Serology Identifies LIPyV as a Feline Rather than a Human Polyomavirus
Abstract
1. Introduction
2. Materials and Methods
2.1. Polyomavirus Serology
2.2. Samples and Study Populations
2.3. Sequence Read Archive Database Search
2.4. Phylogenetic Tree
2.5. Statistics
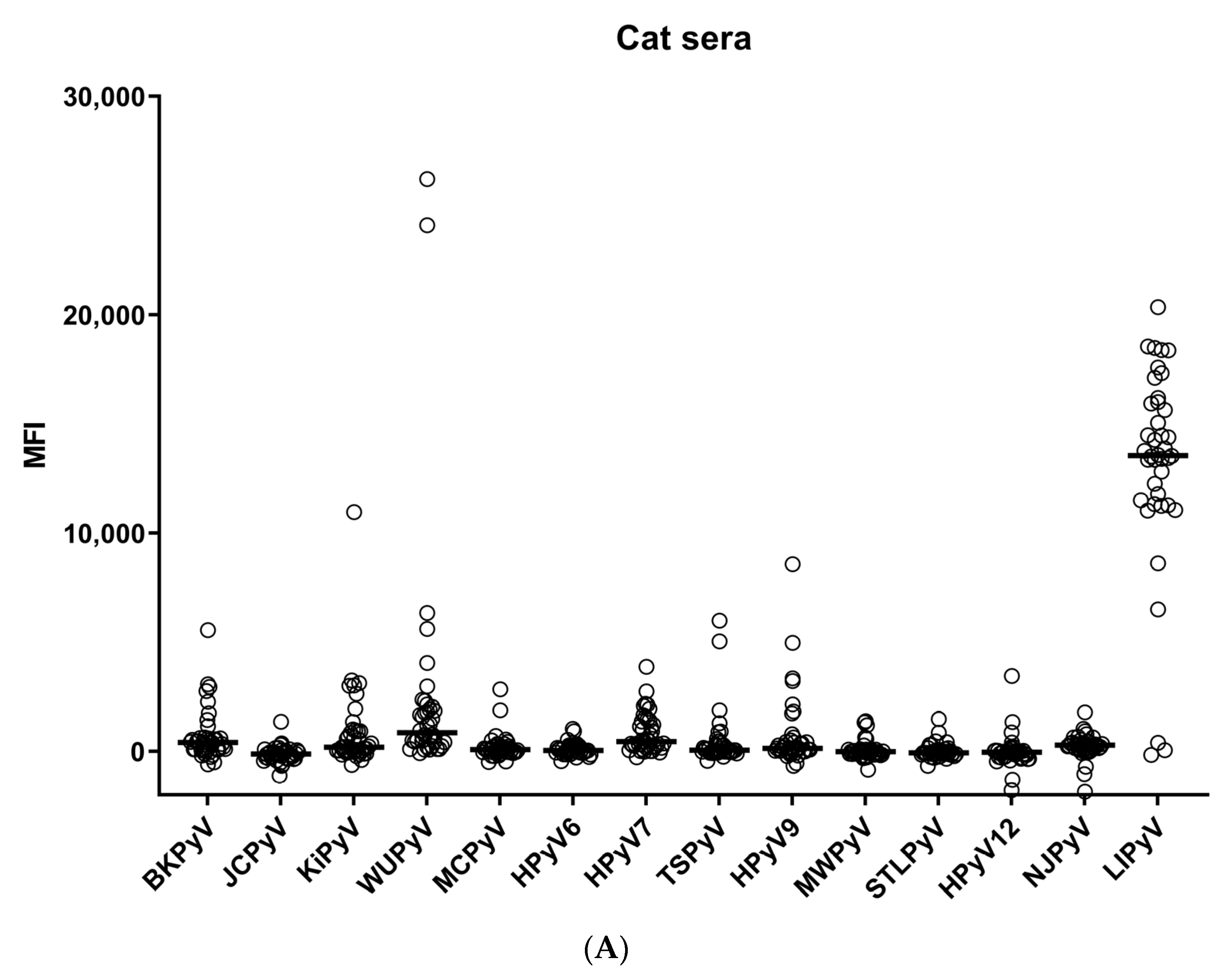
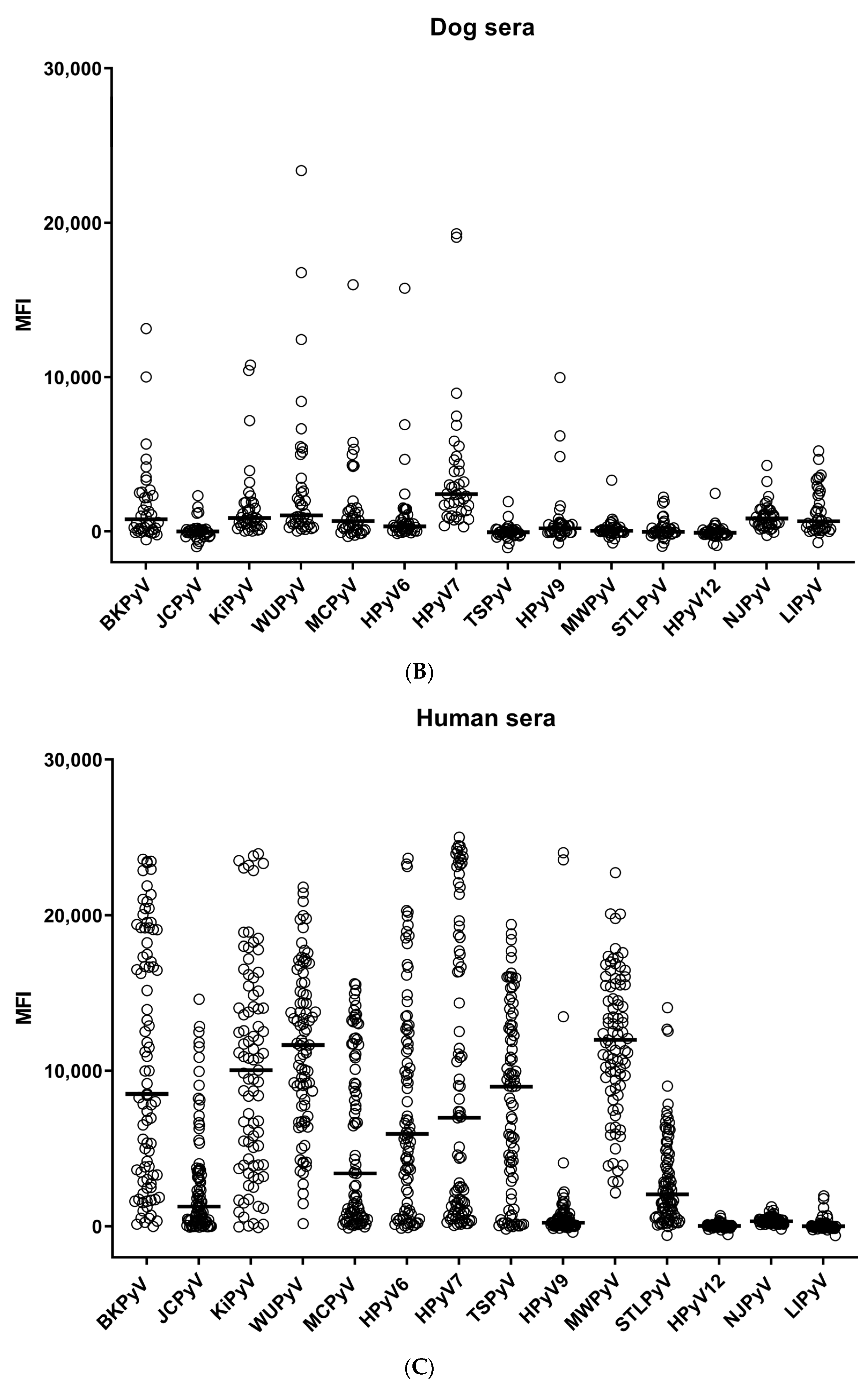
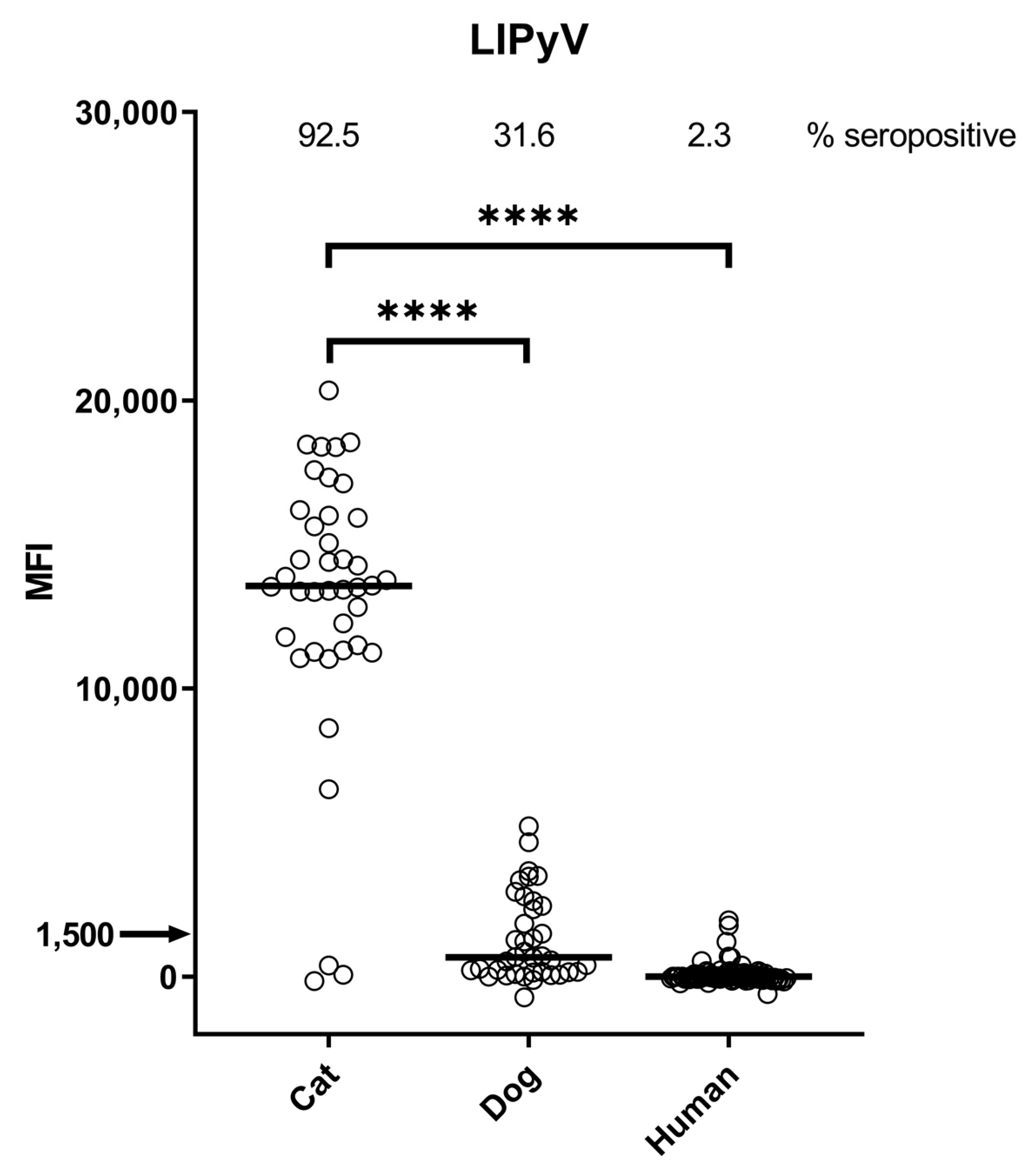
3. Results
4. Discussion
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Moens, U.; Krumbholz, A.; Ehlers, B.; Zell, R.; Johne, R.; Calvignac-Spencer, S.; Lauber, C. Biology, evolution, and medical importance of polyomaviruses: An update. Infect. Genet. Evol. 2017, 54, 18–38. [Google Scholar] [CrossRef] [PubMed]
- Kamminga, S.; Sidorov, I.A.; Tadesse, M.; van der Meijden, E.; de Brouwer, C.; Zaaijer, H.L.; Feltkamp, M.C.; Gorbalenya, A.E. Translating genomic exploration of the family Polyomaviridae into confident human polyomavirus detection. iScience 2022, 25. [Google Scholar] [CrossRef] [PubMed]
- Gardner, S.D.; Field, A.M.; Coleman, D.V.; Hulme, B. New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet 1971, 1, 1253–1257. [Google Scholar] [CrossRef] [PubMed]
- Padgett, B.L.; Walker, D.L.; ZuRhein, G.M.; Eckroade, R.J.; Dessel, B.H. Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet 1971, 1, 1257–1260. [Google Scholar] [CrossRef] [PubMed]
