Fluorescence Microscopy in Adeno-Associated Virus Research
Abstract
1. Introduction
1.1. AAV Genes and Structure
1.2. AAV Life Cycle
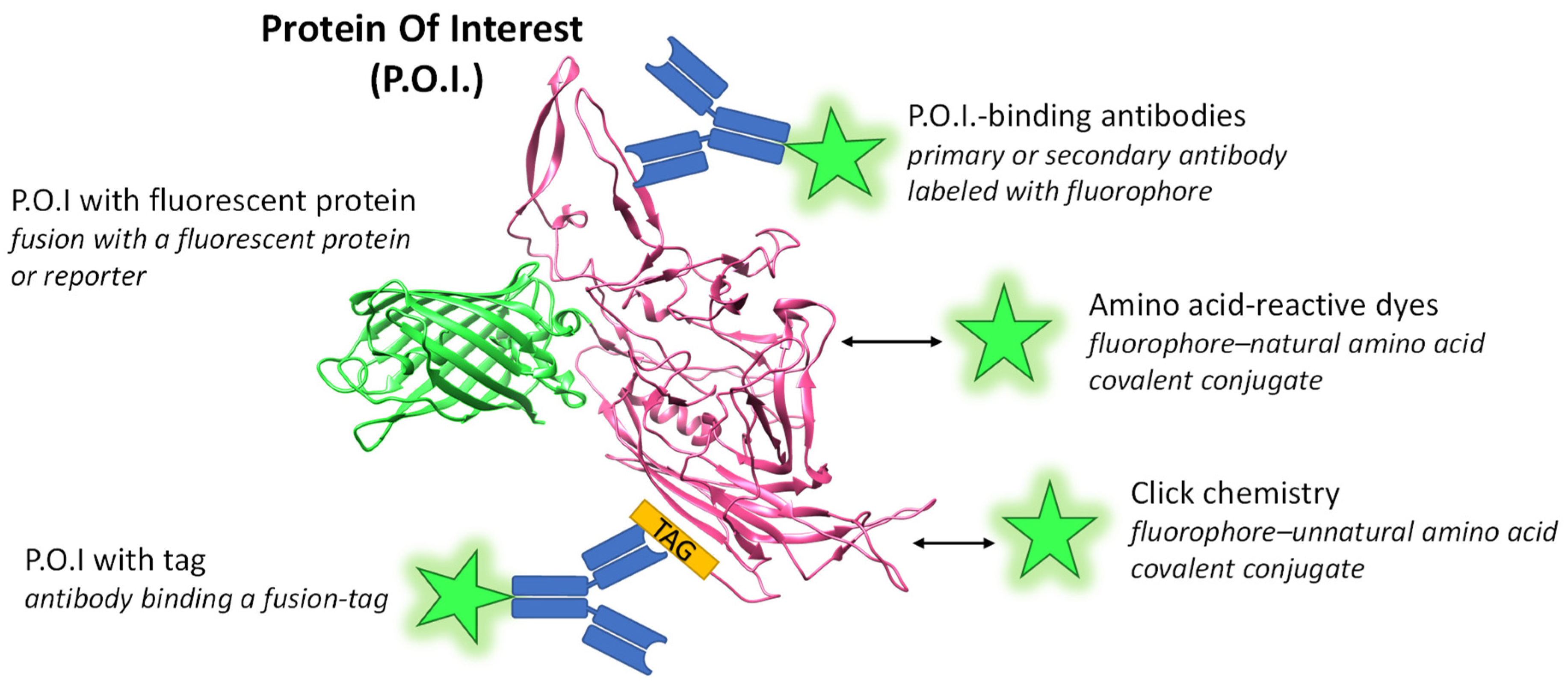
2. Fluorophores and Labeling
2.1. Labeling with Amino Acid-Reactive Dyes
2.2. Labeling with Click Chemistry
2.3. Labeling via Tags
2.4. Labeling with AAV Protein-Binding Antibodies
2.5. Labeling with Genetically-Fused Fluorescent Proteins
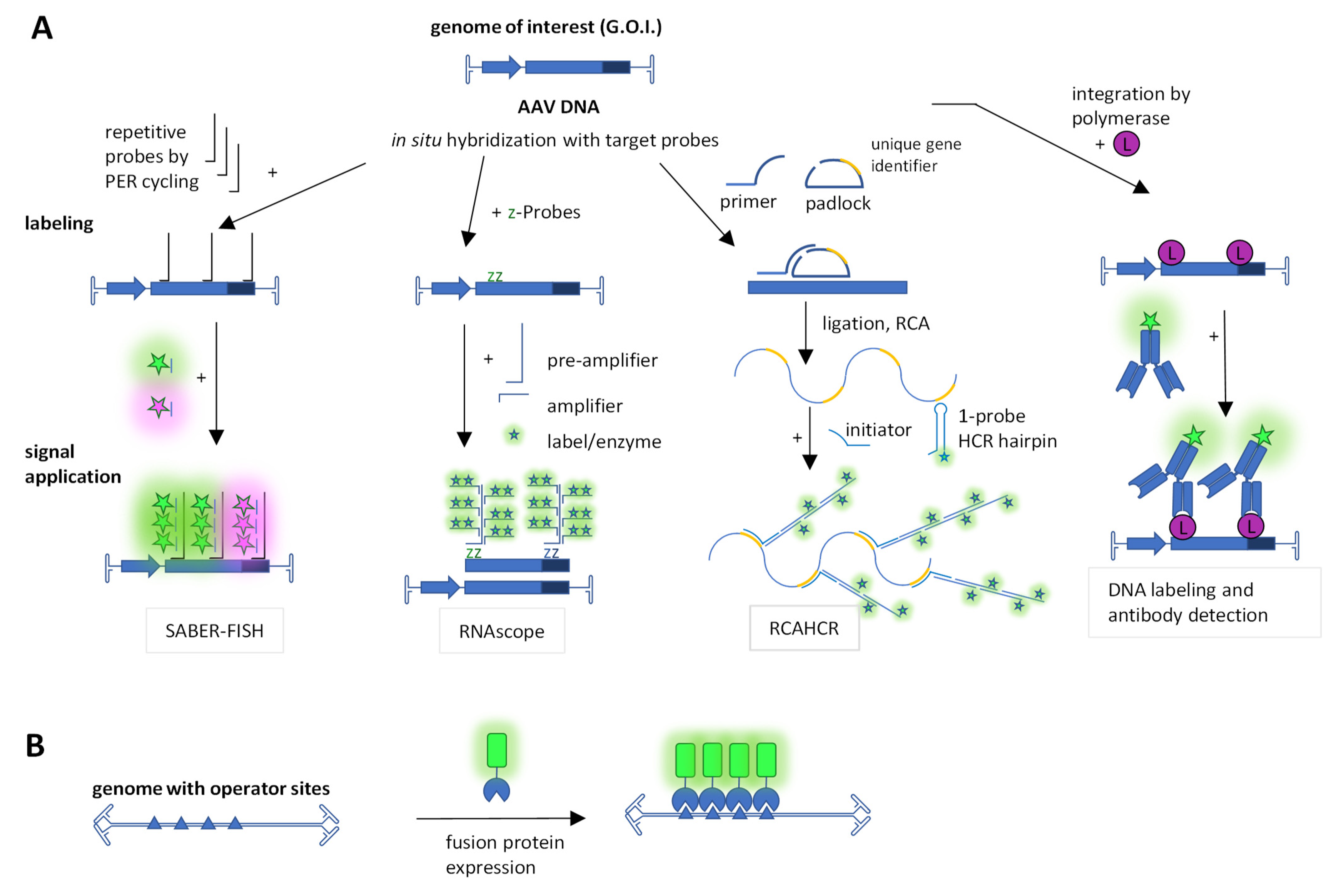
2.6. Staining of DNA and RNA
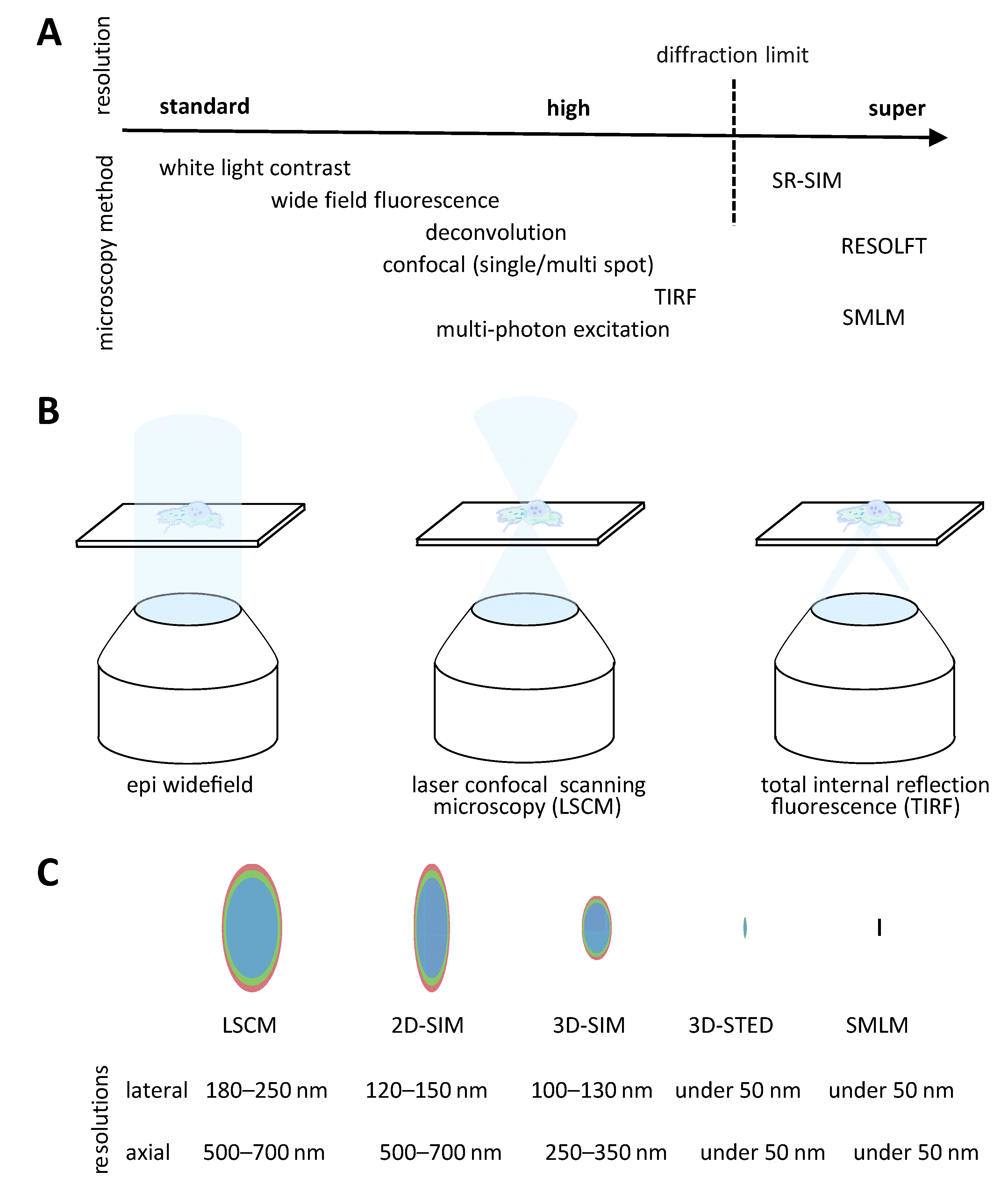
3. Microscopy Methods
3.1. Widefield Microscopy
3.2. Confocal Microscopy
3.3. Total Internal Reflection Fluorescence Microscopy
3.4. Super Resolution Microscopy
3.5. Super-Resolution Microscopy in the Context of AAV
4. Outlook
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Atchison, R.W.; Casto, B.C.; Hammon, W.M. Adenovirus-Associated Defective Virus Particles. Science 1965, 149, 754–756. [Google Scholar] [CrossRef]
- Gao, G.; Vandenberghe, L.H.; Alvira, M.R.; Lu, Y.; Calcedo, R.; Zhou, X.; Wilson, J.M. Clades of Adeno-Associated Viruses Are Widely Disseminated in Human Tissues. J. Virol. 2004, 78, 6381–6388. [Google Scholar] [CrossRef]
- Korneyenkov, M.A.; Zamyatnin, A.A. Next Step in Gene Delivery: Modern Approaches and Further Perspectives of AAV Tropism Modification. Pharmaceutics 2021, 13, 750. [Google Scholar] [CrossRef]
- Xie, Q.; Bu, W.; Bhatia, S.; Hare, J.; Somasundaram, T.; Azzi, A.; Chapman, M.S. The Atomic Structure of Adeno-Associated Virus (AAV-2), a Vector for Human Gene Therapy. Proc. Natl. Acad. Sci. USA 2002, 99, 10405–10410. [Google Scholar] [CrossRef]
- Mietzsch, M.; Jose, A.; Chipman, P.; Bhattacharya, N.; Daneshparvar, N.; McKenna, R.; Agbandje-McKenna, M. Completion of the AAV Structural Atlas: Serotype Capsid Structures Reveals Clade-Specific Features. Viruses 2021, 13, 101. [Google Scholar] [CrossRef]
- Berns, K.I. Parvovirus Replication. Microbiol. Rev. 1990, 54, 316–329. [Google Scholar] [CrossRef]
- Stutika, C.; Gogol-Döring, A.; Botschen, L.; Mietzsch, M.; Weger, S.; Feldkamp, M.; Chen, W.; Heilbronn, R. A Comprehensive RNA Sequencing Analysis of the Adeno-Associated Virus (AAV) Type 2 Transcriptome Reveals Novel AAV Transcripts, Splice Variants, and Derived Proteins. J. Virol. 2016, 90, 1278–1289. [Google Scholar] [CrossRef]
- Trempe, J.P.; Carter, B.J. Alternate MRNA Splicing Is Required for Synthesis of Adeno-Associated Virus VP1 Capsid Protein. J. Virol. 1988, 62, 3356–3363. [Google Scholar] [CrossRef]
- Ogden, P.J.; Kelsic, E.D.; Sinai, S.; Church, G.M. Comprehensive AAV Capsid Fitness Landscape Reveals a Viral Gene and Enables Machine-Guided Design. Science 2019, 366, 1139–1143. [Google Scholar] [CrossRef]
- Elmore, Z.C.; Patrick Havlik, L.; Oh, D.K.; Anderson, L.; Daaboul, G.; Devlin, G.W.; Vincent, H.A.; Asokan, A. The Membrane Associated Accessory Protein Is an Adeno-Associated Viral Egress Factor. Nat. Commun. 2021, 12, 1–13. [Google Scholar] [CrossRef]
- Galibert, L.; Hyvönen, A.; Eriksson, R.A.E.; Mattola, S.; Aho, V.; Salminen, S.; Albers, J.D.; Peltola, S.K.; Weman, S.; Nieminen, T.; et al. Functional Roles of the Membrane-Associated AAV Protein MAAP. Sci. Rep. 2021, 11, 21698. [Google Scholar] [CrossRef] [PubMed]
- Sonntag, F.; Schmidt, K.; Kleinschmidt, J.A. A Viral Assembly Factor Promotes AAV2 Capsid Formation in the Nucleolus. Proc. Natl. Acad. Sci. USA 2010, 107, 10220–10225. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; You, H.; Hermonat, P.L. The X Gene of Adeno-Associated Virus 2 (AAV2) Is Involved in Viral DNA Replication. PLoS ONE 2014, 9, e104596. [Google Scholar] [CrossRef]
- Casto, B.C.; Armstrong, J.A.; Atchison, R.W.; Hammon, W.M. Studies on the Relationship between Adeno-Associated Virus Type 1 (AAV-1) and Adenoviruses. Virology 1967, 33, 452–458. [Google Scholar] [CrossRef] [PubMed]
- Samulski, R.; Zhu, X.; Xiao, X.; Brook, J.; Housman, D.; Epstein, N.; Hunter, L. Targeted Integration of Adeno-Associated Virus (AAV) into Human Chromosome 19. EMBO J. 1991, 10, 3941–3950. [Google Scholar] [CrossRef]
- Chang, L.S.; Shi, Y.; Shenk, T. Adeno-Associated Virus P5 Promoter Contains an Adenovirus E1A-Inducible Element and a Binding Site for the Major Late Transcription Factor. J. Virol. 1989, 63, 3479–3488. [Google Scholar] [CrossRef]
- Chang, L.S.; Shenk, T. The Adenovirus DNA-Binding Protein Stimulates the Rate of Transcription Directed by Adenovirus and Adeno-Associated Virus Promoters. J. Virol. 1990, 64, 2103–2109. [Google Scholar] [CrossRef]
- Ferrari, F.K.; Samulski, T.; Shenk, T.; Samulski, R.J. Second-Strand Synthesis Is a Rate-Limiting Step for Efficient Transduction by Recombinant Adeno-Associated Virus Vectors. J. Virol. 1996, 70, 3227–3234. [Google Scholar] [CrossRef]
- Maurer, A.C.; Weitzman, M.D. Adeno-Associated Virus Genome Interactions Important for Vector Production and Transduction. Hum. Gene Ther. 2020, 31, 499–511. [Google Scholar] [CrossRef]
