Rapid Triage of Children with Suspected COVID-19 Using Laboratory-Based Machine-Learning Algorithms
Abstract
:1. Introduction
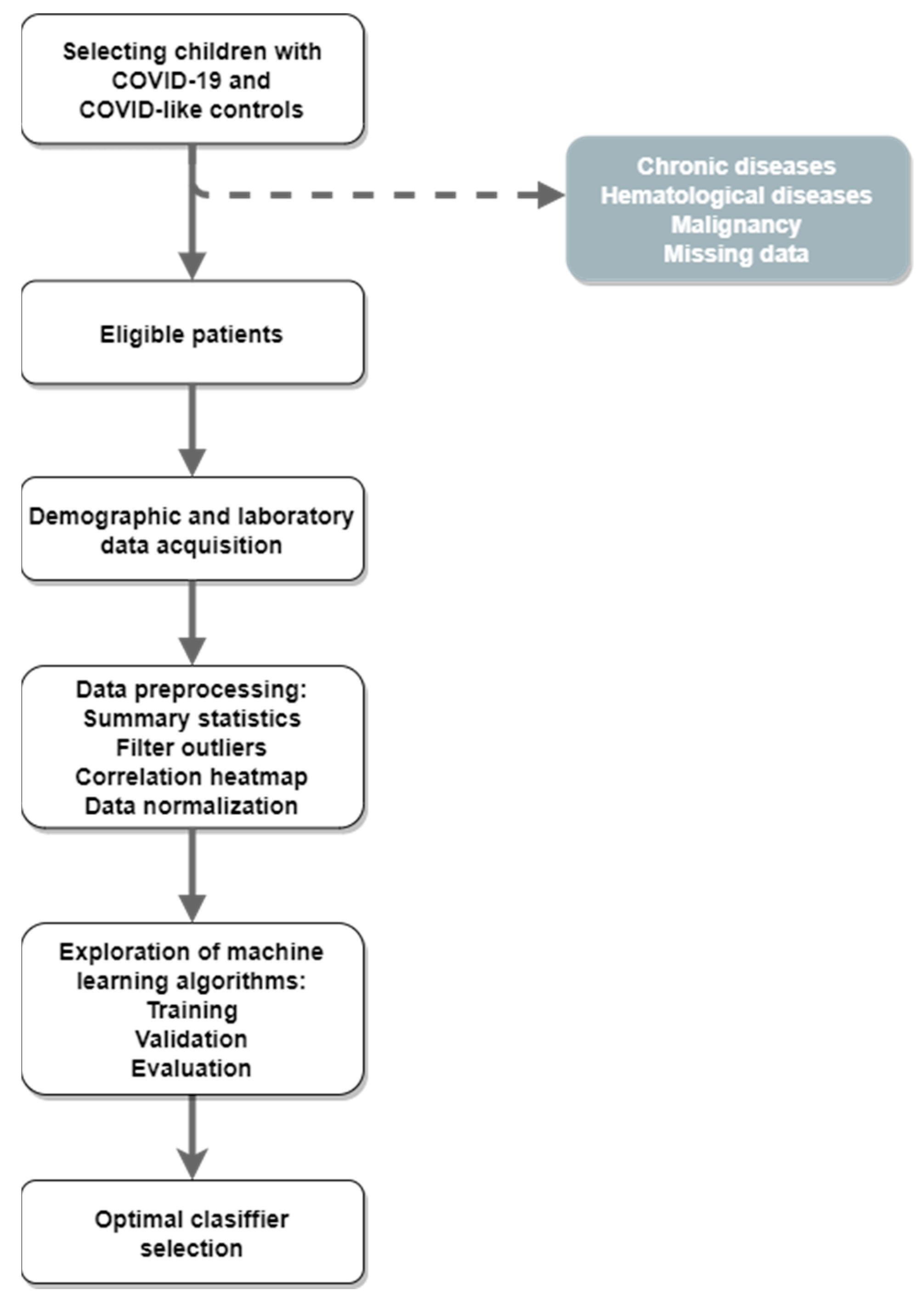
2. Materials and Methods
2.1. Data Acquisition
2.2. Data Preprocessing
2.3. Baseline Statistical Analyses
2.4. Machine-Learning Algorithms
2.5. Ethical Approval
3. Results
3.1. Clinical Laboratory Features
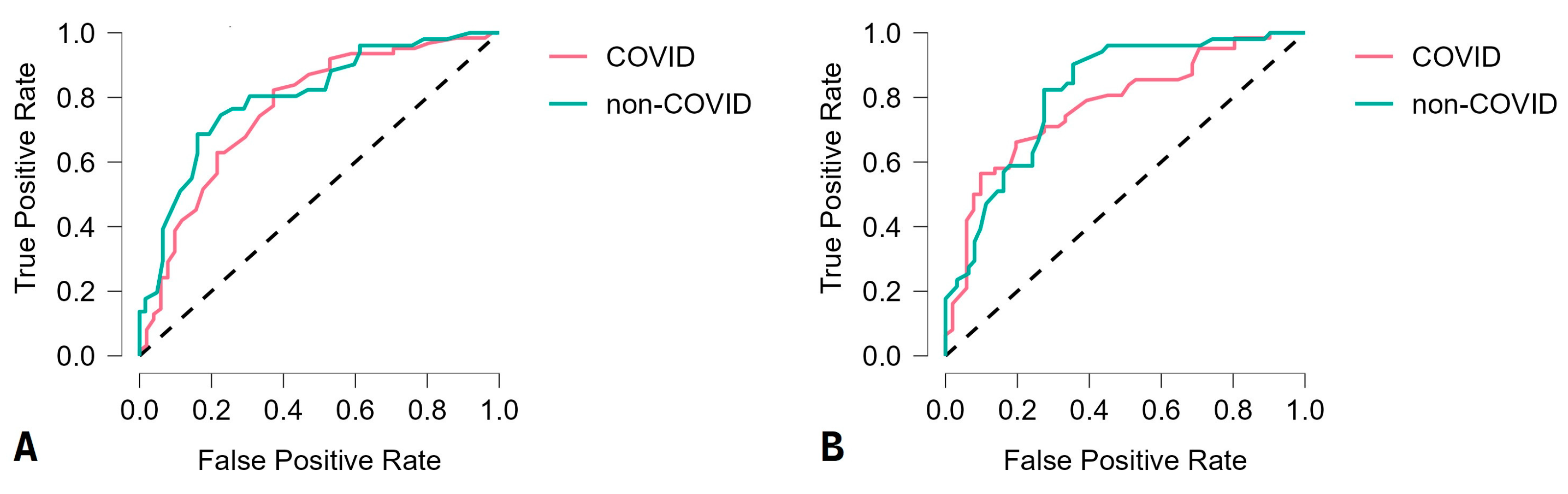
3.2. Machine-Learning Algorithm Performances
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nunziata, F.; Salomone, S.; Catzola, A.; Poeta, M.; Pagano, F.; Punzi, L.; Lo Vecchio, A.; Guarino, A.; Bruzzese, E. Clinical Presentation and Severity of SARS-CoV-2 Infection Compared to Respiratory Syncytial Virus and Other Viral Respiratory Infections in Children Less than Two Years of Age. Viruses 2023, 15, 717. [Google Scholar]
- De Souza, T.H.; Nadal, J.A.; Nogueira, R.J.N.; Pereira, R.M.; Brandão, M.B. Clinical manifestations of children with COVID-19: A systematic review. Pediatr. Pulmonol. 2020, 55, 1892–1899. [Google Scholar]
- Aykac, K.; Cura Yayla, B.C.; Ozsurekci, Y.; Evren, K.; Oygar, P.D.; Gurlevik, S.L.; Coskun, T.; Tasci, O.; Kaya, F.D.; Fidanci, I.; et al. The association of viral load and disease severity in children with COVID-19. J. Med. Virol. 2021, 93, 3077–3083. [Google Scholar] [PubMed]
- Brandi, N.; Ciccarese, F.; Rimondi, M.R.; Balacchi, C.; Modolon, C.; Sportoletti, C.; Renzulli, M.; Coppola, F.; Golfieri, R. An Imaging Overview of COVID-19 ARDS in ICU Patients and Its Complications: A Pictorial Review. Diagnostics 2022, 12, 846. [Google Scholar] [PubMed]
- Pousa, P.A.; Mendonça, T.S.C.; Oliveira, E.A.; Simões-E-Silva, A.C. Extrapulmonary manifestations of COVID-19 in children: A comprehensive review and pathophysiological considerations. J. Pediatr. 2021, 97, 116–139. [Google Scholar]
- Umakanthan, S.; Sahu, P.; Ranade, A.V.; Bukelo, M.M.; Rao, J.S.; Abrahao-Machado, L.F.; Dahal, S.; Kumar, H.; Kv, D. Origin, transmission, diagnosis and management of coronavirus disease 2019 (COVID-19). Postgrad Med. J. 2020, 96, 753–758. [Google Scholar] [PubMed]
- Guan, W.J.; Ni, Z.Y.; Hu, Y.; Liang, W.H.; Ou, C.Q.; He, J.X.; Liu, L.; Shan, H.; Lei, C.L.; Hui, D.S.; et al. Clinical characteristic of Coronavisus disease 2019 China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [PubMed]