- Allander, T.; Andreasson, K.; Gupta, S.; Bjerkner, A.; Bogdanovic, G.; Persson, M.A.A.; Dalianis, T.; Ramqvist, T.; Andersson, B. Identification of a Third Human Polyomavirus. J. Virol. 2007, 81, 4130–4136. [Google Scholar] [CrossRef]
- Gaynor, A.M.; Nissen, M.D.; Whiley, D.M.; Mackay, I.M.; Lambert, S.B.; Wu, G.; Brennan, D.C.; Storch, G.A.; Sloots, T.P.; Wang, D. Identification of a Novel Polyomavirus from Patients with Acute Respiratory Tract Infections. PLoS Pathog. 2007, 3, e64. [Google Scholar] [CrossRef]
- Feng, H.; Shuda, M.; Chang, Y.; Moore, P.S. Clonal Integration of a Polyomavirus in Human Merkel Cell Carcinoma. Science 2008, 319, 1096–1100. [Google Scholar] [CrossRef]
- Wang, D.; Rm, S.; Dv, P.; Ka, P.; AL, M.; Cb, B. Faculty Opinions recommendation of Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe. 2010, 7. [Google Scholar] [CrossRef]
- van der Meijden, E.; Janssens, R.W.A.; Lauber, C.; Bavinck, J.N.B.; Gorbalenya, A.E.; Feltkamp, M.C.W. Discovery of a New Human Polyomavirus Associated with Trichodysplasia Spinulosa in an Immunocompromized Patient. PLoS Pathog. 2010, 6, e1001024. [Google Scholar] [CrossRef]
- Sauvage, V. Human Polyomavirus Related to African Green Monkey Lymphotropic Polyomavirus. Emerg. Infect. Dis. 2011, 17, 1364–1370. [Google Scholar] [CrossRef]
