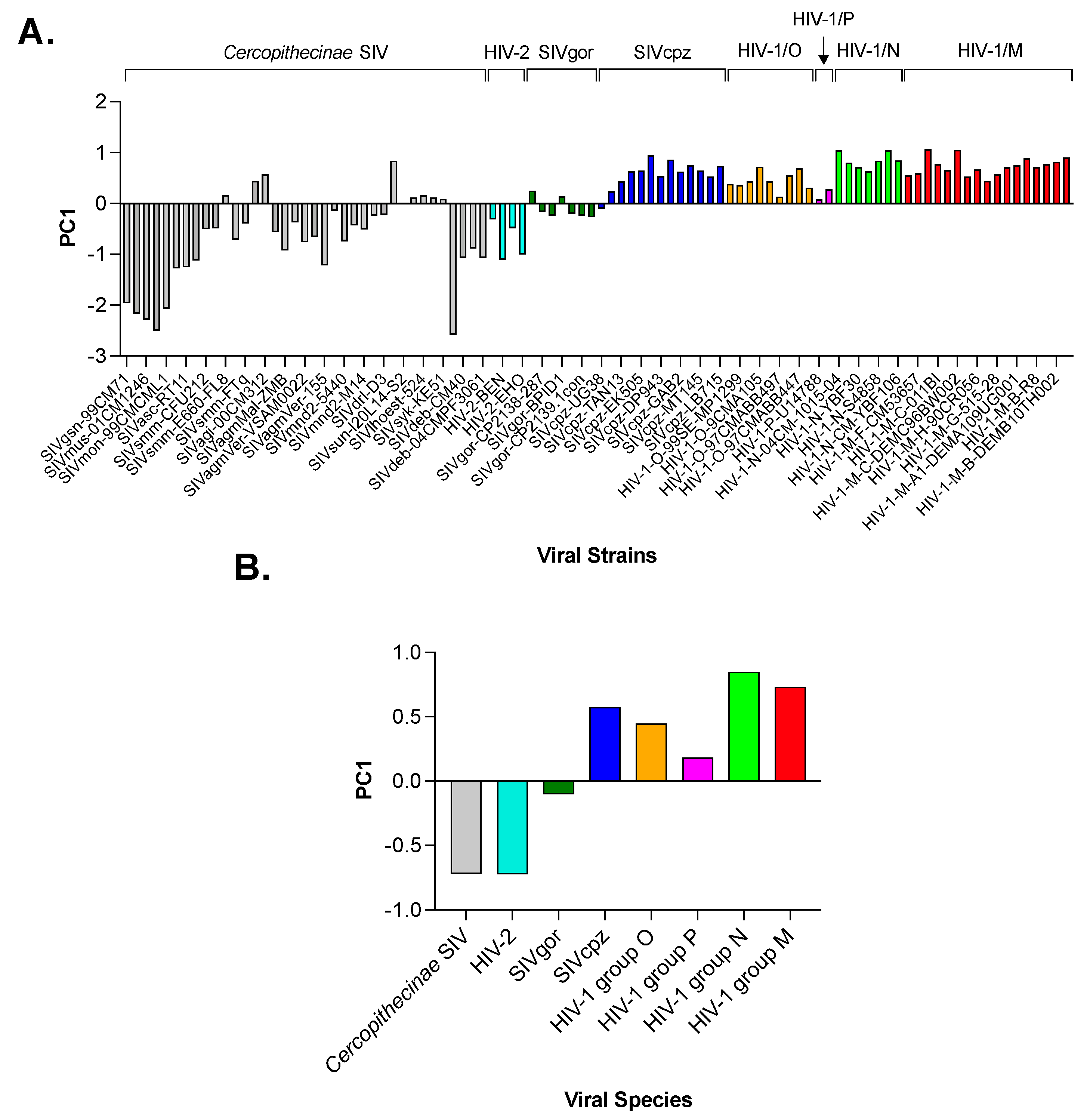
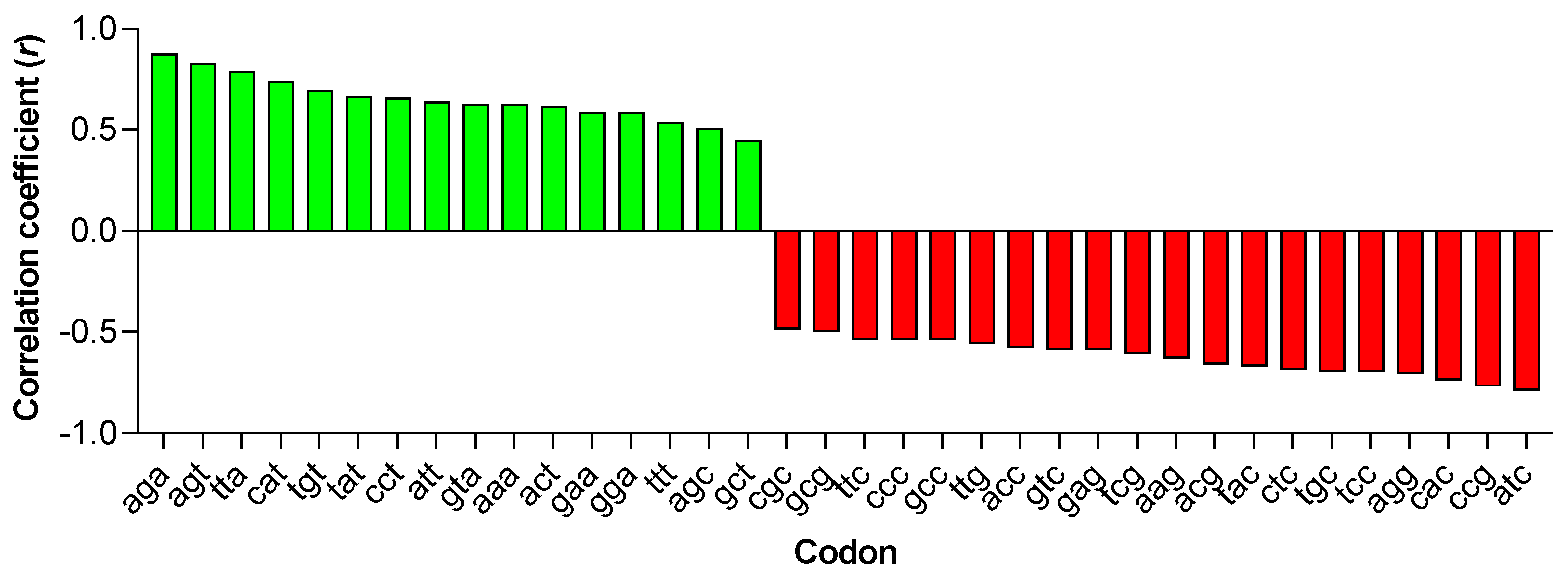
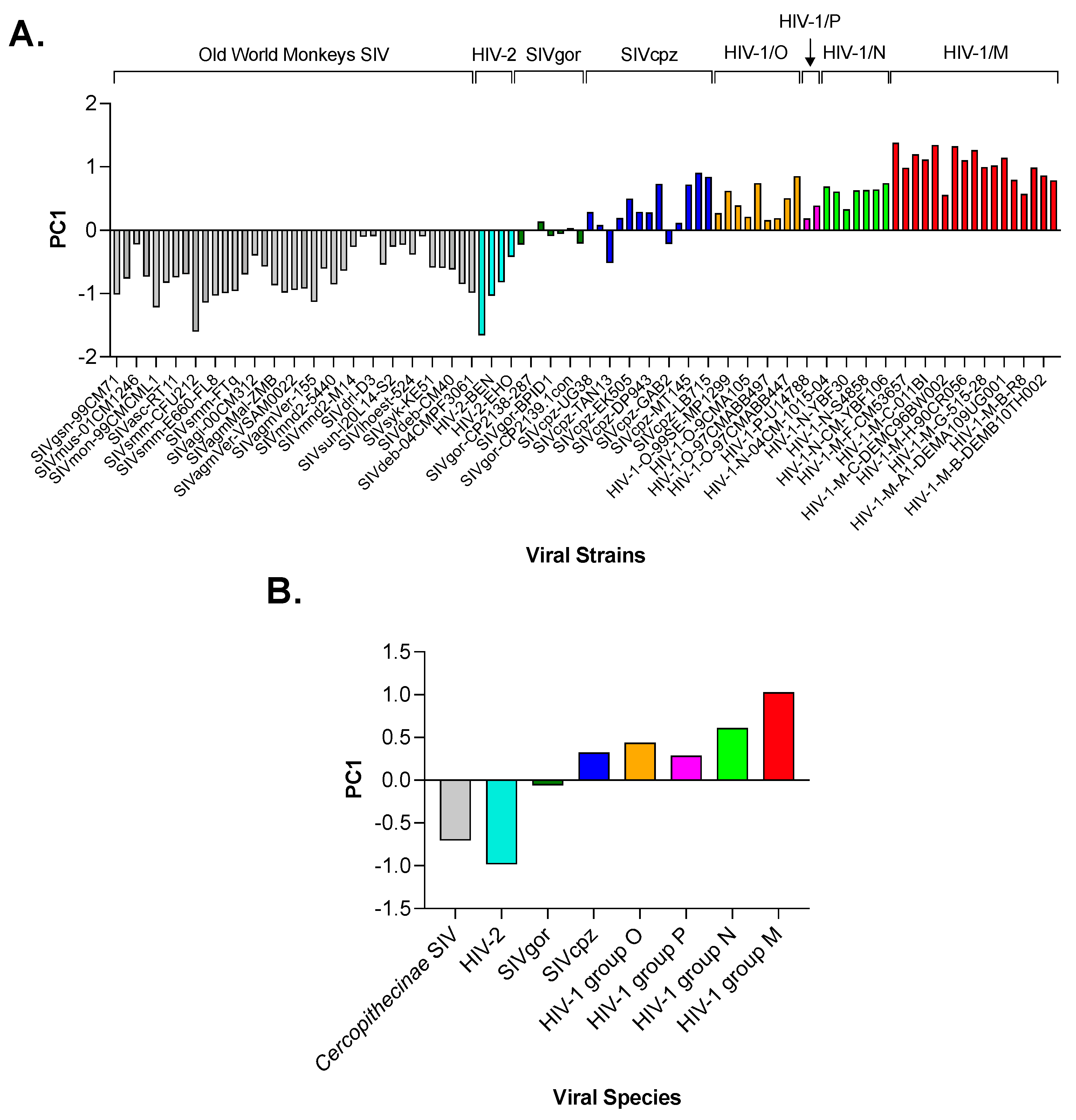
3.2. Principal Component Analysis (PCA) in Primate Lentiviruses Revealed a Trend in the Use of Synonymous and Nonsynonymous Codons
3.3. Principal Component Analysis (PCA) in Primate Lentiviruses Revealed a Trend in the Amino Acid Composition
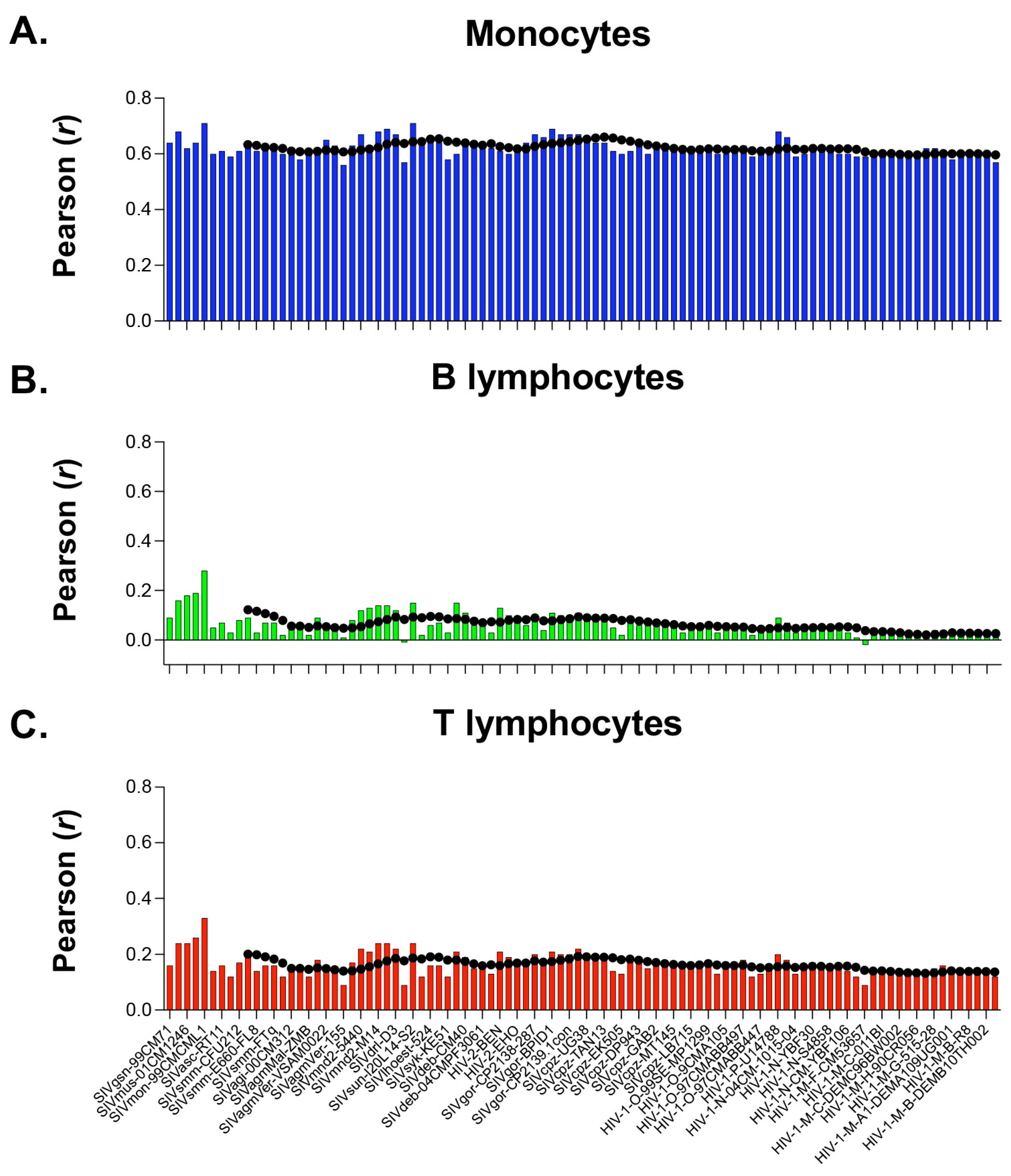
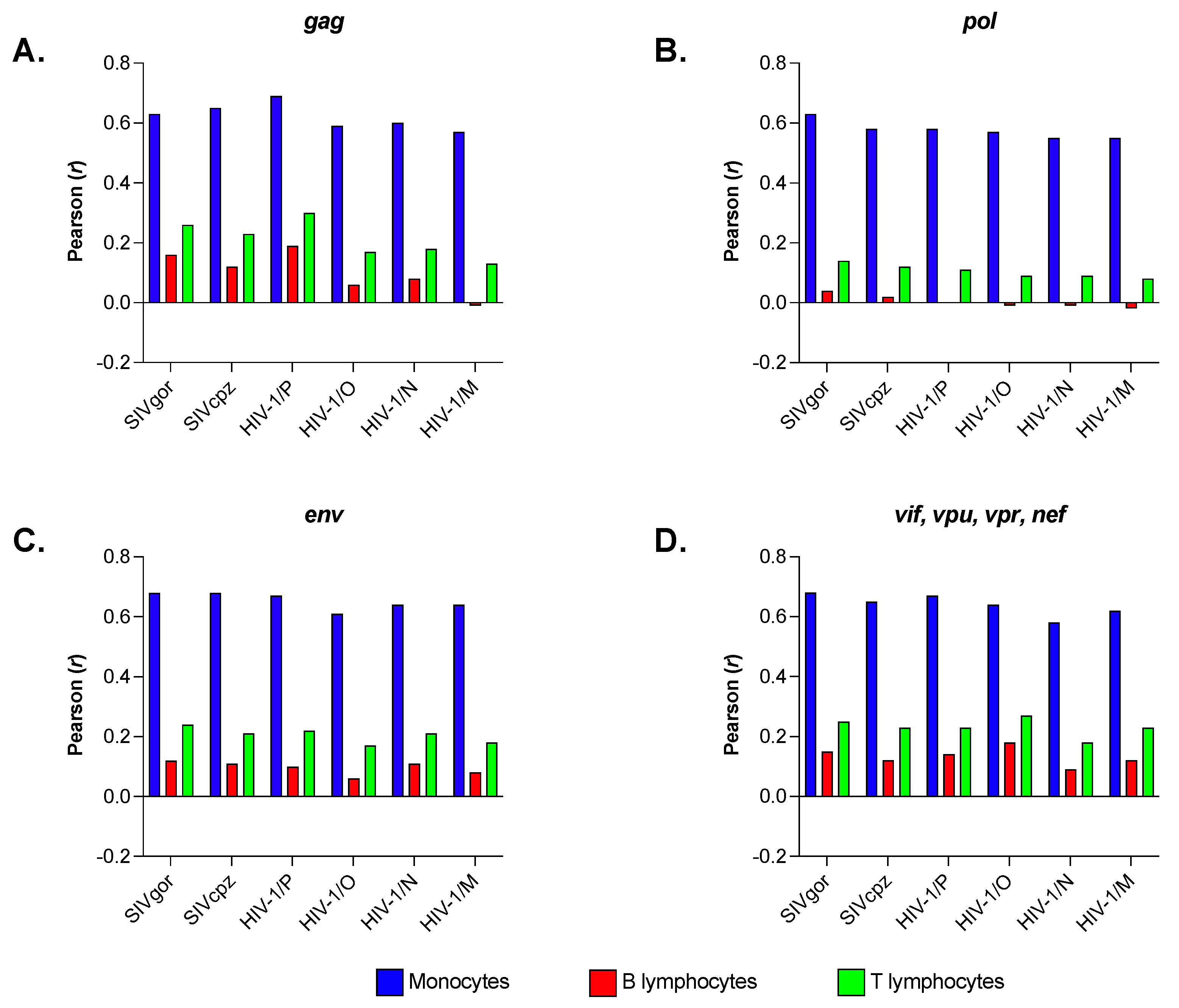
3.4. Primate Lentiviruses Have a Codon Usage Significantly Correlated to That of Human Monocytes (Innate Immunity) but Not to That of Human B and T Lymphocytes (Adaptive Immunity)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- van Hemert, F.J.; Berkhout, B. The tendency of lentiviral open reading frames to become A-rich: Constraints imposed by viral genome organization and cellular tRNA availability. J. Mol. Evol. 1995, 41, 132–140. [Google Scholar] [CrossRef] [PubMed]
- van der Kuyl, A.C.; Berkhout, B. The biased nucleotide composition of the HIV genome: A constant factor in a highly variable virus. Retrovirology 2012, 9, 92. [Google Scholar] [CrossRef] [PubMed]
- van Hemert, F.; van der Kuyl, A.C.; Berkhout, B. On the nucleotide composition and structure of retroviral RNA genomes. Virus Res. 2014, 193, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Kustin, T.; Stern, A. Biased Mutation and Selection in RNA Viruses. Mol. Biol. Evol. 2021, 38, 575–588. [Google Scholar] [CrossRef] [PubMed]