- Qi, K.; Zeng, W.; Ye, M.; Zheng, L.; Song, C.; Hu, S.; Duan, C.; Wei, Y.; Peng, J.; Zhang, W.; et al. Clinical, laboratory, and imaging features of pediatric COVID-19: A systematic review and meta-analysis. Medicine 2021, 100, e25230. [Google Scholar]
- Zanza, C.; Romenskaya, T.; Manetti, A.C.; Franceschi, F.; La Russa, R.; Bertozzi, G.; Maiese, A.; Savioli, G.; Volonnino, G.; Longhitano, Y. Cytokine Storm in COVID-19: Immunopathogenesis and Therapy. Medicina 2022, 58, 144. [Google Scholar]
- Balacchi, C.; Brandi, N.; Ciccarese, F.; Coppola, F.; Lucidi, V.; Bartalena, L.; Parmeggiani, A.; Paccapelo, A.; Golfieri, R. Comparing the first and the second waves of COVID-19 in Italy: Differences in epidemiological features and CT findings using a semi-quantitative score. Emerg. Radiol. 2021, 28, 1055–1061. [Google Scholar]
- Dobrijević, D.; Katanić, J.; Todorović, M.; Vučković, B. Baseline laboratory parameters for preliminary diagnosis of COVID-19 among children: A cross-sectional study. Sao Paulo Med. J. 2022, 140, 691–696. [Google Scholar] [PubMed]
- Khanna, N.N.; Maindarkar, M.A.; Viswanathan, V.; Fernandes, J.F.E.; Paul, S.; Bhagawati, M.; Ahluwalia, P.; Ruzsa, Z.; Sharma, A.; Kolluri, R.; et al. Economics of Artificial Intelligence in Healthcare: Diagnosis vs. Treatment. Healthcare 2022, 10, 2493. [Google Scholar] [PubMed]
- Roy, S.; Meena, T.; Lim, S.J. Demystifying Supervised Learning in Healthcare 4.0: A New Reality of Transforming Diagnostic Medicine. Diagnostics 2022, 12, 2549. [Google Scholar]
- Paul, S.; Riffat, M.; Yasir, A.; Mahim, M.N.; Sharnali, B.Y.; Naheen, I.T.; Rahman, A.; Kulkarni, A. Industry 4.0 Applications for Medical/Healthcare Services. J. Sens. Actuator Netw. 2021, 10, 43. [Google Scholar]
- Dobrijević, D.; Antić, J.; Rakić, G.; Andrijević, L.; Katanić, J.; Pastor, K. Could platelet indices have diagnostic properties in children with COVID-19? J. Clin. Lab. Anal. 2022, 36, e24749. [Google Scholar] [PubMed]
- Kassania, S.H.; Kassanib, P.H.; Wesolowskic, M.J.; Schneidera, K.A.; Detersa, R. Automatic Detection of Coronavirus Disease (COVID-19) in X-ray and CT Images: A Machine Learning Based Approach. Biocybern. Biomed. Eng. 2021, 41, 867–879. [Google Scholar]
- Chadaga, K.; Chakraborty, C.; Prabhu, S.; Umakanth, S.; Bhat, V.; Sampathila, N. Clinical and Laboratory Approach to Diagnose COVID-19 Using Machine Learning. Interdiscip. Sci. Comput. Life Sci. 2022, 14, 452–470. [Google Scholar]