- Scuda, N.; Hofmann, J.; Calvignac-Spencer, S.; Ruprecht, K.; Liman, P.; Kühn, J.; Hengel, H.; Ehlers, B. A Novel Human Polyomavirus Closely Related to the African Green Monkey-Derived Lymphotropic Polyomavirus. J. Virol. 2011, 85, 4586–4590. [Google Scholar] [CrossRef] [PubMed]
- Buck, C.B.; Phan, G.Q.; Raiji, M.T.; Murphy, P.M.; McDermott, D.H.; McBride, A.A. Complete Genome Sequence of a Tenth Human Polyomavirus. J. Virol. 2012, 86, 10887. [Google Scholar] [CrossRef] [PubMed]
- Siebrasse, E.A.; Reyes, A.; Lim, E.S.; Zhao, G.; Mkakosya, R.S.; Manary, M.J.; Gordon, J.I.; Wang, D. Identification of MW Polyomavirus, a Novel Polyomavirus in Human Stool. J. Virol. 2012, 86, 10321–10326. [Google Scholar] [CrossRef] [PubMed]
- Lim, E.S.; Reyes, A.; Antonio, M.; Saha, D.; Ikumapayi, U.N.; Adeyemi, M.; Stine, O.C.; Skelton, R.; Brennan, D.C.; Mkakosya, R.S.; et al. Discovery of STL polyomavirus, a polyomavirus of ancestral recombinant origin that encodes a unique T antigen by alternative splicing. Virology 2013, 436, 295–303. [Google Scholar] [CrossRef]
- Korup, S.; Rietscher, J.; Calvignac-Spencer, S.; Trusch, F.; Hofmann, J.; Moens, U.; Sauer, I.; Voigt, S.; Schmuck, R.; Ehlers, B. Identification of a Novel Human Polyomavirus in Organs of the Gastrointestinal Tract. PLoS ONE 2013, 8, e58021. [Google Scholar] [CrossRef] [PubMed]
