Infection Dynamics and Genomic Mutations of Hepatitis E Virus in Naturally Infected Pigs on a Farrow-to-Finish Farm in Japan: A Survey from 2012 to 2021
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Serum and Fecal Samples from Domestic Pigs
2.2. Environmental Samples
2.3. An Enzyme-Linked Immunosorbent Assay (ELISA) for Detecting Anti-HEV IgG Antibodies
2.4. Qualitative and Quantitative Detection of HEV RNA
2.5. Amplification of the Full-Length HEV Genome
2.6. The Determination and Analysis of Nucleotide Sequences
2.7. Inoculum Preparation for Cell Culture
2.8. Cell Culture and Virus Inoculation
2.9. Western Blotting
2.10. Indirect Immunofluorescence
2.11. Statistical Analyses
3. Results
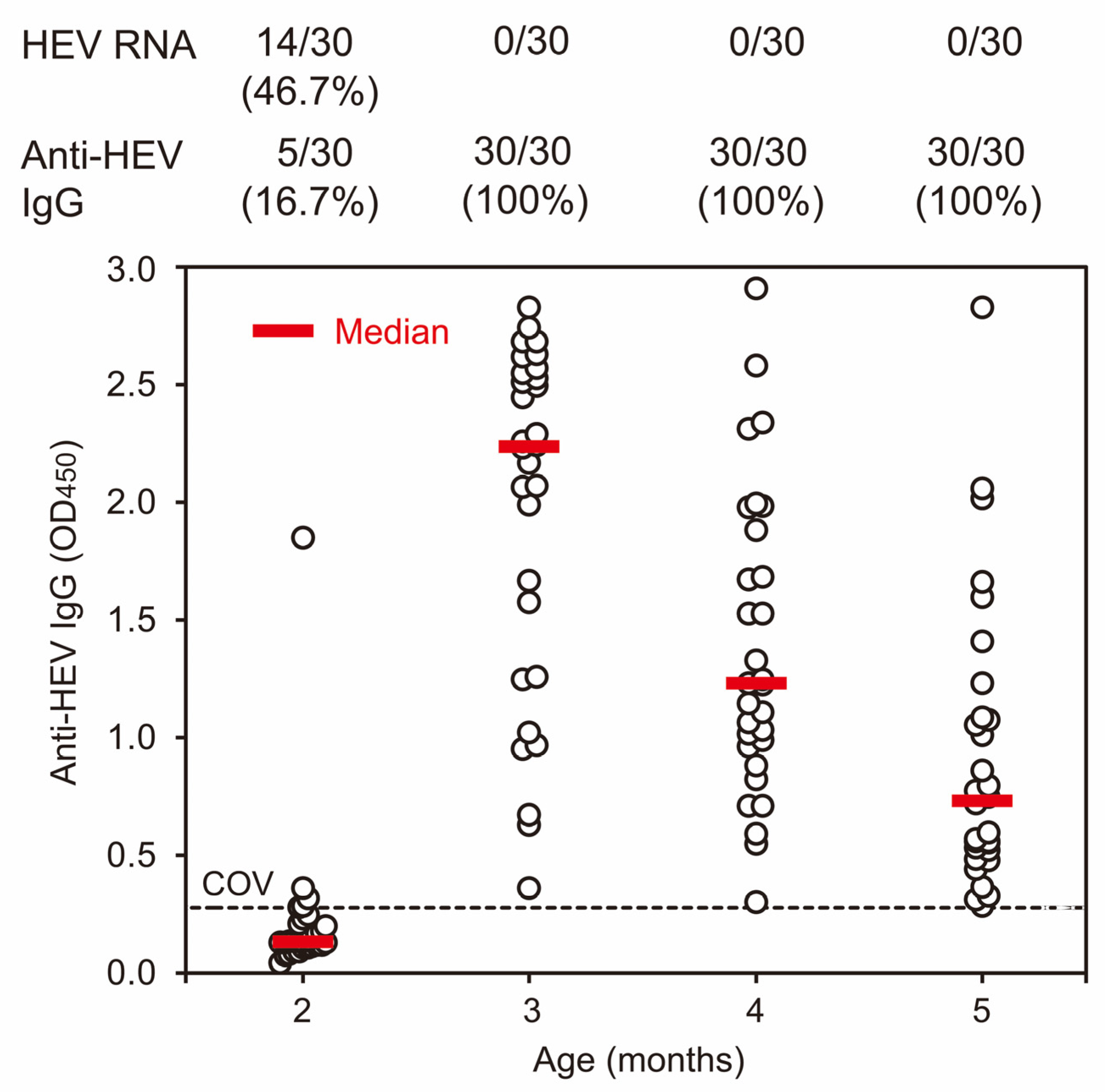
3.1. Prevalence of Anti-HEV IgG Antibodies and HEV RNA among Domestic Pigs on a Swine Farm in 2012
3.2. The Comparison of Kinetics of HEV Infection in Pigs in Weaning and Growing Houses with Varying HEV Contamination Statuses on a Swine Farm in 2013
3.3. Prospective Cohort Studies Conducted to Investigate the Highest HEV Antibody Response and the Initial Occurrence of HEV Viremia in Domestic Pigs on a Swine Farm in 2013, 2014, and 2019
3.4. Consistent Detection of HEV RNA in Environmental Samples Collected from Weaning and Growing Houses on the Investigated Farm during 2016 and 2018
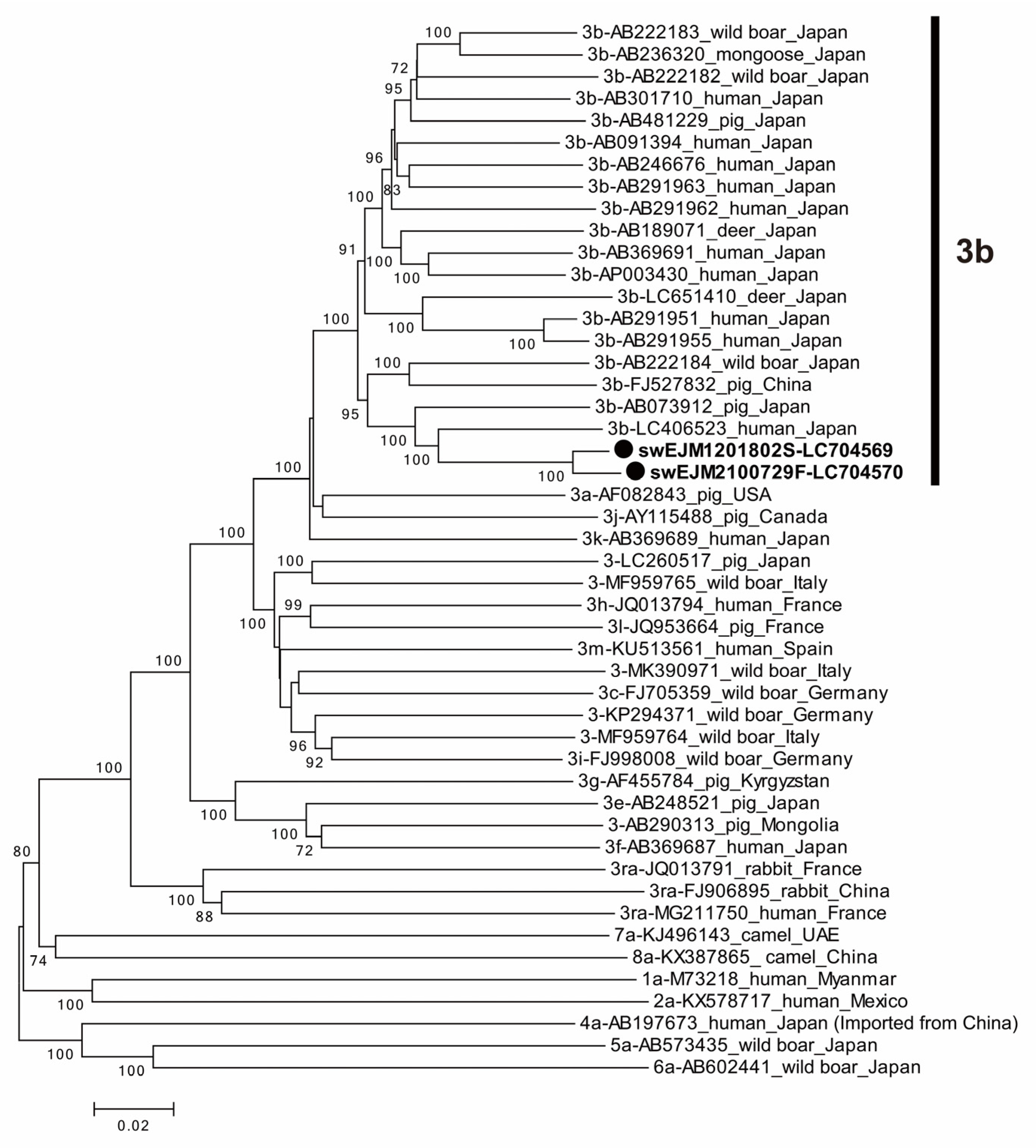
3.5. The Comparison of Partial ORF2 Sequences among 104 HEV Strains Obtained from 2012 and 2021
3.6. The Comparison of Entire Genomic Sequences of Representative HEV Strains from 2012 and 2021
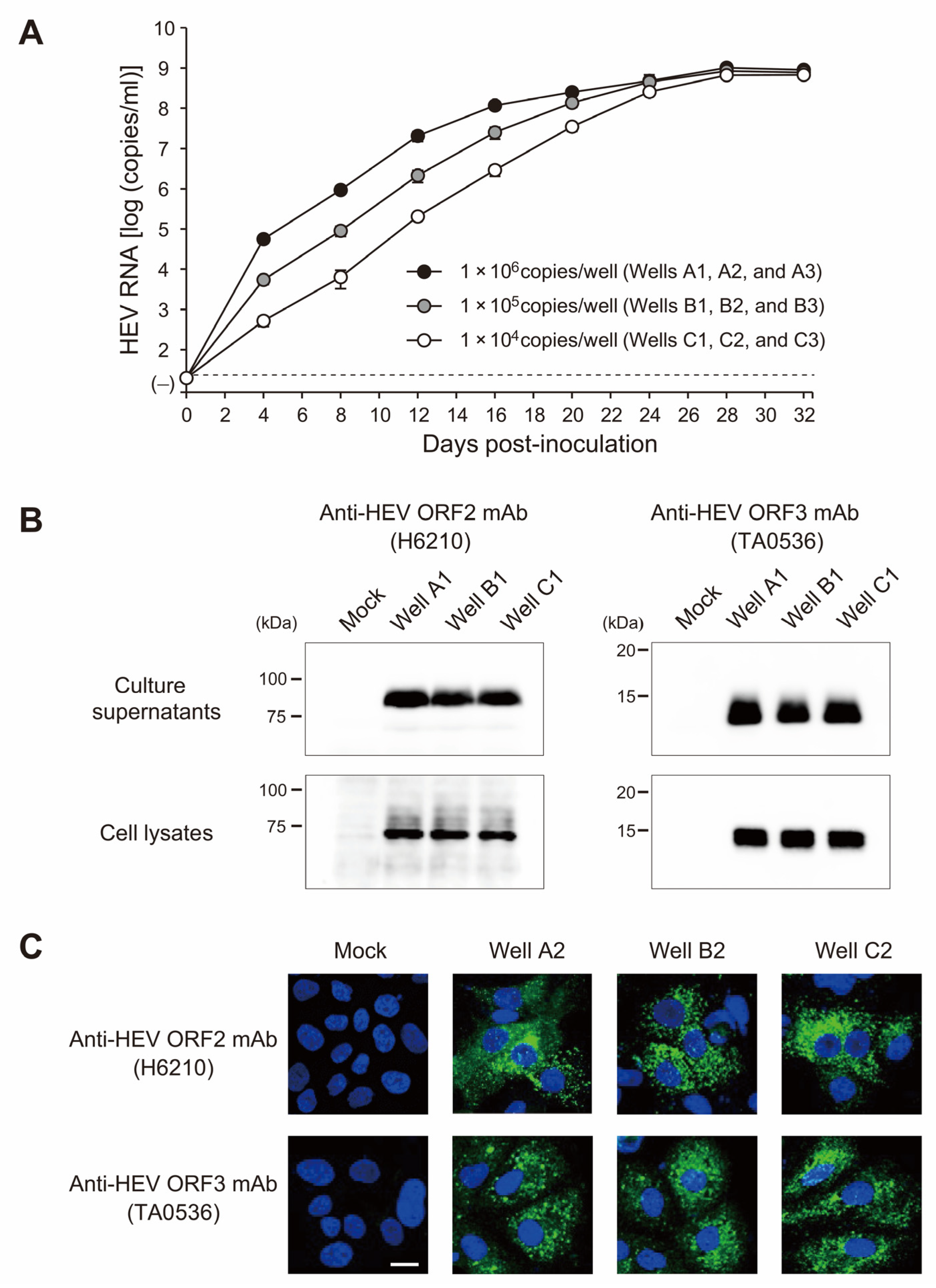