- Summerford, C.; Samulski, R.J. Membrane-Associated Heparan Sulfate Proteoglycan Is a Receptor for Adeno-Associated Virus Type 2 Virions. J. Virol. 1998, 72, 1438–1445. [Google Scholar] [CrossRef]
- Kern, A.; Schmidt, K.; Leder, C.; Müller, O.J.; Wobus, C.E.; Bettinger, K.; Von der Lieth, C.W.; King, J.A.; Kleinschmidt, J.A. Identification of a Heparin-Binding Motif on Adeno-Associated Virus Type 2 Capsids. J. Virol. 2003, 77, 11072–11081. [Google Scholar] [CrossRef]
- Opie, S.R.; Warrington, K.H.; Agbandje-mckenna, M.; Zolotukhin, S.; Muzyczka, N. Identification of Amino Acid Residues in the Capsid Proteins of Adeno-Associated Virus Type 2 That Contribute to Heparan Sulfate Proteoglycan Binding †. Society 2003, 77, 6995–7006. [Google Scholar] [CrossRef] [PubMed]
- Pillay, S.; Meyer, N.L.; Puschnik, A.S.; Davulcu, O.; Diep, J.; Ishikawa, Y.; Jae, L.T.; Wosen, J.E.; Nagamine, C.M.; Chapman, M.S.; et al. An Essential Receptor for Adeno-Associated Virus Infection. Nature 2016, 530, 108–112. [Google Scholar] [CrossRef]
- Iwabuchi, K.; Daida, H.; Matsumoto, K.; Oshimi, K.; Watanabe, M.; Kashiwakura, Y.; Tamayose, K.; Shimada, T.; Nakamura, T.; Hirai, Y. Hepatocyte Growth Factor Receptor Is a Coreceptor for Adeno-Associated Virus Type 2 Infection. J. Virol. 2004, 79, 609–614. [Google Scholar] [CrossRef]
- Akache, B.; Grimm, D.; Pandey, K.; Yant, S.R.; Xu, H.; Kay, M.A. The 37/67-Kilodalton Laminin Receptor Is a Receptor for Adeno-Associated Virus Serotypes 8, 2, 3, and 9. J. Virol. 2006, 80, 9831–9836. [Google Scholar] [CrossRef] [PubMed]
- Asokan, A.; Hamra, J.B.; Govindasamy, L.; Agbandje-McKenna, M.; Samulski, R.J. Adeno-Associated Virus Type 2 Contains an Integrin A5β1 Binding Domain Essential for Viral Cell Entry. J. Virol. 2006, 80, 8961–8969. [Google Scholar] [CrossRef] [PubMed]
- Büning, H.; Perabo, L.; Quadt-Humme, S.; Hallek, M. Recent Developments in Adeno-Associated Virus Vector Technology. J. Gene Med. 2008, 10, 717–733. [Google Scholar] [CrossRef]
- Sanlioglu, S.; Benson, P.K.; Yang, J.; Atkinson, E.M.; Reynolds, T.; Engelhardt, J.F. Endocytosis and Nuclear Trafficking of Adeno-Associated Virus Type 2 Are Controlled by Rac1 and Phosphatidylinositol-3 Kinase Activation. J. Virol. 2000, 74, 9184–9196. [Google Scholar] [CrossRef]
- Nonnenmacher, M.; Weber, T. Adeno-Associated Virus 2 Infection Requires Endocytosis through the CLIC/GEEC Pathway. Cell Host Microbe 2011, 10, 563–576. [Google Scholar] [CrossRef]
- Riyad, J.M.; Weber, T. Intracellular Trafficking of Adeno-Associated Virus (AAV) Vectors: Challenges and Future Directions. Gene Ther. 2021, 28, 683–696. [Google Scholar] [CrossRef]
- Venkatakrishnan, B.; Yarbrough, J.; Domsic, J.; Bennett, A.; Bothner, B.; Kozyreva, O.G.; Samulski, R.J.; Muzyczka, N.; McKenna, R.; Agbandje-McKenna, M. Structure and Dynamics of Adeno-Associated Virus Serotype 1 VP1-Unique N-Terminal Domain and Its Role in Capsid Trafficking. J. Virol. 2013, 87, 4974–4984. [Google Scholar] [CrossRef]
- Penzes, J.J.; Chipman, P.; Bhattacharya, N.; Zeher, A.; Huang, R.; McKenna, R.; Agbandje-McKenna, M. Adeno-Associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking PH and Glycan Attachment. J. Virol. 2021, 95, e00843-21. [Google Scholar] [CrossRef] [PubMed]
- Girod, A.; Wobus, C.E.; Zádori, Z.; Ried, M.; Leike, K.; Tijssen, P.; Kleinschmidt, J.A.; Hallek, M. The VP1 Capsid Protein of Adeno-Associated Virus Type 2 Is Carrying a Phospholipase A2 Domain Required for Virus Infectivity. J. Gen. Virol. 2002, 83, 973–978. [Google Scholar] [CrossRef] [PubMed]
- Stahnke, S.; Lux, K.; Uhrig, S.; Kreppel, F.; Hösel, M.; Coutelle, O.; Ogris, M.; Hallek, M.; Büning, H. Intrinsic Phospholipase A2 Activity of Adeno-Associated Virus Is Involved in Endosomal Escape of Incoming Particles. Virology 2011, 409, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Murakami, M.; Kudo, I. Phospholipase A2. J. Biochem. 2002, 131, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Grieger, J.C.; Snowdy, S.; Samulski, R.J. Separate Basic Region Motifs within the Adeno-Associated Virus Capsid Proteins Are Essential for Infectivity and Assembly. J. Virol. 2006, 80, 5199–5210. [Google Scholar] [CrossRef]
- Popa-Wagner, R.; Porwal, M.; Kann, M.; Reuss, M.; Weimer, M.; Florin, L.; Kleinschmidt, J.A. Impact of VP1-Specific Protein Sequence Motifs on Adeno-Associated Virus Type 2 Intracellular Trafficking and Nuclear Entry. J. Virol. 2012, 86, 9163–9174. [Google Scholar] [CrossRef]
- Dudek, A.M.; Zabaleta, N.; Zinn, E.; Pillay, S.; Zengel, J.; Porter, C.; Franceschini, J.S.; Estelien, R.; Carette, J.E.; Zhou, G.L.; et al. GPR108 Is a Highly Conserved AAV Entry Factor. Mol. Ther. 2020, 28, 367–381. [Google Scholar] [CrossRef]
- Xiao, P.-J.; Samulski, R.J. Cytoplasmic Trafficking, Endosomal Escape, and Perinuclear Accumulation of Adeno-Associated Virus Type 2 Particles Are Facilitated by Microtubule Network. J. Virol. 2012, 86, 10462–10473. [Google Scholar] [CrossRef]