- Mishra, N.; Pereira, M.; Rhodes, R.H.; An, P.; Pipas, J.M.; Jain, K.; Kapoor, A.; Briese, T.; Faust, P.L.; Lipkin, W.I. Identification of a Novel Polyomavirus in a Pancreatic Transplant Recipient with Retinal Blindness and Vasculitic Myopathy. J. Infect. Dis. 2014, 210, 1595–1599. [Google Scholar] [CrossRef]
- Gheit, T.; Dutta, S.; Oliver, J.; Robitaille, A.; Hampras, S.; Combes, J.-D.; McKay-Chopin, S.; Le Calvez-Kelm, F.; Fenske, N.; Cherpelis, B.; et al. Isolation and characterization of a novel putative human polyomavirus. Virology 2017, 506, 45–54. [Google Scholar] [CrossRef]
- Kamminga, S.; van der Meijden, E.; Feltkamp, M.C.W.; Zaaijer, H.L. Seroprevalence of fourteen human polyomaviruses determined in blood donors. PLoS ONE 2018, 13, e0206273. [Google Scholar] [CrossRef]
- Gossai, A.; Waterboer, T.; Nelson, H.H.; Michel, A.; Willhauck-Fleckenstein, M.; Farzan, S.F.; Hoen, A.G.; Christensen, B.C.; Kelsey, K.T.; Marsit, C.J.; et al. Seroepidemiology of Human Polyomaviruses in a US Population. Am. J. Epidemiol. 2015, 183, 61–69. [Google Scholar] [CrossRef]
- Feltkamp, M.C.W.; Kazem, S.; van der Meijden, E.; Lauber, C.; Gorbalenya, A.E. From Stockholm to Malawi: Recent developments in studying human polyomaviruses. J. Gen. Virol. 2013, 94, 482–496. [Google Scholar] [CrossRef]
- ICTV. Taxonomy History-Taxonomy. 2022. Available online: https://talk.ictvonline.org/taxonomy/p/taxonomy-history?taxnode_id=201904426 (accessed on 1 May 2023).