3.7. The swEJM2100729F Strain Efficiently Propagated in Human Hepatoma Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kamar, N.; Izopet, J.; Pavio, N.; Aggarwal, R.; Labrique, A.; Wedemeyer, H.; Dalton, H.R. Hepatitis E virus infection. Nat. Rev. Dis. Prim. 2017, 3, 17086. [Google Scholar] [CrossRef] [PubMed]
- Nagashima, S.; Takahashi, M.; Kobayashi, T.; Tanggis; Nishizawa, T.; Nishiyama, T.; Primadharsini, P.P.; Okamoto, H. Characterization of the Quasi-Enveloped Hepatitis E Virus Particles Released by the Cellular Exosomal Pathway. J. Virol. 2017, 91, e00822-17. [Google Scholar] [CrossRef] [PubMed]
- Debing, Y.; Moradpour, D.; Neyts, J.; Gouttenoire, J. Update on hepatitis E virology: Implications for clinical practice. J. Hepatol. 2016, 65, 200–212. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Gorbalenya, A.E.; Purdy, M.A.; Rozanov, M.N.; Reyes, G.R.; Bradley, D.W. Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: Delineation of an additional group of positive-strand RNA plant and animal viruses. Proc. Natl. Acad. Sci. USA 1992, 89, 8259–8263. [Google Scholar] [CrossRef]
- Panda, S.K.; Varma, S.P. Hepatitis E: Molecular virology and pathogenesis. J. Clin. Exp. Hepatol. 2013, 3, 114–124. [Google Scholar] [CrossRef]
- Montpellier, C.; Wychowski, C.; Sayed, I.M.; Meunier, J.C.; Saliou, J.M.; Ankavay, M.; Bull, A.; Pillez, A.; Abravanel, F.; Helle, F.; et al. Hepatitis E Virus Lifecycle and Identification of 3 Forms of the ORF2 Capsid Protein. Gastroenterology 2018, 154, 211–223.e8. [Google Scholar] [CrossRef]
- Yin, X.; Ying, D.; Lhomme, S.; Tang, Z.; Walker, C.M.; Xia, N.; Zheng, Z.; Feng, Z. Origin, antigenicity, and function of a secreted form of ORF2 in hepatitis E virus infection. Proc. Natl. Acad. Sci. USA 2018, 115, 4773–4778. [Google Scholar] [CrossRef]