- Liu, Y.; Joo, K.; Wang, P. Endocytic Processing of Adeno-Associated Virus Type 8 Vectors for Transduction of Target Cells. Gene Ther. 2013, 20, 308–317. [Google Scholar] [CrossRef]
- Mattola, S.; Hakanen, S.; Salminen, S.; Aho, V.; Mäntylä, E.; Ihalainen, T.O.; Kann, M.; Vihinen-Ranta, M. Concepts to Reveal Parvovirus–Nucleus Interactions. Viruses 2021, 13, 1306. [Google Scholar] [CrossRef] [PubMed]
- Snyder, R.O.; Moullier, P. Adeno-Associated Virus; Snyder, R.O., Moullier, P., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2011; Volume 807, ISBN 978-1-61779-369-1, ISBN 978-1-61779-370-7. [Google Scholar]
- Wistuba, A.; Kern, A.; Weger, S.; Grimm, D.; Kleinschmidt, J.A. Subcellular Compartmentalization of Adeno-Associated Virus Type 2 Assembly. J. Virol. 1997, 71, 1341–1352. [Google Scholar] [CrossRef]
- Nicolson, S.C.; Samulski, R.J. Recombinant Adeno-Associated Virus Utilizes Host Cell Nuclear Import Machinery To Enter the Nucleus. J. Virol. 2014, 88, 4132–4144. [Google Scholar] [CrossRef]
- Sonntag, F.; Bleker, S.; Leuchs, B.; Fischer, R.; Kleinschmidt, J. Adeno-Associated Virus Type 2 Capsids with Externalized VP1/VP2 Trafficking Domains Are Generated Prior to Passage through the Cytoplasm and Are Maintained until Uncoating Occurs in the Nucleus. J. Virol. 2006, 80, 11040–11054. [Google Scholar] [CrossRef] [PubMed]
- Sonntag, F.; Köther, K.; Schmidt, K.; Weghofer, M.; Raupp, C.; Nieto, K.; Kuck, A.; Gerlach, B.; Böttcher, B.; Müller, O.J.; et al. The Assembly-Activating Protein Promotes Capsid Assembly of Different Adeno-Associated Virus Serotypes. J. Virol. 2011, 85, 12686–12697. [Google Scholar] [CrossRef]
- Maurer, A.C.; Pacouret, S.; Cepeda Diaz, A.K.; Blake, J.; Andres-Mateos, E.; Vandenberghe, L.H. The Assembly-Activating Protein Promotes Stability and Interactions between AAV’s Viral Proteins to Nucleate Capsid Assembly. Cell Rep. 2018, 23, 1817–1830. [Google Scholar] [CrossRef] [PubMed]
- Cassell, G.D.; Weitzman, M.D. Characterization of a Nuclear Localization Signal in the C-Terminus of the Adeno-Associated Virus Rep68/78 Proteins. Virology 2004, 327, 206–214. [Google Scholar] [CrossRef] [PubMed]
- King, J.A.; Dubielzig, R.; Grimm, D.; Kleinschmidt, J.A. DNA Helicase-Mediated Packaging of Adeno-Associated Virus Type 2 Genomes into Preformed Capsids. EMBO J. 2001, 20, 3282–3291. [Google Scholar] [CrossRef]
- Bartlett, J.S.; Samulski, R.J.; McCown, T.J. Selective and Rapid Uptake of Adeno-Associated Virus Type 2 in Brain. Hum. Gene Ther. 1998, 9, 1181–1186. [Google Scholar] [CrossRef]
- Bartlett, J.S.; Samulski, R.J. Fluorescent Viral Vectors: A New Technique for the Pharmacological Analysis of Gene Therapy. Nat. Med. 1998, 4, 635–637. [Google Scholar] [CrossRef]
- Bartlett, J.S.; Wilcher, R.; Samulski, R.J. Infectious Entry Pathway of Adeno-Associated Virus and Adeno-Associated Virus Vectors. J. Virol. 2000, 74, 2777–2785. [Google Scholar] [CrossRef] [PubMed]
- Seisenberger, G.; Ried, M.U.; Endreß, T.; Büning, H.; Hallek, M.; Bräuchle, C. Real-Time Single-Molecule Imaging of the Infection Pathway of an Adeno-Associated Virus. Science 2001, 294, 1929–1932. [Google Scholar] [CrossRef]
- Dalkara, D.; Kolstad, K.D.; Caporale, N.; Visel, M.; Klimczak, R.R.; Schaffer, D.V.; Flannery, J.G. Inner Limiting Membrane Barriers to AAV-Mediated Retinal Transduction From the Vitreous. Mol. Ther. 2009, 17, 2096–2102. [Google Scholar] [CrossRef] [PubMed]
- Mével, M.; Bouzelha, M.; Leray, A.; Pacouret, S.; Guilbaud, M.; Penaud-Budloo, M.; Alvarez-Dorta, D.; Dubreil, L.; Gouin, S.G.; Combal, J.P.; et al. Chemical Modification of the Adeno-Associated Virus Capsid to Improve Gene Delivery. Chem. Sci. 2020, 11, 1122–1131. [Google Scholar] [CrossRef]
- Büning, H.; Bolyard, C.M.; Hallek, M.; Bartlett, J.S. Modification and Labeling of AAV Vector Particles. In Adeno-Associated Virus: Methods and Protocols; Humana Press/Springer Science: New York, NY, USA, 2011; Volume 807, pp. 273–300. ISBN 9781617793707. [Google Scholar]
- Xiao, W.; Warrington, K.H.; Hearing, P.; Hughes, J.; Muzyczka, N. Adenovirus-Facilitated Nuclear Translocation of Adeno-Associated Virus Type 2. J. Virol. 2002, 76, 11505–11517. [Google Scholar] [CrossRef] [PubMed]
- Joo, K.; Fang, Y.; Liu, Y.; Xiao, L.; Gu, Z.; Tai, A.; Lee, C.; Tang, Y.; Wang, P. Enhanced Real-Time Monitoring of Adeno-Associated Virus Trafficking by Virus-Quantum Dot Conjugates. ACS Nano 2011, 5, 3523–3535. [Google Scholar] [CrossRef]
- Cui, M.; Lu, Y.; Tang, C.; Zhang, R.; Wang, J.; Si, Y.; Cheng, S.; Ding, W. A Generic Method for Fast and Sensitive Detection of Adeno-Associated Viruses Using Modified AAV Receptor Recombinant Proteins. Molecules 2019, 24, 3973. [Google Scholar] [CrossRef]
- Zhang, C.; Zhou, X.; Yao, T.; Tian, Z.; Zhou, D. Precision Fluorescent Labeling of an Adeno-Associated Virus Vector to Monitor the Viral Infection Pathway. Biotechnol. J. 2018, 13, e1700374. [Google Scholar] [CrossRef]