- Li, Y.; Huang, H.; Lan, T.; Wang, W.; Zhang, J.; Zheng, M.; Cao, L.; Sun, W.; Lu, H. First detection and complete genome analysis of the Lyon IARC polyomavirus in China from samples of diarrheic cats. Virus Genes 2021, 1–5. [Google Scholar] [CrossRef]
- Fahsbender, E.; Altan, E.; Estrada, M.; Seguin, M.A.; Young, P.; Leutenegger, C.M.; Delwart, E. Lyon-IARC Polyomavirus DNA in Feces of Diarrheic Cats. Genome Announc. 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Keinonen, A.; Koskenmies, S.; Pitkänen, S.; Fyhrquist, N.; Sadeghi, M.; Mäkisalo, H.; Söderlund-Venermo, M.; Hedman, K. Occurrence of newly discovered human polyomaviruses in skin of liver transplant recipients and their relation with squamous cell carcinoma in situ and actinic keratosis–a single-center cohort study. Transpl. Int. 2019, 32, 516–522. [Google Scholar] [CrossRef] [PubMed]
- Buck, C.B.; Van Doorslaer, K.; Peretti, A.; Geoghegan, E.M.; Tisza, M.J.; An, P.; Katz, J.P.; Pipas, J.M.; McBride, A.A.; Camus, A.C.; et al. The Ancient Evolutionary History of Polyomaviruses. PLoS Pathog. 2016, 12, e1005574. [Google Scholar] [CrossRef] [PubMed]
- Kraberger, S.; Serieys, L.E.K.; Riley, S.P.D.; Schmidlin, K.; Newkirk, E.S.; Squires, J.R.; Buck, C.B.; Varsani, A. Novel polyomaviruses identified in fecal samples from four carnivore species. Arch. Virol. 2023, 168, 18. [Google Scholar] [CrossRef] [PubMed]
- Kamminga, S.; van der Meijden, E.; Wunderink, H.F.; Touzé, A.; Zaaijer, H.L.; Feltkamp, M.C.W. Development and Evaluation of a Broad Bead-Based Multiplex Immunoassay To Measure IgG Seroreactivity against Human Polyomaviruses. J. Clin. Microbiol. 2018, 56. [Google Scholar] [CrossRef] [PubMed]
- van der Meijden, E.; Wunderink, H.F.; Brouwer, C.S.v.d.B.-D.; Zaaijer, H.L.; Rotmans, J.I.; Bavinck, J.N.B.; Feltkamp, M.C. Human Polyomavirus 9 Infection in Kidney Transplant Patients. Emerg. Infect. Dis. 2014, 20, 991–999. [Google Scholar] [CrossRef]
- Buchfink, B.; Reuter, K.; Drost, H.-G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods 2021, 18, 366–368. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.-M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- Lemoine, F.; Correia, D.; Lefort, V.; Doppelt-Azeroual, O.; Mareuil, F.; Cohen-Boulakia, S.; Gascuel, O. NGPhylogeny.fr: New generation phylogenetic services for non-specialists. Nucleic Acids Res. 2019, 47, W260–W265. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Parry, J.V.; Gardner, S.D. Human exposure to bovine polyomavirus: A zoonosis? Arch. Virol. 1986, 87, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Carr, M.; Gonzalez, G.; Sasaki, M.; Dool, S.E.; Ito, K.; Ishii, A.; Hang’ombe, B.M.; Mweene, A.S.; Teeling, E.C.; Hall, W.W.; et al. Identification of the same polyomavirus species in different African horseshoe bat species is indicative of short-range host-switching events. J. Gen. Virol. 2017, 98, 2771–2785. [Google Scholar] [CrossRef]
- Bouvard, V.; Baan, R.A.; Grosse, Y.; Lauby-Secretan, B.; El Ghissassi, F.; Benbrahim-Tallaa, L.; Guha, N.; Straif, K. Carcinogenicity of malaria and of some polyomaviruses. Lancet Oncol. 2012, 13, 339–340. [Google Scholar] [CrossRef]