- Horvat, M.; Horvat, Z.; Pastor, K. Multivariate analysis of water quality parameters in Lake Palic, Serbia. Env. Monit. Assess 2021, 193, 410. [Google Scholar]
- Syeda, H.B.; Syed, M.; Sexton, K.W.; Syed, S.; Begum, S.; Syed, F.; Prior, F.; Jr, F.Y. Role of Machine Learning Techniques to Tackle the COVID-19 Crisis: Systematic Review. JMIR Med. Inform. 2021, 9, e23811. [Google Scholar]
- Isaacs, D. Artificial intelligence in health care. J. Paediatr. Child. Health 2020, 56, 1493–1495. [Google Scholar]
- Witten, I.H.; Frank, E.; Hall, M.A.; Pal, C.H. Data Mining: Practical Machine Learning Tools and Techniques, 4th ed.; Elsevier: Amsterdam, The Netherlands, 2016. [Google Scholar]
- Le, D.N.; Parvathy, V.S.; Gupta, D.; Khanna, A.; Rodrigues, J.; Shankar, K. IoT enabled depthwise separable convolution neural network with deep support vector machine for COVID-19 diagnosis and classification. Int. J. Mach. Learn. Cyber 2021, 12, 3235–3248. [Google Scholar]
- Maia, M.; Pimentel, J.S.; Pereira, I.S.; Gondim, J.; Barreto, M.E.; Ara, A. Convolutional Support Vector Models: Prediction of Coronavirus Disease Using Chest X-rays. Information 2020, 11, 548. [Google Scholar]
- Guhathakurata, S.; Kundu, S.; Chakraborty, A.; Banerjee, J.S. A novel approach to predict COVID-19 using support vector machine. In Data Science for COVID-19; Academic Press: Cambridge, MA, USA, 2021; pp. 351–364. [Google Scholar]
- Gao, Y.; Cai, G.Y.; Fang, W.; Li, H.Y.; Wang, S.Y.; Chen, L.; Yu, Y.; Liu, D.; Xu, S.; Cui, P.-F.; et al. Machine learning based early warning system enables accurate mortality risk prediction for COVID-19. Nat. Commun. 2020, 11, 5033. [Google Scholar] [PubMed]
- Wan, T.K.; Huang, R.X.; Tulu, T.W.; Liu, J.D.; Vodencarevic, A.; Wong, C.W.; Chan, K.-H.K. Identifying Predictors of COVID-19 Mortality Using Machine Learning. Life 2022, 12, 547. [Google Scholar] [PubMed]
- Inokuchi, R.; Iwagami, M.; Sun, Y.; Sakamoto, A.; Tamiya, N. Machine learning models predicting undertriage in telephone triage. Ann. Med. 2022, 54, 2990–2997. [Google Scholar] [PubMed]
- Dobrijević, D.; Andrijević, L.; Antić, J.; Rakić, G.; Pastor, K. Hemogram-based decision tree models for discriminating COVID-19 from RSV in infants. J. Clin. Lab. Anal. 2023, 37, e24862. [Google Scholar]
- Gupta, V.K.; Gupta, A.; Kumar, D.; Sardana, A. Prediction of COVID-19 Confirmed, Death, and Cured Cases in India Using Random Forest Model. Big Data Min. Anal. 2021, 4, 116–123. [Google Scholar]