- Katrekar, D.; Moreno, A.M.; Chen, G.; Worlikar, A.; Mali, P. Oligonucleotide Conjugated Multi-Functional Adeno-Associated Viruses. Sci. Rep. 2018, 8, 3589. [Google Scholar] [CrossRef]
- Chandran, J.S.; Sharp, P.S.; Karyka, E.; Aves-Cruzeiro, J.M.D.C.; Coldicott, I.; Castelli, L.; Hautbergue, G.; Collins, M.O.; Azzouz, M. Site Specific Modification of Adeno-Associated Virus Enables Both Fluorescent Imaging of Viral Particles and Characterization of the Capsid Interactome. Sci. Rep. 2017, 7, 14766. [Google Scholar] [CrossRef]
- Stachler, M.D.; Chen, I.; Ting, A.Y.; Bartlett, J.S. Site-Specific Modification of AAV Vector Particles with Biophysical Probes and Targeting Ligands Using Biotin Ligase. Mol. Ther. 2008, 16, 1467–1473. [Google Scholar] [CrossRef] [PubMed]
- Hoque, M.; Shimizu, N.; Ishizu, K.; Yajima, H.; Arisaka, F.; Suzuki, K.; Watanabe, H.; Handa, H. Chimeric Virus-like Particle Formation of Adeno-Associated Virus. Biochem. Biophys. Res. Commun. 1999, 266, 371–376. [Google Scholar] [CrossRef]
- Terpe, K. Overview of Tag Protein Fusions: From Molecular and Biochemical Fundamentals to Commercial Systems. Appl. Microbiol. Biotechnol. 2003, 60, 523–533. [Google Scholar] [CrossRef]
- Li, H.; Zhang, F.L.; Shi, W.J.; Bai, X.J.; Jia, S.Q.; Zhang, C.G.; Ding, W.D. Immobilization of FLAG-Tagged Recombinant Adeno-Associated Virus 2 onto Tissue Engineering Scaffolds for the Improvement of Transgene Delivery in Cell Transplants. PLoS ONE 2015, 10, e0129013. [Google Scholar] [CrossRef] [PubMed]
- Earley, L.F.; Kawano, Y.; Adachi, K.; Sun, X.-X.; Dai, M.-S.; Nakai, H. Identification and Characterization of Nuclear and Nucleolar Localization Signals in the Adeno-Associated Virus Serotype 2 Assembly-Activating Protein. J. Virol. 2015, 89, 3038–3048. [Google Scholar] [CrossRef] [PubMed]
- Earley, L.F.; Powers, J.M.; Adachi, K.; Baumgart, J.T.; Meyer, N.L.; Xie, Q.; Chapman, M.S.; Nakai, H. Adeno-Associated Virus (AAV) Assembly-Activating Protein Is Not an Essential Requirement for Capsid Assembly of AAV Serotypes 4, 5, and 11. J. Virol. 2017, 91, e01980-16. [Google Scholar] [CrossRef]
- Wobus, C.E.; Hügle-Dörr, B.; Girod, A.; Petersen, G.; Hallek, M.; Kleinschmidt, J.A. Monoclonal Antibodies against the Adeno-Associated Virus Type 2 (AAV-2) Capsid: Epitope Mapping and Identification of Capsid Domains Involved in AAV-2–Cell Interaction and Neutralization of AAV-2 Infection. J. Virol. 2000, 74, 9281–9293. [Google Scholar] [CrossRef] [PubMed]
- Tseng, Y.-S.; Gurda, B.L.; Chipman, P.; McKenna, R.; Afione, S.; Chiorini, J.A.; Muzyczka, N.; Olson, N.H.; Baker, T.S.; Kleinschmidt, J.; et al. Adeno-Associated Virus Serotype 1 (AAV1)- and AAV5-Antibody Complex Structures Reveal Evolutionary Commonalities in Parvovirus Antigenic Reactivity. J. Virol. 2015, 89, 1794–1808. [Google Scholar] [CrossRef]
- Meyer, N.L.; Chapman, M.S. Adeno-Associated Virus (AAV) Cell Entry: Structural Insights. Trends Microbiol. 2022, 30, 432–451. [Google Scholar] [CrossRef]
- Mietzsch, M.; Pénzes, J.J.; Agbandje-McKenna, M. Twenty-Five Years of Structural Parvovirology. Viruses 2019, 11, 362. [Google Scholar] [CrossRef]
- Stagg, S.M.; Yoshioka, C.; Davulcu, O.; Chapman, M.S. Cryo-Electron Microscopy of Adeno-Associated Virus. Chem. Rev. 2022, 122, 14018–14054. [Google Scholar] [CrossRef]
- Gurda, B.L.; DiMattia, M.A.; Miller, E.B.; Bennett, A.; McKenna, R.; Weichert, W.S.; Nelson, C.D.; Chen, W.; Muzyczka, N.; Olson, N.H.; et al. Capsid Antibodies to Different Adeno-Associated Virus Serotypes Bind Common Regions. J. Virol. 2013, 87, 9111–9124. [Google Scholar] [CrossRef]
- McCraw, D.M.; O’Donnell, J.K.; Taylor, K.A.; Stagg, S.M.; Chapman, M.S. Structure of Adeno-Associated Virus-2 in Complex with Neutralizing Monoclonal Antibody A20. Virology 2012, 431, 40–49. [Google Scholar] [CrossRef]
- Jose, A.; Mietzsch, M.; Smith, J.K.; Kurian, J.; Chipman, P.; McKenna, R.; Chiorini, J.; Agbandje-McKenna, M. High-Resolution Structural Characterization of a New Adeno-Associated Virus Serotype 5 Antibody Epitope toward Engineering Antibody-Resistant Recombinant Gene Delivery Vectors. J. Virol. 2019, 93, e01394-18. [Google Scholar] [CrossRef] [PubMed]
- Bennett, A.D.; Wong, K.; Lewis, J.; Tseng, Y.; Smith, J.K.; Chipman, P.; McKenna, R.; Samulski, R.J.; Kleinschmidt, J.; Agbandje-McKenna, M. AAV6 K531 Serves a Dual Function in Selective Receptor and Antibody ADK6 Recognition. Virology 2018, 518, 369–376. [Google Scholar] [CrossRef]
- Gurda, B.L.; Raupp, C.; Popa-Wagner, R.; Naumer, M.; Olson, N.H.; Ng, R.; McKenna, R.; Baker, T.S.; Kleinschmidt, J.A.; Agbandje-McKenna, M. Mapping a Neutralizing Epitope onto the Capsid of Adeno-Associated Virus Serotype 8. J. Virol. 2012, 86, 7739–7751. [Google Scholar] [CrossRef] [PubMed]