- Berkhout, B.; van Hemert, F.J. The unusual nucleotide content of the HIV RNA genome results in a biased amino acid composition of HIV proteins. Nucleic Acids Res. 1994, 22, 1705–1711. [Google Scholar] [CrossRef] [PubMed]
- Berkhout, B.; Grigoriev, A.; Bakker, M.; Lukashov, V.V. Codon and amino acid usage in retroviral genomes is consistent with virus-specific nucleotide pressure. AIDS Res. Hum. Retroviruses 2002, 18, 133–141. [Google Scholar] [CrossRef]
- Cho, M.; Min, X.; Son, H.S. Analysis of evolutionary and genetic patterns in structural genes of primate lentiviruses. Genes Genom. 2022, 44, 773–791. [Google Scholar] [CrossRef]
- Karlin, S.; Doerfler, W.; Cardon, L.R. Why is CpG suppressed in the genomes of virtually all small eukaryotic viruses but not in those of large eukaryotic viruses? J. Virol. 1994, 68, 2889–2897. [Google Scholar] [CrossRef]
- Cheng, X.; Virk, N.; Chen, W.; Ji, S.; Ji, S.; Sun, Y.; Wu, X. CpG usage in RNA viruses: Data and hypotheses. PLoS ONE 2013, 8, e74109. [Google Scholar] [CrossRef]
- Subramanian, K.; Payne, B.; Feyertag, F.; Alvarez-Ponce, D. The Codon Statistics Database: A Database of Codon Usage Bias. Mol. Biol. Evol. 2022, 39. [Google Scholar] [CrossRef]