- Dobrijević, D.; Antić, J.; Rakić, G.; Katanić, J.; Andrijević, L.; Pastor, K. Clinical hematochemical parameters in differential diagnosis between pediatric SARS-CoV-2 and Influenza virus infection: An automated machine learning approach. Children 2023, 10, 761. [Google Scholar]
- Wang, J.; Yu, H.; Hua, Q.; Jing, S.; Liu, Z.; Peng, X.; Cao, C.; Luo, Y. A descriptive study of random forest algorithm for predicting COVID-19 patients outcome. PeerJ 2020, 8, e9945. [Google Scholar]
- Cornelius, E.; Akman, O.; Hrozencik, D. COVID-19 Mortality Prediction Using Machine Learning-Integrated Random Forest Algorithm under Varying Patient Frailty. Mathematics 2021, 9, 2043. [Google Scholar]
- Barbosa, V.A.F.; Gomes, J.C.; de Santana, M.A.; de Lima, C.L.; Calado, R.B.; Bertoldo Júnior, C.R.; Albuquerque, J.E.D.A.; de Souza, R.G.; de Araújo, R.J.E.; Júnior, L.A.R.M.; et al. Covid-19 rapid test by combining a Random Forest-based web system and blood tests. J. Biomol. Struct. Dyn. 2022, 40, 11948–11967. [Google Scholar] [PubMed]
- Iwendi, C.; Bashir, A.K.; Peshkar, A.; Sujatha, R.; Chatterjee, J.M.; Pasupuleti, S.; Mishra, R.; Pillai, S.; Jo, O. COVID-19 Patient Health Prediction Using Boosted Random Forest Algorithm. Front. Public Health 2020, 8, 357. [Google Scholar]
- Çubukçu, H.C.; Topcu, D.İ.; Bayraktar, N.; Gülşen, M.; Sarı, N.; Arslan, A.H. Detection of COVID-19 by Machine Learning Using Routine Laboratory Tests. Am. J. Clin. Pathol. 2022, 157, 758–766. [Google Scholar] [PubMed]
- Thimoteo, L.M.; Vellasco, M.M.; Amaral, J.; Figueiredo, K.; Yokoyama, C.L.; Marques, E. Explainable Artificial Intelligence for COVID-19 Diagnosis Through Blood Test Variables. J. Control Autom. Electr. Syst. 2022, 33, 625–644. [Google Scholar]
- Van Kasteren, P.B.; van der Veer, B.; van den Brink, S.; Wijsman, L.; de Jonge, J.; van den Brandt, A.; Molenkamp, R.; Reusken, C.B.; Meijer, A. Comparison of seven commercial RT-PCR diagnostic kits for COVID-19. J. Clin. Virol. 2020, 128, 104412. [Google Scholar]
- Yamayoshi, S.; Sakai-Tagawa, Y.; Koga, M.; Akasaka, O.; Nakachi, I.; Koh, H.; Maeda, K.; Adachi, E.; Saito, M.; Nagai, H.; et al. Comparison of Rapid Antigen Tests for COVID-19. Viruses 2020, 12, 1420. [Google Scholar]
- Scohy, A.; Anantharajah, A.; Bodéus, M.; Kabamba-Mukadi, B.; Verroken, A.; Rodriguez-Villalobos, H. Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis. J. Clin. Virol. 2020, 129, 104455. [Google Scholar] [PubMed]