- Giles, A.R.; Govindasamy, L.; Somanathan, S.; Wilson, J.M. Mapping an Adeno-Associated Virus 9-Specific Neutralizing Epitope To Develop Next-Generation Gene Delivery Vectors. J. Virol. 2018, 92, e01011-18. [Google Scholar] [CrossRef] [PubMed]
- Wistuba, A.; Weger, S.; Kern, A.; Kleinschmidt, J.A. Intermediates of Adeno-Associated Virus Type 2 Assembly: Identification of Soluble Complexes Containing Rep and Cap Proteins. J. Virol. 1995, 69, 5311–5319. [Google Scholar] [CrossRef] [PubMed]
- Kuck, D.; Kern, A.; Kleinschmidt, J.A. Development of AAV Serotype-Specific ELISAs Using Novel Monoclonal Antibodies. J. Virol. Methods 2006, 140, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Brown, K.E. A 110-KDa Nuclear Shuttle Protein, Nucleolin, Specifically Binds to Adeno-Associated Virus Type 2 (AAV-2) Capsid. Virology 1999, 257, 373–382. [Google Scholar] [CrossRef]
- Johnson, J.S.; Samulski, R.J. Enhancement of Adeno-Associated Virus Infection by Mobilizing Capsids into and out of the Nucleolus. J. Virol. 2009, 83, 2632–2644. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.S.; Li, C.; DiPrimio, N.; Weinberg, M.S.; McCown, T.J.; Samulski, R.J. Mutagenesis of Adeno-Associated Virus Type 2 Capsid Protein VP1 Uncovers New Roles for Basic Amino Acids in Trafficking and Cell-Specific Transduction. J. Virol. 2010, 84, 8888–8902. [Google Scholar] [CrossRef]
- Rossi, A.; Dupaty, L.; Aillot, L.; Zhang, L.; Gallien, C.; Hallek, M.; Odenthal, M.; Adriouch, S.; Salvetti, A.; Büning, H. Vector Uncoating Limits Adeno-Associated Viral Vector-Mediated Transduction of Human Dendritic Cells and Vector Immunogenicity. Sci. Rep. 2019, 9, 3631. [Google Scholar] [CrossRef]
- Berry, G.E.; Asokan, A. Chemical Modulation of Endocytic Sorting Augments Adeno-Associated Viral Transduction. J. Biol. Chem. 2016, 291, 939–947. [Google Scholar] [CrossRef] [PubMed]
- Madigan, V.J.; Yuziuk, J.A.; Chiarella, A.M.; Tyson, T.O.; Meganck, R.M.; Elmore, Z.C.; Tse, L.V.; Hathaway, N.A.; Asokan, A. Ring Finger Protein 121 Is a Potent Regulator of Adeno-Associated Viral Genome Transcription. PLoS Pathog. 2019, 15, e1007988. [Google Scholar] [CrossRef]
- Madigan, V.J.; Berry, G.E.; Tyson, T.O.; Nardone-White, D.; Ark, J.; Elmore, Z.C.; Murlidharan, G.; Vincent, H.A.; Asokan, A. The Golgi Calcium ATPase Pump Plays an Essential Role in Adeno-Associated Virus Trafficking and Transduction. J. Virol. 2020, 94, e01604-20. [Google Scholar] [CrossRef] [PubMed]
- Le, D.T.; Radukic, M.T.; Müller, K.M. Adeno-Associated Virus Capsid Protein Expression in Escherichia Coli and Chemically Defined Capsid Assembly. Sci. Rep. 2019, 9, 18631. [Google Scholar] [CrossRef] [PubMed]
- Le, D.T.; Radukic, M.T.; Teschner, K.; Becker, L.; Müller, K.M. Synthesis and Concomitant Assembly of Adeno-Associated Virus-like Particles in Escherichia Coli. ACS Synth. Biol. 2022, 11, 3601–3607. [Google Scholar] [CrossRef]
- Hunter, L.A.; Samulski, R.J. Colocalization of Adeno-Associated Virus Rep and Capsid Proteins in the Nuclei of Infected Cells. J. Virol. 1992, 66, 317–324. [Google Scholar] [CrossRef]
- Stracker, T.H.; Cassell, G.D.; Ward, P.; Loo, Y.-M.; van Breukelen, B.; Carrington-Lawrence, S.D.; Hamatake, R.K.; van der Vliet, P.C.; Weller, S.K.; Melendy, T.; et al. The Rep Protein of Adeno-Associated Virus Type 2 Interacts with Single-Stranded DNA-Binding Proteins That Enhance Viral Replication. J. Virol. 2004, 78, 441–453. [Google Scholar] [CrossRef]
- Slanina, H.; Weger, S.; Stow, N.D.; Kuhrs, A.; Heilbronn, R. Role of the Herpes Simplex Virus Helicase-Primase Complex during Adeno-Associated Virus DNA Replication. J. Virol. 2006, 80, 5241–5250. [Google Scholar] [CrossRef]
- Heilbronn, R.; Engstler, M.; Weger, S.; Krahn, A.; Schetter, C.; Boshart, M. SsDNA-Dependent Colocalization of Adeno-Associated Virus Rep and Herpes Simplex Virus ICP8 in Nuclear Replication Domains. Nucleic Acids Res. 2003, 31, 6206–6213. [Google Scholar] [CrossRef]
- Bevington, J.M.; Needham, P.G.; Verrill, K.C.; Collaco, R.F.; Basrur, V.; Trempe, J.P. Adeno-Associated Virus Interactions with B23/Nucleophosmin: Identification of Sub-Nucleolar Virion Regions. Virology 2007, 357, 102–113. [Google Scholar] [CrossRef]
- Tsien, R.Y. The Green Fluorescent Protein. Annu. Rev. Biochem. 1998, 67, 509–544. [Google Scholar] [CrossRef]
- Rodriguez, E.A.; Campbell, R.E.; Lin, J.Y.; Lin, M.Z.; Miyawaki, A.; Palmer, A.E.; Shu, X.; Zhang, J.; Tsien, R.Y. The Growing and Glowing Toolbox of Fluorescent and Photoactive Proteins. Trends Biochem. Sci. 2017, 42, 111–129. [Google Scholar] [CrossRef]
- Lambert, T.J. FPbase: A Community-Editable Fluorescent Protein Database. Nat. Methods 2019, 16, 277–278. [Google Scholar] [CrossRef] [PubMed]