- Gouttenoire, J.; Pollan, A.; Abrami, L.; Oechslin, N.; Mauron, J.; Matter, M.; Oppliger, J.; Szkolnicka, D.; Dao Thi, V.L.; van der Goot, F.G.; et al. Palmitoylation mediates membrane association of hepatitis E virus ORF3 protein and is required for infectious particle secretion. PLoS Pathog. 2018, 14, e1007471. [Google Scholar] [CrossRef]
- Purdy, M.A.; Drexler, J.F.; Meng, X.J.; Norder, H.; Okamoto, H.; Van der Poel, W.H.M.; Reuter, G.; de Souza, W.M.; Ulrich, R.G.; Smith, D.B. ICTV Virus Taxonomy Profile: Hepeviridae 2022. J. Gen. Virol. 2022, 103, 001778. [Google Scholar] [CrossRef]
- Heemelaar, S.; Hangula, A.L.; Chipeio, M.L.; Josef, M.; Stekelenburg, J.; van den Akker, T.H.; Pischke, S.; Mackenzie, S.B.P. Maternal and fetal outcomes of pregnancies complicated by acute hepatitis E and the impact of HIV status: A cross-sectional study in Namibia. Liver Int. 2022, 42, 50–58. [Google Scholar] [CrossRef]
- Nelson, K.E.; Labrique, A.B.; Kmush, B.L. Epidemiology of Genotype 1 and 2 Hepatitis E Virus Infections. Cold Spring Harb. Perspect. Med. 2019, 9, a031732. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.H.; Tan, B.H.; Teo, E.C.Y.; Lim, S.G.; Dan, Y.Y.; Wee, A.; Aw, P.P.K.; Zhu, Y.; Hibberd, M.L.; Tan, C.K.; et al. Chronic Infection With Camelid Hepatitis E Virus in a Liver Transplant Recipient Who Regularly Consumes Camel Meat and Milk. Gastroenterology 2016, 150, 355–357.e3. [Google Scholar] [CrossRef] [PubMed]
- Sridhar, S.; Yip, C.C.Y.; Lo, K.H.Y.; Wu, S.; Situ, J.; Chew, N.F.S.; Leung, K.H.; Chan, H.S.Y.; Wong, S.C.Y.; Leung, A.W.S.; et al. Hepatitis E Virus Species C Infection in Humans, Hong Kong. Clin. Infect. Dis. 2022, 75, 288–296. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Meng, X.J. Hepatitis E virus: Host tropism and zoonotic infection. Curr. Opin. Microbiol. 2021, 59, 8–15. [Google Scholar] [CrossRef]