- Pedersen, A.K.; Wiuf, C.; Christiansen, F.B. A codon-based model designed to describe lentiviral evolution. Mol. Biol. Evol. 1998, 15, 1069–1081. [Google Scholar] [CrossRef]
- Vidyavijayan, K.K.; Hassan, S.; Precilla, L.K.; Ashokkumar, M.; Chandrasekeran, P.; Swaminathan, S.; Hanna, L.E. Biased Nucleotide Composition and Differential Codon Usage Pattern in HIV-1 and HIV-2. AIDS Res. Hum. Retroviruses 2017, 33, 298–307. [Google Scholar] [CrossRef] [PubMed]
- Sharp, P.M. What can AIDS virus codon usage tell us? Nature 1986, 324, 114. [Google Scholar] [CrossRef] [PubMed]
- Ruzman, M.A.; Ripen, A.M.; Mirsafian, H.; Ridzwan, N.F.; Mohamad, S.B. Analysis of synonymous codon usage bias in human monocytes, B, and T lymphocytes based on transcriptome data. Gene Rep. 2021, 23, 101034. [Google Scholar] [CrossRef]
- Kumar, A.; Abbas, W.; Herbein, G. HIV-1 latency in monocytes/macrophages. Viruses 2014, 6, 1837–1860. [Google Scholar] [CrossRef] [PubMed]
- Veenhuis, R.T.; Abreu, C.M.; Costa, P.A.G.; Ferreira, E.A.; Ratliff, J.; Pohlenz, L.; Shirk, E.N.; Rubin, L.H.; Blankson, J.N.; Gama, L.; et al. Monocyte-derived macrophages contain persistent latent HIV reservoirs. Nat. Microbiol. 2023, 8, 833–844. [Google Scholar] [CrossRef] [PubMed]
- Wright, F. The ‘effective number of codons’ used in a gene. Gene 1990, 87, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Sharp, P.M.; Li, W.H. The codon Adaptation Index—A measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987, 15, 1281–1295. [Google Scholar] [CrossRef] [PubMed]