- Albert, E.; Torres, I.; Bueno, F.; Huntley, D.; Molla, E.; Fernández-Fuentes, M.Á.; Martínez, M.; Poujois, S.; Forqué, L.; Valdivia, A.; et al. Field evaluation of a rapid antigen test (Panbio™ COVID-19 Ag Rapid Test Device) for COVID-19 diagnosis in primary healthcare centres. Clin. Microbiol. Infect. 2021, 27, e7–e472. [Google Scholar]
- Goodman-Meza, D.; Rudas, A.; Chiang, J.N.; Adamson, P.C.; Ebinger, J.; Sun, N.; Botting, P.; Fulcher, J.A.; Saab, F.G.; Brook, R.; et al. A machine learning algorithm to increase COVID-19 inpatient diagnostic capacity. PLoS ONE 2020, 15, e0239474. [Google Scholar]
- Assaf, D.; Gutman, Y.; Neuman, Y.; Segal, G.; Amit, S.; Gefen-Halevi, S.; Shilo, N.; Epstein, A.; Mor-Cohen, R.; Biber, A.; et al. Utilization of machine-learning models to accurately predict the risk for critical COVID-19. Intern. Emerg. Med. 2020, 15, 1435–1443. [Google Scholar]
- Zhang, R.K.; Xiao, Q.; Zhu, S.L.; Lin, H.Y.; Tang, M. Using different machine learning models to classify patients into mild and severe cases of COVID-19 based on multivariate blood testing. J. Med. Virol. 2022, 94, 357–365. [Google Scholar] [PubMed]
- Sztefko, K.; Beba, J.; Mamica, K.; Tomasik, P. Blood loss from laboratory diagnostic tests in children. Clin. Chem. Lab. Med. 2013, 51, 1623–1626. [Google Scholar] [PubMed]
- Tajbakhsh, A.; Jaberi, K.R.; Hayat, S.M.G.; Sharifi, M.; Johnston, T.P.; Guest, P.C.; Jafari, M.; Sahebkar, A. Age-Specific Differences in the Severity of COVID-19 Between Children and Adults: Reality and Reasons. Adv. Exp. Med. Biol. 2021, 1327, 63–78. [Google Scholar]
- Wong, L.S.Y.; Loo, E.X.L.; Kang, A.Y.H.; Lau, H.X.; Tambyah, P.A.; Tham, E.H. Age-Related Differences in Immunological Responses to SARS-CoV-2. J. Allergy Clin. Immunol. Pract. 2020, 8, 3251–3258. [Google Scholar]
- Carobene, A.; Milella, F.; Famiglini, L.; Cabitza, F. How is test laboratory data used and characterised by machine learning models? A systematic review of diagnostic and prognostic models developed for COVID-19 patients using only laboratory data. Clin. Chem. Lab. Med. 2022, 60, 1887–1901. [Google Scholar]
- Lippi, G. Machine learning in laboratory diagnostics: Valuable resources or a big hoax? Diagnosis 2019, 8, 133–135. [Google Scholar]
- Núñez, I.; Belaunzarán-Zamudio, P.F.; Caro-Vega, Y. Result Turnaround Time of RT-PCR for SARS-CoV-2 is the Main Cause of COVID-19 Diagnostic Delay: A Country-Wide Observational Study of Mexico and Colombia. Rev. Investig. Clin. 2022, 74, 071–080. [Google Scholar]