- Smith, D.B.; Izopet, J.; Nicot, F.; Simmonds, P.; Jameel, S.; Meng, X.J.; Norder, H.; Okamoto, H.; van der Poel, W.H.M.; Reuter, G.; et al. Update: Proposed reference sequences for subtypes of hepatitis E virus (species Orthohepevirus A). J. Gen. Virol. 2020, 101, 692–698. [Google Scholar] [CrossRef]
- Meng, X.J.; Purcell, R.H.; Halbur, P.G.; Lehman, J.R.; Webb, D.M.; Tsareva, T.S.; Haynes, J.S.; Thacker, B.J.; Emerson, S.U. A novel virus in swine is closely related to the human hepatitis E virus. Proc. Natl. Acad. Sci. USA 1997, 94, 9860–9865. [Google Scholar] [CrossRef]
- Meng, X.J. From barnyard to food table: The omnipresence of hepatitis E virus and risk for zoonotic infection and food safety. Virus Res. 2011, 161, 23–30. [Google Scholar] [CrossRef]
- Salines, M.; Andraud, M.; Rose, N. From the epidemiology of hepatitis E virus (HEV) within the swine reservoir to public health risk mitigation strategies: A comprehensive review. Vet. Res. 2017, 48, 31. [Google Scholar] [CrossRef]
- Sooryanarain, H.; Meng, X.J. Swine hepatitis E virus: Cross-species infection, pork safety and chronic infection. Virus Res. 2020, 284, 197985. [Google Scholar] [CrossRef]
- Takahashi, M.; Okamoto, H. Features of hepatitis E virus infection in humans and animals in Japan. Hepatol. Res. 2014, 44, 43–58. [Google Scholar] [CrossRef]
- Yotsuyanagi, H. Hepatitis E, 2014–2021. Infect. Agents Surveill. Rep. (IASR) 2021, 42, 271–274. (In Japanese). Available online: www.niid.go.jp/niid/ja/iasr-vol42/10847-idx502.html (accessed on 5 July 2023).