- Hotelling, H. Relations between two sets of variates. Biometrika 1936, 28, 321–377. [Google Scholar] [CrossRef]
- Morrison, D.F. Multivariate Statistical Methods; McGraw-Hill: New York, NY, USA, 1976. [Google Scholar]
- Ringner, M. What is principal component analysis? Nat. Biotechnol. 2008, 26, 303–304. [Google Scholar] [CrossRef] [PubMed]
- Pavesi, A.; Romerio, F. Extending the Coding Potential of Viral Genomes with Overlapping Antisense ORFs: A Case for the De Novo Creation of the Gene Encoding the Antisense Protein ASP of HIV-1. Viruses 2022, 14, 146. [Google Scholar] [CrossRef] [PubMed]
- Le, S.Y.; Malim, M.H.; Cullen, B.R.; Maizel, J.V. A highly conserved RNA folding region coincident with the Rev response element of primate immunodeficiency viruses. Nucleic Acids Res. 1990, 18, 1613–1623. [Google Scholar] [CrossRef] [PubMed]
- Mirsafian, H.; Ripen, A.M.; Manaharan, T.; Mohamad, S.B.; Merican, A.F. Toward a Reference Gene Catalog of Human Primary Monocytes. OMICS 2016, 20, 627–634. [Google Scholar] [CrossRef] [PubMed]
- Hotelling, H. The selection of variates for use in prediction with some comments on the problem of nuisance parameters. Ann. Math. Stat. 1940, 11, 271–283. [Google Scholar] [CrossRef]
- Dawson, B.; Trapp, R.G. Basic and Clinical Biostatistics, 3rd ed.; McGraw-Hill: New York, NY, USA, 2001. [Google Scholar]