- Church, M.; Cruz, F.D.; Estrada, M.; Leutenegger, C.; Pesavento, P.; Woolard, K. Exposure to raccoon polyomavirus (RacPyV) in free-ranging North American raccoons (Procyon lotor). Virology 2016, 489, 292–299. [Google Scholar] [CrossRef] [PubMed]

| Name (Acronym) | Species | Disease or Symptom Associated with Infection | Seroprevalence, % 1 | Year and Specimen of Virus Discovery (Reference) |
|---|---|---|---|---|
| BK polyomavirus (BKPyV) | Betapolyomavirus hominis | Transplant nephropathy; Hemorrhagic cystitis | 99 | 1971, Urine [3] |
| JC polyomavirus (JCPyV) | Betapolyomavirus secuhominis | Progressive multifocal leukoencephalopathy (PML) | 62 | 1971, Brain [4] |
| Karolinska Institutet polyomavirus (KIPyV) | Betapolyomavirus tertihominis | Respiratory illness | 92 | 2007, Nasopharynx [5] |
| Washington University polyomavirus (WUPyV) | Betapolyomavirus quartihominis | Respiratory illness | 99 | 2007, Nasopharynx [6] |
| Merkel cell polyomavirus (MCPyV) | Alphapolyomavirus quintihominis | Merkel cell carcinoma | 82 | 2008, Skin [7] |
| Human polyomavirus 6 (HPyV6) | Deltapolyomavirus sextihominis | Pruritic and dyskeratotic dermatosis | 83 | 2010, Skin [8] |
| Human polyomavirus 7 (HPyV7) | Deltapolyomavirus septihominis | Pruritic and dyskeratotic dermatosis | 71 | 2010, Skin [8] |
| Trichodysplasia spinulosa polyomavirus (TSPyV) | Alphapolyomavirus octihominis | Trichodysplasia spinulosa | 79 | 2010, Skin [9] |
| Human polyomavirus 9 (HPyV9) | Alphapolyomavirus nonihominis | None | 19 | 2011, Serum [10,11] |
| Malawi polyomavirus (MWPyV) | Deltapolyomavirus decihominis | None | 100 | 2012, Feces [12,13] |
| Saint Louis polyomavirus (STLPyV) | Deltapolyomavirus undecihominis | None | 65 | 2012, Feces [14] |
| Human polyomavirus 12 (HPyV12) | Sorex araneus polyomavirus 1 | None | 4 | 2013, Liver [15] |
| New Jersey polyomavirus (NJPyV) | Alphapolyomavirus terdecihominis | Vasculitis, myositis, retinitis | 5 | 2014, Muscle [16] |
| Lyon IARC polyomavirus (LIPyV) | Alphapolyomavirus quardecihominis | None | 6 | 2017, Skin [17] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamminga, S.; van der Meijden, E.; Pesavento, P.; Buck, C.B.; Feltkamp, M.C.W. Serology Identifies LIPyV as a Feline Rather than a Human Polyomavirus. Viruses 2023, 15, 1546. https://doi.org/10.3390/v15071546
Kamminga S, van der Meijden E, Pesavento P, Buck CB, Feltkamp MCW. Serology Identifies LIPyV as a Feline Rather than a Human Polyomavirus. Viruses. 2023; 15(7):1546. https://doi.org/10.3390/v15071546
Chicago/Turabian StyleKamminga, Sergio, Els van der Meijden, Patricia Pesavento, Christopher B. Buck, and Mariet C. W. Feltkamp. 2023. "Serology Identifies LIPyV as a Feline Rather than a Human Polyomavirus" Viruses 15, no. 7: 1546. https://doi.org/10.3390/v15071546
APA StyleKamminga, S., van der Meijden, E., Pesavento, P., Buck, C. B., & Feltkamp, M. C. W. (2023). Serology Identifies LIPyV as a Feline Rather than a Human Polyomavirus. Viruses, 15(7), 1546. https://doi.org/10.3390/v15071546