- Kogoj, R.; Korva, M.; Knap, N.; Resman Rus, K.; Pozvek, P.; Avšič-Županc, T.; Poljak, M. Comparative Evaluation of Six SARS-CoV-2 Real-Time RT-PCR Diagnostic Approaches Shows Substantial Genomic Variant-Dependent Intra- and Inter-Test Variability, Poor Interchangeability of Cycle Threshold and Complementary Turn-Around Times. Pathogens 2022, 11, 462. [Google Scholar]
- Minhas, N.; Gurav, Y.K.; Sambhare, S.; Potdar, V.; Choudhary, M.L.; Bhardwaj, S.D.; Abraham, P. Cost-analysis of real time RT-PCR test performed for COVID-19 diagnosis at India’s national reference laboratory during the early stages of pandemic mitigation. PLoS ONE 2023, 18, e0277867. [Google Scholar]
- Alyasseri, Z.A.A.; Al-Betar, M.A.; Doush, I.A.; Awadallah, M.A.; Abasi, A.K.; Makhadmeh, S.N.; Alomari, O.A.; Abdulkareem, K.H.; Adam, A.; Damasevicius, R. Review on COVID-19 diagnosis models based on machine learning and deep learning approaches. Expert Syst. 2022, 39, e12759. [Google Scholar] [PubMed]
- Heidari, A.; Jafari Navimipour, N.; Unal, M.; Toumaj, S. The COVID-19 epidemic analysis and diagnosis using deep learning: A systematic literature review and future directions. Comput. Biol. Med. 2022, 141, 105141. [Google Scholar] [PubMed]
| Laboratory Parameter a | COVID Group (n = 280) | Non-COVID Group (n = 286) | Overall (n = 566) | p-Value | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | p-Value | OR (95% CI) | p-Value | |||||
| WBC (109) | 7.9 (5.6–11.8) | 10.9 (7.9–15.2) | 9.4 (6.6–13.9) | <0.001 | 1.088 (1.055–1.123) | <0.001 | 1.052 (1.016–1.089) | 0.004 |
| RBC (1012) | 4.5 (4.1–4.9) | 4.4 (4.1–4.8) | 4.5 (4.1–4.8) | 0.359 | NA | NA | NA | NA |
| MCV (fL) | 80.7 (77.2–85.1) | 79.9 (76.1–84.9) | 80.1 (76.4–85) | 0.130 | NA | NA | NA | NA |
| MCHC (g/L) | 340 (331–347) | 344 (332–352) | 342 (331.2–350) | 0.003 | 1.039 (1.024–1.056) | <0.001 | 1.029 (1.014–1.044) | <0.001 |
| PLT (109) | 300 (217–386.2) | 342 (239.5–403.5) | 315 (230–392.8) | 0.028 | 1.001 (0.999–1.003) | 0.189 | NA | NA |
| MPV (fL) | 7.8 (7.1–8.5) | 7.4 (6.8–8) | 7.5 (7–8.3) | <0.001 | 1.028 (1.002–1.054) | 0.031 | 1.001 (0.996–1.007) | 0.387 |
| PCT (%) | 0.24 (0.18–0.3) | 0.23 (0.18–0.3) | 0.23 (0.18–0.3) | 0.943 | NA | NA | NA | NA |