- Snedecor, G.W.; Cochran, W.G. Statistical Methods, 6th ed.; Iowa State University Press: Ames, IA, USA, 1967; p. 593. [Google Scholar]
- RoyChoudhury, S.; Mukherjee, D. Complex codon usage pattern and compositional features of retroviruses. Comput. Math. Methods Med. 2013, 2013, 848123. [Google Scholar] [CrossRef]
- Pandit, A.; Sinha, S. Differential trends in the codon usage patterns in HIV-1 genes. PLoS ONE 2011, 6, e28889. [Google Scholar] [CrossRef]
- Koning, F.A.; Newman, E.N.; Kim, E.Y.; Kunstman, K.J.; Wolinsky, S.M.; Malim, M.H. Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J. Virol. 2009, 83, 9474–9485. [Google Scholar] [CrossRef] [PubMed]
- Doehle, B.P.; Schafer, A.; Cullen, B.R. Human APOBEC3B is a potent inhibitor of HIV-1 infectivity and is resistant to HIV-1 Vif. Virology 2005, 339, 281–288. [Google Scholar] [CrossRef]
- OhAinle, M.; Kerns, J.A.; Li, M.M.; Malik, H.S.; Emerman, M. Antiretroelement activity of APOBEC3H was lost twice in recent human evolution. Cell Host Microbe 2008, 4, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Berkhout, B.; van Hemert, F.J. Silent codon positions in the A-rich HIV RNA genome that do not easily become A: Restrictions imposed by the RNA sequence and structure. Virus Evol. 2022, 8, veac072. [Google Scholar] [CrossRef] [PubMed]