- Takahashi, M.; Tanaka, T.; Azuma, M.; Kusano, E.; Aikawa, T.; Shibayama, T.; Yazaki, Y.; Mizuo, H.; Inoue, J.; Okamoto, H. Prolonged fecal shedding of hepatitis E virus (HEV) during sporadic acute hepatitis E: Evaluation of infectivity of HEV in fecal specimens in a cell culture system. J. Clin. Microbiol. 2007, 45, 3671–3679. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, H.; Akahane, Y.; Ukita, M.; Fukuda, M.; Tsuda, F.; Miyakawa, Y.; Mayumi, M. Fecal excretion of a nonenveloped DNA virus (TTV) associated with posttransfusion non-A-G hepatitis. J. Med. Virol. 1998, 56, 128–132. [Google Scholar] [CrossRef]
- Mizuo, H.; Suzuki, K.; Takikawa, Y.; Sugai, Y.; Tokita, H.; Akahane, Y.; Itoh, K.; Gotanda, Y.; Takahashi, M.; Nishizawa, T.; et al. Polyphyletic strains of hepatitis E virus are responsible for sporadic cases of acute hepatitis in Japan. J. Clin. Microbiol. 2002, 40, 3209–3218. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Nishizawa, T.; Tanaka, T.; Tsatsralt-Od, B.; Inoue, J.; Okamoto, H. Correlation between positivity for immunoglobulin A antibodies and viraemia of swine hepatitis E virus observed among farm pigs in Japan. J. Gen. Virol. 2005, 86, 1807–1813. [Google Scholar] [CrossRef]
- Inoue, J.; Takahashi, M.; Yazaki, Y.; Tsuda, F.; Okamoto, H. Development and validation of an improved RT-PCR assay with nested universal primers for detection of hepatitis E virus strains with significant sequence divergence. J. Virol. Methods 2006, 137, 325–333. [Google Scholar] [CrossRef]
- Takahashi, M.; Hoshino, Y.; Tanaka, T.; Takahashi, H.; Nishizawa, T.; Okamoto, H. Production of monoclonal antibodies against hepatitis E virus capsid protein and evaluation of their neutralizing activity in a cell culture system. Arch. Virol. 2008, 153, 657–666. [Google Scholar] [CrossRef]
- Takahashi, M.; Nishizono, A.; Kawakami, M.; Fukui, E.; Isogai, E.; Matsuoka, H.; Yamamoto, S.; Mizuo, H.; Nagashima, S.; Murata, K.; et al. Identification of hepatitis E virus in wild sika deer in Japan. Virus Res. 2022, 308, 198645. [Google Scholar] [CrossRef]
- Okamoto, H.; Takahashi, M.; Nishizawa, T.; Fukai, K.; Muramatsu, U.; Yoshikawa, A. Analysis of the complete genome of indigenous swine hepatitis E virus isolated in Japan. Biochem. Biophys. Res. Commun. 2001, 289, 929–936. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Jirintai, S.; Manglai, D.; Takahashi, M.; Nagashima, S.; Kobayashi, T.; Nishizawa, T.; Okamoto, H. Molecular analysis of hepatitis E virus from farm rabbits in Inner Mongolia, China and its successful propagation in A549 and PLC/PRF/5 cells. Virus Res. 2012, 170, 126–137. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Takahashi, M.; Kusano, E.; Okamoto, H. Development and evaluation of an efficient cell-culture system for Hepatitis E virus. J. Gen. Virol. 2007, 88, 903–911. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Yamada, K.; Hoshino, Y.; Takahashi, H.; Ichiyama, K.; Tanaka, T.; Okamoto, H. Monoclonal antibodies raised against the ORF3 protein of hepatitis E virus (HEV) can capture HEV particles in culture supernatant and serum but not those in feces. Arch. Virol. 2008, 153, 1703–1713. [Google Scholar] [CrossRef] [PubMed]
- Johne, R.; Dremsek, P.; Reetz, J.; Heckel, G.; Hess, M.; Ulrich, R.G. Hepeviridae: An expanding family of vertebrate viruses. Infect. Genet. Evol. 2014, 27, 212–229. [Google Scholar] [CrossRef]
- Kobayashi, T.; Takahashi, M.; Tanggis; Mulyanto; Jirintai, S.; Nagashima, S.; Nishizawa, T.; Okamoto, H. Characterization and epitope mapping of monoclonal antibodies raised against rat hepatitis E virus capsid protein: An evaluation of their neutralizing activity in a cell culture system. J. Virol. Methods 2016, 233, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Motoya, T.; Umezawa, M.; Goto, K.; Doi, I.; Nagata, N.; Ikeda, Y.; Sakuta, A.; Sasaki, N.; Ishii, K. High prevalence of hepatitis E virus infection among domestic pigs in Ibaraki Prefecture, Japan. BMC Vet. Res. 2019, 15, 87. [Google Scholar] [CrossRef]
- Takahashi, M.; Nishizawa, T.; Miyajima, H.; Gotanda, Y.; Iita, T.; Tsuda, F.; Okamoto, H. Swine hepatitis E virus strains in Japan form four phylogenetic clusters comparable with those of Japanese isolates of human hepatitis E virus. J. Gen. Virol. 2003, 84, 851–862. [Google Scholar] [CrossRef]