- Fraefel, C.; Bittermann, A.G.; Büeler, H.; Heid, I.; Bächi, T.; Ackermann, M. Spatial and Temporal Organization of Adeno-Associated Virus DNA Replication in Live Cells. J. Virol. 2004, 78, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Glauser, D.L.; Saydam, O.; Balsiger, N.A.; Heid, I.; Linden, R.M.; Ackermann, M.; Fraefel, C. Four-Dimensional Visualization of the Simultaneous Activity of Alternative Adeno-Associated Virus Replication Origins. J. Virol. 2005, 79, 12218–12230. [Google Scholar] [CrossRef] [PubMed]
- Lux, K.; Goerlitz, N.; Schlemminger, S.; Perabo, L.; Goldnau, D.; Endell, J.; Leike, K.; Kofler, D.M.; Finke, S.; Hallek, M. Green Fluorescent Protein-Tagged Adeno-Associated Virus Particles Allow the Study of Cytosolic and Nuclear Trafficking. J. Virol. 2005, 79, 11776–11787. [Google Scholar] [CrossRef] [PubMed]
- Judd, J.; Wei, F.; Nguyen, P.Q.; Tartaglia, L.J.; Agbandje-McKenna, M.; Silberg, J.J.; Suh, J. Random Insertion of MCherry Into VP3 Domain of Adeno-Associated Virus Yields Fluorescent Capsids With No Loss of Infectivity. Mol. Ther. Nucleic Acids 2012, 1, e54. [Google Scholar] [CrossRef]
- Weitzman, M.D.; Fisher, K.J.; Wilson, J.M. Recruitment of Wild-Type and Recombinant Adeno-Associated Virus into Adenovirus Replication Centers. J. Virol. 1996, 70, 1845–1854. [Google Scholar] [CrossRef] [PubMed]
- Cervelli, T.; Palacios, J.A.; Zentilin, L.; Mano, M.; Schwartz, R.A.; Weitzman, M.D.; Giacca, M. Processing of Recombinant AAV Genomes Occurs in Specific Nuclear Structures That Overlap with Foci of DNA-Damage-Response Proteins. J. Cell Sci. 2008, 121, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, R.A.; Palacios, J.A.; Cassell, G.D.; Adam, S.; Giacca, M.; Weitzman, M.D. The Mre11/Rad50/Nbs1 Complex Limits Adeno-Associated Virus Transduction and Replication. J. Virol. 2007, 81, 12936–12945. [Google Scholar] [CrossRef] [PubMed]
- Dewar, R.L.; Highbarger, H.C.; Sarmiento, M.D.; Todd, J.A.; Vasudevachari, M.B.; Davey, R.T.; Kovacs, J.A.; Salzman, N.P.; Lane, H.C.; Urdea, M.S. Application of Branched DNA Signal Amplification to Monitor Human Immunodeficiency Virus Type 1 Burden in Human Plasma. J. Infect. Dis. 1994, 170, 1172–1179. [Google Scholar] [CrossRef]
- Kishi, J.Y.; Lapan, S.W.; Beliveau, B.J.; West, E.R.; Zhu, A.; Sasaki, H.M.; Saka, S.K.; Wang, Y.; Cepko, C.L.; Yin, P. SABER Amplifies FISH: Enhanced Multiplexed Imaging of RNA and DNA in Cells and Tissues. Nat. Methods 2019, 16, 533–544. [Google Scholar] [CrossRef]
- Wang, S.K.; Lapan, S.W.; Hong, C.M.; Krause, T.B.; Cepko, C.L. In Situ Detection of Adeno-Associated Viral Vector Genomes with SABER-FISH. Mol. Ther.-Methods Clin. Dev. 2020, 19, 376–386. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Flanagan, J.; Su, N.; Wang, L.C.; Bui, S.; Nielson, A.; Wu, X.; Vo, H.T.; Ma, X.J.; Luo, Y. RNAscope: A Novel in Situ RNA Analysis Platform for Formalin-Fixed, Paraffin-Embedded Tissues. J. Mol. Diagn. 2012, 14, 22–29. [Google Scholar] [CrossRef]
- Zhao, J.; Yue, Y.; Patel, A.; Wasala, L.; Karp, J.F.; Zhang, K.; Duan, D.; Lai, Y. High-Resolution Histological Landscape of AAV DNA Distribution in Cellular Compartments and Tissues Following Local and Systemic Injection. Mol. Ther.-Methods Clin. Dev. 2020, 18, 856–868. [Google Scholar] [CrossRef]
- Sutter, S.O.; Lkharrazi, A.; Schraner, E.M.; Michaelsen, K.; Meier, A.F.; Marx, J.; Vogt, B.; Büning, H.; Fraefel, C. Adeno-Associated Virus Type 2 (AAV2) Uncoating Is a Stepwise Process and Is Linked to Structural Reorganization of the Nucleolus. PLoS Pathog. 2022, 18, e1010187. [Google Scholar] [CrossRef]
- Wang, X.; Allen, W.E.; Wright, M.A.; Sylwestrak, E.L.; Samusik, N.; Vesuna, S.; Evans, K.; Liu, C.; Ramakrishnan, C.; Liu, J.; et al. Three-Dimensional Intact-Tissue Sequencing of Single-Cell Transcriptional States. Science 2018, 361, eaat5691. [Google Scholar] [CrossRef]
- Banér, J.; Nilsson, M.; Mendel-Hartvig, M.; Landegren, U. Signal Amplification of Padlock Probes by Rolling Circle Replication. Nucleic Acids Res. 1998, 26, 5073–5078. [Google Scholar] [CrossRef]
- Kishi, J.Y.; Schaus, T.E.; Gopalkrishnan, N.; Xuan, F.; Yin, P. Programmable Autonomous Synthesis of Single-Stranded DNA. Nat. Chem. 2018, 10, 155–164. [Google Scholar] [CrossRef]
- Jang, M.J.; Coughlin, G.M.; Jackson, C.R.; Chen, X.; Chuapoco, M.R.; Vendemiatti, J.L.; Wang, A.Z.; Gradinaru, V. Spatial Transcriptomics for Profiling the Tropism of Viral Vectors in Tissues. Nat. Biotechnol. 2023. [Google Scholar] [CrossRef] [PubMed]
- Schermelleh, L.; Ferrand, A.; Huser, T.; Eggeling, C.; Sauer, M.; Biehlmaier, O.; Drummen, G.P.C. Super-Resolution Microscopy Demystified. Nat. Cell Biol. 2019, 21, 72–84. [Google Scholar] [CrossRef]
- Combs, C.A. Shroff H. Fluorescence Microscopy: A Concise Guide to Current Imaging Methods. Curr. Protoc. Neeurosci. 2017, 79, 2.1.1–2.1.25. [Google Scholar] [CrossRef]
- Pawley, J. Handbook of Biological Confocal Microscopy, 3rd ed.; Springer: New York, NY, USA, 2006; ISBN 978-0-387-45524-2. [Google Scholar]
- Lippincott-Schwartz, J.; Snapp, E.L.; Phair, R.D. The Development and Enhancement of FRAP as a Key Tool for Investigating Protein Dynamics. Biophys. J. 2018, 115, 1146–1155. [Google Scholar] [CrossRef]