- Sharp, P.M.; Hahn, B.H. The evolution of HIV-1 and the origin of AIDS. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010, 365, 2487–2494. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.J.; Marthas, M. Simian Immunodeficiency Virus: Animal Models of Disease. In Encyclopedia of Virology; Mahy, B.W.J., Van Regenmortel, M.H.V., Eds.; Elsevier: Amsterdam, The Netherlands, 2008; pp. 594–603. [Google Scholar]
- Mayrose, I.; Stern, A.; Burdelova, E.O.; Sabo, Y.; Laham-Karam, N.; Zamostiano, R.; Bacharach, E.; Pupko, T. Synonymous site conservation in the HIV-1 genome. BMC Evol. Biol. 2013, 13, 164. [Google Scholar] [CrossRef] [PubMed]
- Bazykin, G.A.; Dushoff, J.; Levin, S.A.; Kondrashov, A.S. Bursts of nonsynonymous substitutions in HIV-1 evolution reveal instances of positive selection at conservative protein sites. Proc. Natl. Acad. Sci. USA 2006, 103, 19396–19401. [Google Scholar] [CrossRef] [PubMed]
- Snoeck, J.; Fellay, J.; Bartha, I.; Douek, D.C.; Telenti, A. Mapping of positive selection sites in the HIV-1 genome in the context of RNA and protein structural constraints. Retrovirology 2011, 8, 87. [Google Scholar] [CrossRef] [PubMed]
- Wallis, Z.K.; Williams, K.C. Monocytes in HIV and SIV Infection and Aging: Implications for Inflamm-Aging and Accelerated Aging. Viruses 2022, 14, 409. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Chaillon, A.; Gianella, S.; Wong, L.M.; Li, D.; Simermeyer, T.L.; Porrachia, M.; Ignacio, C.; Woodworth, B.; Zhong, D.; et al. Brain microglia serve as a persistent HIV reservoir despite durable antiretroviral therapy. J. Clin. Investig. 2023, 133, e167417. [Google Scholar] [CrossRef]
- van Weringh, A.; Ragonnet-Cronin, M.; Pranckeviciene, E.; Pavon-Eternod, M.; Kleiman, L.; Xia, X. HIV-1 modulates the tRNA pool to improve translation efficiency. Mol. Biol. Evol. 2011, 28, 1827–1834. [Google Scholar] [CrossRef] [PubMed]
- Real, F.; Zhu, A.; Huang, B.; Belmellat, A.; Sennepin, A.; Vogl, T.; Ransy, C.; Revol, M.; Arrigucci, R.; Lombes, A.; et al. S100A8-mediated metabolic adaptation controls HIV-1 persistence in macrophages in vivo. Nat. Commun. 2022, 13, 5956. [Google Scholar] [CrossRef] [PubMed]
- Ganor, Y.; Real, F.; Sennepin, A.; Dutertre, C.A.; Prevedel, L.; Xu, L.; Tudor, D.; Charmeteau, B.; Couedel-Courteille, A.; Marion, S.; et al. HIV-1 reservoirs in urethral macrophages of patients under suppressive antiretroviral therapy. Nat. Microbiol. 2019, 4, 633–644. [Google Scholar] [CrossRef] [PubMed]
- Graziano, F.; Desdouits, M.; Garzetti, L.; Podini, P.; Alfano, M.; Rubartelli, A.; Furlan, R.; Benaroch, P.; Poli, G. Extracellular ATP induces the rapid release of HIV-1 from virus containing compartments of human macrophages. Proc. Natl. Acad. Sci. USA 2015, 112, E3265–E3273. [Google Scholar] [CrossRef]