- Kanai, Y.; Tsujikawa, M.; Yunoki, M.; Nishiyama, S.; Ikuta, K.; Hagiwara, K. Long-term shedding of hepatitis E virus in the feces of pigs infected naturally, born to sows with and without maternal antibodies. J. Med. Virol. 2010, 82, 69–76. [Google Scholar] [CrossRef]
- Sovijit, W.; Taesuji, M.; Rattanamas, K.; Punyadarsaniya, D.; Mamom, T.; Nguyen, H.T.; Ruenphet, S. In vitro cytotoxicity and virucidal efficacy of potassium hydrogen peroxymonosulfate compared to quaternary ammonium compound under various concentrations, exposure times and temperatures against African swine fever virus. Vet. World 2021, 14, 2936–2940. [Google Scholar] [CrossRef]
- Meester, M.; Tobias, T.J.; Bouwknegt, M.; Kusters, N.E.; Stegeman, J.A.; van der Poel, W.H.M. Infection dynamics and persistence of hepatitis E virus on pig farms—A review. Porc. Health Manag. 2021, 7, 16. [Google Scholar] [CrossRef]
- de Deus, N.; Casas, M.; Peralta, B.; Nofrarias, M.; Pina, S.; Martin, M.; Segales, J. Hepatitis E virus infection dynamics and organic distribution in naturally infected pigs in a farrow-to-finish farm. Vet. Microbiol. 2008, 132, 19–28. [Google Scholar] [CrossRef]
- Leblanc, D.; Ward, P.; Gagne, M.J.; Poitras, E.; Muller, P.; Trottier, Y.L.; Simard, C.; Houde, A. Presence of hepatitis E virus in a naturally infected swine herd from nursery to slaughter. Int. J. Food Microbiol. 2007, 117, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Feng, R.; Zhao, C.; Li, M.; Harrison, T.J.; Qiao, Z.; Feng, Y.; Ma, Z.; Wang, Y. Infection dynamics of hepatitis E virus in naturally infected pigs in a Chinese farrow-to-finish farm. Infect. Genet. Evol. 2011, 11, 1727–1731. [Google Scholar] [CrossRef]
- Salines, M.; Dumarest, M.; Andraud, M.; Mahe, S.; Barnaud, E.; Cineux, M.; Eveno, E.; Eono, F.; Dorenlor, V.; Grasland, B.; et al. Natural viral co-infections in pig herds affect hepatitis E virus (HEV) infection dynamics and increase the risk of contaminated livers at slaughter. Transbound. Emerg. Dis. 2019, 66, 1930–1945. [Google Scholar] [CrossRef] [PubMed]
- Andraud, M.; Casas, M.; Pavio, N.; Rose, N. Early-life hepatitis e infection in pigs: The importance of maternally-derived antibodies. PLoS ONE 2014, 9, e105527. [Google Scholar] [CrossRef] [PubMed]
- Pavio, N.; Meng, X.J.; Renou, C. Zoonotic hepatitis E: Animal reservoirs and emerging risks. Vet. Res. 2010, 41, 46. [Google Scholar] [CrossRef] [PubMed]
- Ianiro, G.; Monini, M.; Ammendolia, M.G.; De Sabato, L.; Ostanello, F.; Vaccari, G.; Di Bartolo, I. In Vitro Replication of Swine Hepatitis E Virus (HEV): Production of Cell-Adapted Strains. Animals 2023, 13, 276. [Google Scholar] [CrossRef] [PubMed]
- Meester, M.; Bouwknegt, M.; Hakze-van der Honing, R.; Vernooij, H.; Houben, M.; van Oort, S.; van der Poel, W.H.M.; Stegeman, A.; Tobias, T. Repeated cross-sectional sampling of pigs at slaughter indicates varying age of hepatitis E virus infection within and between pig farms. Vet. Res. 2022, 53, 50. [Google Scholar] [CrossRef]
- Withenshaw, S.M.; Grierson, S.S.; Smith, R.P. Study of Animal Mixing and the Dynamics of Hepatitis E Virus Infection on a Farrow-to-Finish Pig Farm. Animals 2022, 12, 272. [Google Scholar] [CrossRef]
- van Tong, H.; Hoan, N.X.; Wang, B.; Wedemeyer, H.; Bock, C.T.; Velavan, T.P. Hepatitis E Virus Mutations: Functional and Clinical Relevance. EBioMedicine 2016, 11, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Toyota, J.; Karino, Y.; Kang, J.H.; Maekubo, H.; Abe, N.; Mishiro, S. Estimation of the mutation rate of hepatitis E virus based on a set of closely related 7.5-year-apart isolates from Sapporo, Japan. Hepatol. Res. 2004, 29, 212–215. [Google Scholar] [CrossRef] [PubMed]
- Nagashima, S.; Kobayashi, T.; Tanaka, T.; Tanggis; Jirintai, S.; Takahashi, M.; Nishizawa, T.; Okamoto, H. Analysis of adaptive mutations selected during the consecutive passages of hepatitis E virus produced from an infectious cDNA clone. Virus Res. 2016, 223, 170–180. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo, F.R.; Tanaka, T.; Takahashi, H.; Ichiyama, K.; Hoshino, Y.; Yamada, K.; Inoue, J.; Takahashi, M.; Okamoto, H. Mutational events during the primary propagation and consecutive passages of hepatitis E virus strain JE03-1760F in cell culture. Virus Res. 2008, 137, 86–96. [Google Scholar] [CrossRef]