- Ihalainen, T.O.; Niskanen, E.A.; Jylhävä, J.; Paloheimo, O.; Dross, N.; Smolander, H.; Langowski, J.; Timonen, J.; Vihinen-Ranta, M. Parvovirus Induced Alterations in Nuclear Architecture and Dynamics. PLoS ONE 2009, 4, e5948. [Google Scholar] [CrossRef]
- Ihalainen, T.O.; Willman, S.F.; Niskanen, E.A.; Paloheimo, O.; Smolander, H.; Laurila, J.P.; Kaikkonen, M.U.; Vihinen-Ranta, M. Distribution and Dynamics of Transcription-Associated Proteins during Parvovirus Infection. J. Virol. 2012, 86, 13779–13784. [Google Scholar] [CrossRef] [PubMed]
- Reilly, W.M.; Obara, C.J. Advances in Confocal Microscopy and Selected Applications. In Confocal Microscopy: Methods in Molecular Biology; Brzostowski, J., Sohn, H., Eds.; Humana: New York, NY, USA, 2021; Volume 2304. [Google Scholar] [CrossRef]
- Mäntylä, E.; Chacko, J.V.; Aho, V.; Parrish, C.R.; Shahin, V.; Kann, M.; Digman, M.A.; Gratton, E.; Vihinen-Ranta, M. Viral Highway to Nucleus Exposed by Image Correlation Analyses. Sci. Rep. 2018, 8, 1152. [Google Scholar] [CrossRef]
- Stout, A.L.; Axelrod, D. Evanescent Field Excitation of Fluorescence by Epi-Illumination Microscopy. Appl. Opt. 1989, 28, 5237. [Google Scholar] [CrossRef] [PubMed]
- Tokunaga, M.; Imamoto, N.; Sakata-Sogawa, K. Highly Inclined Thin Illumination Enables Clear Single-Molecule Imaging in Cells. Nat. Methods 2008, 5, 159–161. [Google Scholar] [CrossRef]
- Sahl, S.J.; Hell, S.W. High Resolution Imaging in Microscopy and Ophthalmology; Bille, J.F., Ed.; Springer International Publishing: Cham, Switzerland, 2019; ISBN 978-3-030-16637-3. [Google Scholar]
- Gustafsson, N.; Culley, S.; Ashdown, G.; Owen, D.M.; Pereira, P.M.; Henriques, R. Fast Live-Cell Conventional Fluorophore Nanoscopy with ImageJ through Super-Resolution Radial Fluctuations. Nat. Commun. 2016, 7, 12471. [Google Scholar] [CrossRef]
- Dertinger, T.; Colyera, R.; Iyer, G.; Weiss, S.; Enderlein, J. Fast, Background-Free, 3D Super-Resolution Optical Fluctuation Imaging (SOFI). Proc. Natl. Acad. Sci. USA 2009, 106, 22287–22292. [Google Scholar] [CrossRef] [PubMed]
- Witte, R.; Andriasyan, V.; Georgi, F.; Yakimovich, A.; Greber, U. Concepts in Light Microscopy of Viruses. Viruses 2018, 10, 202. [Google Scholar] [CrossRef]
- Robb, N.C. Virus Morphology: Insights from Super-Resolution Fluorescence Microscopy. Biochim. Biophys. Acta-Mol. Basis Dis. 2022, 1868, 166347. [Google Scholar] [CrossRef] [PubMed]
- Jouvenet, N.; Bieniasz, P.D.; Simon, S.M. Imaging the Biogenesis of Individual HIV-1 Virions in Live Cells. Nature 2008, 454, 236–240. [Google Scholar] [CrossRef]
- Hübner, W.; Chen, P.; Del Portillo, A.; Liu, Y.; Gordon, R.E.; Chen, B.K. Sequence of Human Immunodeficiency Virus Type 1 (HIV-1) Gag Localization and Oligomerization Monitored with Live Confocal Imaging of a Replication-Competent, Fluorescently Tagged HIV-1. J. Virol. 2007, 81, 12596–12607. [Google Scholar] [CrossRef] [PubMed]
- Nakane, S.; Iwamoto, A.; Matsuda, Z. The V4 and V5 Variable Loops of HIV-1 Envelope Glycoprotein Are Tolerant to Insertion of Green Fluorescent Protein and Are Useful Targets for Labeling. J. Biol. Chem. 2015, 290, 15279–15291. [Google Scholar] [CrossRef]
- Wang, L.; Sandmeyer, A.; Hübner, W.; Li, H.; Huser, T.; Chen, B.K. A Replication-Competent HIV Clone Carrying GFP-Env Reveals Rapid Env Recycling at the HIV-1 T Cell Virological Synapse. Viruses 2021, 14, 38. [Google Scholar] [CrossRef]
- Hübner, W.; McNerney, G.P.; Chen, P.; Dale, B.M.; Gordon, R.E.; Chuang, F.Y.S.; Li, X.; Asmuth, D.M.; Huser, T.; Chen, B.K. Quantitative 3D Video Microscopy of HIV Transfer across T Cell Virological Synapses. Science 2009, 323, 1743–1747. [Google Scholar] [CrossRef]
- Nikolaitchik, O.A.; Dilley, K.A.; Fu, W.; Gorelick, R.J.; Tai, S.-H.S.; Soheilian, F.; Ptak, R.G.; Nagashima, K.; Pathak, V.K.; Hu, W.-S. Dimeric RNA Recognition Regulates HIV-1 Genome Packaging. PLoS Pathog. 2013, 9, e1003249. [Google Scholar] [CrossRef] [PubMed]
- Sandmeyer, A.; Wang, L.; Hübner, W.; Müller, M.; Chen, B.K.; Huser, T. Cost-Effective High-Speed, Three-Dimensional Live-Cell Imaging of HIV-1 Transfer at the T Cell Virological Synapse. iScience 2022, 25, 105468. [Google Scholar] [CrossRef]
- Laine, R.F.; Goodfellow, G.; Young, L.J.; Travers, J.; Carroll, D.; Dibben, O.; Bright, H.; Kaminski, C.F. Structured Illumination Microscopy Combined with Machine Learning Enables the High Throughput Analysis and Classification of Virus Structure. Elife 2018, 13, e40183. [Google Scholar] [CrossRef] [PubMed]
- Horsington, J.; Turnbull, L.; Whitchurch, C.B.; Newsome, T.P. Sub-Viral Imaging of Vaccinia Virus Using Super-Resolution Microscopy. J. Virol. Methods 2012, 186, 132–136. [Google Scholar] [CrossRef] [PubMed]