- Rodrigues, V.; Ruffin, N.; San-Roman, M.; Benaroch, P. Myeloid Cell Interaction with HIV: A Complex Relationship. Front. Immunol. 2017, 8, 1698. [Google Scholar] [CrossRef] [PubMed]
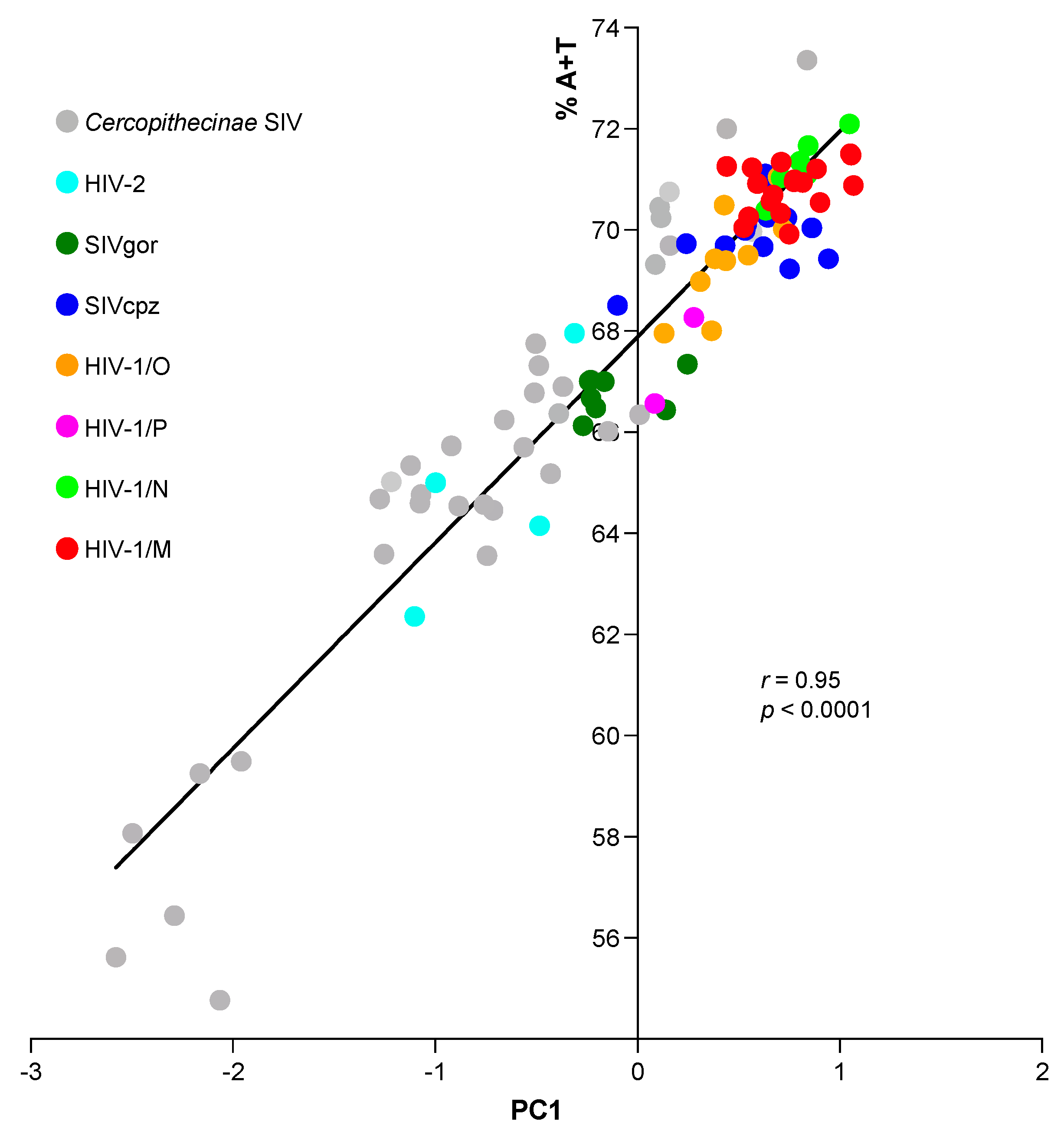
| Primate Lentiviruses | A | T | G | C | A + T | G + C |
|---|---|---|---|---|---|---|
| PC1 vs. first codon position | 0.83 | −0.75 | 0.27 | −0.76 | 0.60 | −0.60 |
| PC1 vs. second codon position | −0.32 | 0.52 | 0.74 | −0.51 | 0.11 | −0.11 |
| PC1 vs. third codon position | 0.83 | 0.85 | −0.88 | −0.84 | 0.95 | −0.95 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pavesi, A.; Romerio, F. Different Patterns of Codon Usage and Amino Acid Composition across Primate Lentiviruses. Viruses 2023, 15, 1580. https://doi.org/10.3390/v15071580
Pavesi A, Romerio F. Different Patterns of Codon Usage and Amino Acid Composition across Primate Lentiviruses. Viruses. 2023; 15(7):1580. https://doi.org/10.3390/v15071580
Chicago/Turabian StylePavesi, Angelo, and Fabio Romerio. 2023. "Different Patterns of Codon Usage and Amino Acid Composition across Primate Lentiviruses" Viruses 15, no. 7: 1580. https://doi.org/10.3390/v15071580
APA StylePavesi, A., & Romerio, F. (2023). Different Patterns of Codon Usage and Amino Acid Composition across Primate Lentiviruses. Viruses, 15(7), 1580. https://doi.org/10.3390/v15071580