- Okamoto, H.; Kojima, M.; Okada, S.; Yoshizawa, H.; Iizuka, H.; Tanaka, T.; Muchmore, E.E.; Peterson, D.A.; Ito, Y.; Mishiro, S. Genetic drift of hepatitis C virus during an 8.2-year infection in a chimpanzee: Variability and stability. Virology 1992, 190, 894–899. [Google Scholar] [CrossRef]
- Sari, G.; van de Garde, M.D.B.; van Schoonhoven, A.; Voermans, J.J.C.; van der Eijk, A.A.; de Man, R.A.; Boonstra, A.; Vanwolleghem, T.; Pas, S.D. Hepatitis E Virus Shows More Genomic Alterations in Cell Culture than In Vivo. Pathogens 2019, 8, 255. [Google Scholar] [CrossRef]
- Takahashi, H.; Tanaka, T.; Jirintai, S.; Nagashima, S.; Takahashi, M.; Nishizawa, T.; Mizuo, H.; Yazaki, Y.; Okamoto, H. A549 and PLC/PRF/5 cells can support the efficient propagation of swine and wild boar hepatitis E virus (HEV) strains: Demonstration of HEV infectivity of porcine liver sold as food. Arch. Virol. 2012, 157, 235–246. [Google Scholar] [CrossRef]


| Year | Study Type | Sample | Age in Months a (Days after Birth) | Place (House) | Number. of Pigs | Number (%) of Pigs with: | |
|---|---|---|---|---|---|---|---|
| Anti-HEV IgG | HEV RNA | ||||||
| 2012 | Cross-sectional | Serum | 2 (60–63) | Weaning | 30 | 5 (16.7) | 14 (46.7) |
| 3 (90–93) | Growing | 30 | 30 (100) | 0 | |||
| 4 (121–126) | Growing | 30 | 30 (100) | 0 | |||
| 5 (149–155) | Growing | 30 | 30 (100) | 0 | |||
| Subtotal | 120 | 95 (79.2) | 14 (11.7) | ||||
| 2013 | Cohort (Study group) | Serum | 1 (19–23) | Farrowing | 19 | 11 (57.9) | 0 |
| 3 (82–86) | HEV-free (A) b | 19 | 0 | 0 | |||
| 4 (110–114) | HEV-free (A) | 19 | 1 (5.3) | 1 (5.3) | |||
| 5 (138–142) | HEV-free (A) | 19 | 16 (84.2) | 1 (5.3) | |||
| 6 (166–170) | HEV-free (A) | 19 | 18 (94.7) | 0 | |||
| Subtotal | 19 | 18 (94.7) | 2 (10.5) | ||||
| Cohort (Control group) | Serum | 1 (19–23) | Farrowing | 26 | 18 (69.2) | 0 | |
| 3 (82–86) | Growing | 26 | 15 (57.7) | 10 (38.5) | |||
| 4 (110–114) | Growing | 26 | 26 (100) | 1 (3.8) | |||
| 5 (138–142) | Growing | 26 | 26 (100) | 0 | |||
| 6 (166–170) | Growing | 25 | 23 (92.0) | 0 | |||
| Subtotal | 26 | 26 (100) | 10 (38.5) | ||||
| 2014 | Cohort | Serum | 1 (22–27) | Farrowing | 69 | 23 (33.3) | 0 |
| 2 (53–60) | Weaning | 69 | 1 (1.4) | 2 (2.9) | |||
| 3 (83–91) | Growing | 69 | 54 (78.3) | 39 (56.5) | |||
| 4 (113–123) | Growing | 69 | 69 (100) | 3 (4.3) | |||
| 5 (144–156) | Growing | 69 | 68 (98.6) | 0 | |||
| 6 (173–207) | Growing | 69 | 67 (97.1) | 0 | |||
| Subtotal | 69 | 69 (100) | 40 (58.0) | ||||
| 2016 | Cohort | Serum | 2 (58–59) | HEV-free (B) c | 3 | 1 (33.3) | 2 (66.7) |
| 3 (85–86) | HEV-free (B) | 3 | 3 (100) | 2 (66.7) | |||
| Subtotal | 3 | 3 (100) | 2 (66.7) | ||||
| 2019 | Cohort | Serum | 1 (24–28) | Farrowing | 11 | 6 (54.5) | 0 |
| 2 (54–58) | Growing d | 11 | 2 (18.2) | 0 | |||
| 3 (88–92) | Growing | 11 | 2 (18.2) | 7 (63.6) | |||
| 4 (117–121) | Growing | 11 | 7 (63.6) | 2 (18.2) | |||
| 5 (144–148) | Growing | 11 | 10 (90.9) | 0 | |||
| 6 (173–177) | Growing | 11 | 10 (90.9) | 0 | |||
| Subtotal | 11 | 11 (100) | 9 (81.8) | ||||
| 2021 | Cross-sectional | Feces | 2 e | Weaning | 10 | - | 8 (80.0) |
| 3 e | Growing | 10 | - | 10 (100) | |||
| Subtotal | 20 | - | 18 (90.0) | ||||
| Total | 268 | 222 (89.5) f | 95 (35.4) | ||||
| Year | Number of Pigs Studied | Months after Birth | p-Value | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | ||||
| Number (%) of pigs by month with the highest OD value of anti-HEV IgG | |||||||||
| 2013 a | 26 | 18 (69.2) | NA b | 7 (26.9) | 15 (57.7) | 2 (7.7) | 2 (7.7) | (a) 0.9760 (c) 0.1583 | |
| 2014 a | 69 | 23 (33.3) | 0 | 31 (44.9) | 31 (44.9) | 5 (7.2) | 2 (2.9) | ||
| With c | 50 | 23 (46.0) | 0 | 20 (40.0) | 24 (48.0) | 4 (8.0) | 2 (4.0) | ||
| Without c | 19 | 0 | 0 | 11 (57.9) | 7 (36.8) | 1 (5.3) | 0 | ||
| 2019 a | 11 | 6 (54.5) | 0 | 2 (18.2) | 5 (45.5) | 3 (27.3) | 1 (9.1) | ||
| Number (%) of pigs by month with the first appearance of HEV viremia | |||||||||