| PDW (%) | 13.8 (11.7–16.3) | 12.9 (11.7–14.6) | 13.4 (11.7–15.5) | 0.010 | 1.427 (1.283–1.587) | <0.001 | 1.183 (1.090–1.283) | <0.001 |
| LYM# (109) | 2.3 (1.4–3.9) | 2.8 (1.9–4.9) | 2.6 (1.6–4.4) | <0.001 | 1.054 (0.956–1.161) | 0.291 | NA | NA |
| EOS# (109) | 0.07 (0.03–0.13) | 0.1 (0.05–0.15) | 0.09 (0.04–0.15) | 0.889 | NA | NA | NA | NA |
| AST (µkat/L) | 0.62 (0.46–0.82) | 0.57 (0.44–0.45) | 0.58 (0.45–0.79) | 0.048 | 0.821 (0.621–1.085) | 0.165 | NA | NA |
| GGT (µkat/L) | 0.24 (0.18–0.47) | 0.26 (0.19–0.56) | 0.25 (0.18–0.52) | 0.124 | NA | NA | NA | NA |
| LDH (µkat/L) | 4.39 (3.55–5.1) | 4.68 (3.73–5.44) | 4.4 (3.7–5.3) | 0.017 | 1.107 (0.992–1.234) | 0.068 | NA | NA |
| CRP (mg/L) | 5.5 (1.2–30) | 13.2 (2.7–71.6) | 9.7 (1.6–53.7) | <0.001 | 1.002 (0.999–1.005) | 0.171 | NA | NA |
| Classifier | Accuracy (%) | Sensitivity (%) | Specificity (%) | Positive Predictive Value (%) | Negative Predictive Value (%) | F1 Score (%) |
|---|---|---|---|---|---|---|
| Random forest | 85.0 | 86.0 | 83.9 | 84.5 | 85.5 | 85.2 |
| Support vector machine | 82.1 | 84.8 | 79.4 | 80.0 | 84.4 | 82.3 |
| Linear discriminant analysis | 78.8 | 81.1 | 76.7 | 75.4 | 82.1 | 78.1 |
| Neural network | 76.1 | 72.6 | 80.4 | 81.8 | 70.7 | 76.9 |
| k-nearest neighbors | 73.5 | 71.0 | 76.5 | 78.6 | 68.4 | 74.6 |
| Decision tree | 68.1 | 65.5 | 70.7 | 67.9 | 68.3 | 66.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dobrijević, D.; Vilotijević-Dautović, G.; Katanić, J.; Horvat, M.; Horvat, Z.; Pastor, K. Rapid Triage of Children with Suspected COVID-19 Using Laboratory-Based Machine-Learning Algorithms. Viruses 2023, 15, 1522. https://doi.org/10.3390/v15071522
Dobrijević D, Vilotijević-Dautović G, Katanić J, Horvat M, Horvat Z, Pastor K. Rapid Triage of Children with Suspected COVID-19 Using Laboratory-Based Machine-Learning Algorithms. Viruses. 2023; 15(7):1522. https://doi.org/10.3390/v15071522
Chicago/Turabian StyleDobrijević, Dejan, Gordana Vilotijević-Dautović, Jasmina Katanić, Mirjana Horvat, Zoltan Horvat, and Kristian Pastor. 2023. "Rapid Triage of Children with Suspected COVID-19 Using Laboratory-Based Machine-Learning Algorithms" Viruses 15, no. 7: 1522. https://doi.org/10.3390/v15071522
APA StyleDobrijević, D., Vilotijević-Dautović, G., Katanić, J., Horvat, M., Horvat, Z., & Pastor, K. (2023). Rapid Triage of Children with Suspected COVID-19 Using Laboratory-Based Machine-Learning Algorithms. Viruses, 15(7), 1522. https://doi.org/10.3390/v15071522