| 2013 d | 26 | 0 | NA | 10 (38.5) | 0 | 0 | 0 | (d) 0.0837 (e) 0.4081 | |
| 2014 d | 69 | 0 | 2 (2.9) | 38 (55.1) | 0 | 0 | 0 | ||
| With e | 50 | 0 | 1 (2.0) | 29 (58.0) | 0 | 0 | 0 | ||
| Without e | 19 | 0 | 1 (5.3) | 9 (47.4) | 0 | 0 | 0 | ||
| 2019 d | 11 | 0 | 0 | 7 (63.6) | 2 (18.2) | 0 | 0 | ||
| Place | Slurry in the Manure-Pit of Each House (Copies/g) a | Samples in Pens | Dust Samples from Filters in Four Rooms (Copies/Filter) e | |||
|---|---|---|---|---|---|---|
| Feces on the Floor b | Floor Swab (Copies/cm2) c | Wall Swab (Copies/cm2) c | Feed Samples from/around the Trough in One Each Pen of Four Rooms (Copies/g) d | |||
| Farrowing house | (–) f | 0/10 | (–) | (–) | NT g | NT |
| Weaning house | 2.7 × 105 | 5/5 (100%) h | 4.9 × 102 | 2.2 × 102 | (R1) 4.5 × 101/7.3 × 102 (R2) 1.8 × 101/7.7 × 102 (R3) 3.7 × 103/1.8 × 105 (R4) 1.6 × 103/9.9 × 104 | (R1) 3.0 × 104 (R2) 3.4 × 103 (R3) 7.5 × 105 (R4) 8.0 × 105 |
| Growing house | 7.4 × 105 | 2/3 (67%) i | (+) j | (+) j | NT | NT |
| Sow house | (–) | NT | NT | NT | NT | NT |
| Boar house | (–) | NT | NT | NT | NT | NT |
| Year | Number of Strains Compared | Nucleotide Sequence Identity (%) | Number (%) of Strains with the Mutation at the Indicated Nucleotide Position a: | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean ± SD (Median) | Range | 5972 T/C | 6005 T/C | 6035 T/C | 6102 T/C | 6116 T/C | 6194 T/A | 6266 T/C | 6285 T/C | 6287 G/A | 6318 T/C | 6344 T/C | ||
| 2012 | 13 | 99.6 ± 0.1 (99.7) | 99.5–99.7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2013 | 12 | 99.0 ± 0.5 (99.0) | 98.0–99.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2014 | 40 | 98.6 ± 0.5 (98.7) | 97.8–99.2 | 0 | 0 | 5 (12.5) | 19 (47.5) | 1(2.5) | 0 | 19 (47.5) | 25 (62.5) | 0 | 0 | 0 |
| 2016 | 2 | 98.0 ± 1.0 (98.0) | 97.3–98.7 | 0 | 0 | 0 | 2 (100) | 0 | 0 | 0 | 2 (100) | 0 | 0 | 0 |
| 2017 | 7 | 98.2 ± 0.4 (98.0) | 98.0–99.2 | 0 | 0 | 0 | 7 (100) | 6 (85.7) | 0 | 0 | 2 (28.6) | 0 | 3 (42.9) | 0 |
| 2018 | 2 | 97.5 ± 0.0 (97.5) | 97.5 | 0 | 0 | 0 | 2 (100) | 2 (100) | 0 | 2 (100) | 2 (100) | 2 (100) | 2 (100) | 2 (100) |
| 2019 | 9 | 96.9 ± 0.4 (97.0) | 96.3–97.3 | 9 (100) | 9 (100) | 9 (100) | 9 (100) | 9 (100) | 0 | 9 (100) | 9 (100) | 9 (100) | 9 (100) | 9 (100) |
| 2021 | 18 | 97.0 ± 0.2 (97.0) | 96.6–97.3 | 18 (100) | 18 (100) | 18 (100) | 18 (100) | 18 (100) | 18 (100) | 18 (100) | 18 (100) | 18 (100) | 11 (61.1) | 18 (100) |
| Region | Identity (%) | |
|---|---|---|
| Nucleotides | Amino Acids | |
| Entire genome a | 97.9 (7071/7226) | - b |
| 5′UTR | 100 (25/25) | - |
| ORF1 | 98.0 (5006/5109) | 99.6 (1696/1703) c |
| ORF2 | 97.5 (1930/1980) | 99.7 (658/660) d |
| ORF3 | 99.1 (336/339) | 99.1 (112/113) e |
| 3′UTR a | 97.3 (73/75) | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takahashi, M.; Kunita, S.; Nishizawa, T.; Ohnishi, H.; Primadharsini, P.P.; Nagashima, S.; Murata, K.; Okamoto, H. Infection Dynamics and Genomic Mutations of Hepatitis E Virus in Naturally Infected Pigs on a Farrow-to-Finish Farm in Japan: A Survey from 2012 to 2021. Viruses 2023, 15, 1516. https://doi.org/10.3390/v15071516
Takahashi M, Kunita S, Nishizawa T, Ohnishi H, Primadharsini PP, Nagashima S, Murata K, Okamoto H. Infection Dynamics and Genomic Mutations of Hepatitis E Virus in Naturally Infected Pigs on a Farrow-to-Finish Farm in Japan: A Survey from 2012 to 2021. Viruses. 2023; 15(7):1516. https://doi.org/10.3390/v15071516
Chicago/Turabian StyleTakahashi, Masaharu, Satoshi Kunita, Tsutomu Nishizawa, Hiroshi Ohnishi, Putu Prathiwi Primadharsini, Shigeo Nagashima, Kazumoto Murata, and Hiroaki Okamoto. 2023. "Infection Dynamics and Genomic Mutations of Hepatitis E Virus in Naturally Infected Pigs on a Farrow-to-Finish Farm in Japan: A Survey from 2012 to 2021" Viruses 15, no. 7: 1516. https://doi.org/10.3390/v15071516
APA StyleTakahashi, M., Kunita, S., Nishizawa, T., Ohnishi, H., Primadharsini, P. P., Nagashima, S., Murata, K., & Okamoto, H. (2023). Infection Dynamics and Genomic Mutations of Hepatitis E Virus in Naturally Infected Pigs on a Farrow-to-Finish Farm in Japan: A Survey from 2012 to 2021. Viruses, 15(7), 1516. https://doi.org/10.3390/v15071516