- Chojnacki, J.; Staudt, T.; Glass, B.; Bingen, P.; Engelhardt, J.; Anders, M.; Schneider, J.; Müller, B.; Hell, S.W.; Kräusslich, H.-G. Maturation-Dependent HIV-1 Surface Protein Redistribution Revealed by Fluorescence Nanoscopy. Science 2012, 338, 524–528. [Google Scholar] [CrossRef]
- Muranyi, W.; Malkusch, S.; Müller, B.; Heilemann, M.; Kräusslich, H.-G. Super-Resolution Microscopy Reveals Specific Recruitment of HIV-1 Envelope Proteins to Viral Assembly Sites Dependent on the Envelope C-Terminal Tail. PLoS Pathog. 2013, 9, e1003198. [Google Scholar] [CrossRef] [PubMed]
- Laine, R.F.; Albecka, A.; van de Linde, S.; Rees, E.J.; Crump, C.M.; Kaminski, C.F. Structural Analysis of Herpes Simplex Virus by Optical Super-Resolution Imaging. Nat. Commun. 2015, 6, 5980. [Google Scholar] [CrossRef]
- Chojnacki, J.; Waithe, D.; Carravilla, P.; Huarte, N.; Galiani, S.; Enderlein, J.; Eggeling, C. Envelope Glycoprotein Mobility on HIV-1 Particles Depends on the Virus Maturation State. Nat. Commun. 2017, 8, 545. [Google Scholar] [CrossRef] [PubMed]
- Sakin, V.; Paci, G.; Lemke, E.A.; Müller, B. Labeling of Virus Components for Advanced, Quantitative Imaging Analyses. FEBS Lett. 2016, 590, 1896–1914. [Google Scholar] [CrossRef] [PubMed]
- Sakin, V.; Hanne, J.; Dunder, J.; Anders-Össwein, M.; Laketa, V.; Nikić, I.; Kräusslich, H.-G.; Lemke, E.A.; Müller, B. A Versatile Tool for Live-Cell Imaging and Super-Resolution Nanoscopy Studies of HIV-1 Env Distribution and Mobility. Cell Chem. Biol. 2017, 24, 635–645.e5. [Google Scholar] [CrossRef]
- Zou, W.; Wang, Z.; Xiong, M.; Chen, A.Y.; Xu, P.; Ganaie, S.S.; Badawi, Y.; Kleiboeker, S.; Nishimune, H.; Ye, S.Q.; et al. Human Parvovirus B19 Utilizes Cellular DNA Replication Machinery for Viral DNA Replication. J. Virol. 2018, 92, e01881-17. [Google Scholar] [CrossRef] [PubMed]
- Castelletto, S.; Boretti, A. Viral Particle Imaging by Super-Resolution Fluorescence Microscopy. Chem. Phys. Impact 2021, 2, 100013. [Google Scholar] [CrossRef]
- Arista-Romero, M.; Pujals, S.; Albertazzi, L. Towards a Quantitative Single Particle Characterization by Super Resolution Microscopy: From Virus Structures to Antivirals Design. Front. Bioeng. Biotechnol. 2021, 9, 647874. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Eng, E.T.; Law, K.; Gordon, R.E.; Rice, W.J.; Chen, B.K. Visualization of HIV T Cell Virological Synapses and Virus-Containing Compartments by Three-Dimensional Correlative Light and Electron Microscopy. J. Virol. 2017, 91, e01605-16. [Google Scholar] [CrossRef] [PubMed]
- Xiao, P.-J.; Li, C.; Neumann, A.; Samulski, R.J. Quantitative 3D Tracing of Gene-Delivery Viral Vectors in Human Cells and Animal Tissues. Mol. Ther. 2012, 20, 317–328. [Google Scholar] [CrossRef]
- Ma, J.; Yang, W. Three-Dimensional Distribution of Transient Interactions in the Nuclear Pore Complex Obtained from Single-Molecule Snapshots. Proc. Natl. Acad. Sci. USA 2010, 107, 7305–7310. [Google Scholar] [CrossRef]
- Junod, S.L.; Saredy, J.; Yang, W. Nuclear Import of Adeno-Associated Viruses Imaged by High-Speed Single-Molecule Microscopy. Viruses 2021, 13, 167. [Google Scholar] [CrossRef]
- Kelich, J.M.; Ma, J.; Dong, B.; Wang, Q.; Chin, M.; Magura, C.M.; Xiao, W.; Yang, W. Super-Resolution Imaging of Nuclear Import of Adeno-Associated Virus in Live Cells. Mol. Ther.-Methods Clin. Dev. 2015, 2, 15047. [Google Scholar] [CrossRef]
- Wegner, W.; Steffens, H.; Gregor, C.; Wolf, F.; Willig, K.I. Environmental Enrichment Enhances Patterning and Remodeling of Synaptic Nanoarchitecture as Revealed by STED Nanoscopy. Elife 2022, 11, e73603. [Google Scholar] [CrossRef] [PubMed]
- Willig, K.I.; Steffens, H.; Gregor, C.; Herholt, A.; Rossner, M.J.; Hell, S.W. Nanoscopy of Filamentous Actin in Cortical Dendrites of a Living Mouse. Biophys. J. 2014, 106, L01–L03. [Google Scholar] [CrossRef] [PubMed]
- Cole, D.; Young, G.; Weigel, A.; Sebesta, A.; Kukura, P. Label-Free Single-Molecule Imaging with Numerical-Aperture-Shaped Interferometric Scattering Microscopy. ACS Photonics 2017, 4, 211–216. [Google Scholar] [CrossRef]
- Wu, D.; Hwang, P.; Li, T.; Piszczek, G. Rapid Characterization of Adeno-Associated Virus (AAV) Gene Therapy Vectors by Mass Photometry. Gene Ther. 2022, 29, 691–697. [Google Scholar] [CrossRef]
- Balzarotti, F.; Eilers, Y.; Gwosch, K.C.; Gynnå, A.H.; Westphal, V.; Stefani, F.D.; Elf, J.; Hell, S.W. Nanometer Resolution Imaging and Tracking of Fluorescent Molecules with Minimal Photon Fluxes. Science 2017, 355, 606–612. [Google Scholar] [CrossRef] [PubMed]
- Deguchi, T.; Iwanski, M.K.; Schentarra, E.-M.; Heidebrecht, C.; Schmidt, L.; Heck, J.; Weihs, T.; Schnorrenberg, S.; Hoess, P.; Liu, S.; et al. Direct Observation of Motor Protein Stepping in Living Cells Using MINFLUX. Science 2023, 379, 1010–1015. [Google Scholar] [CrossRef] [PubMed]
- Weber, M.; von der Emde, H.; Leutenegger, M.; Gunkel, P.; Sambandan, S.; Khan, T.A.; Keller-Findeisen, J.; Cordes, V.C.; Hell, S.W. MINSTED Nanoscopy Enters the Ångström Localization Range. Nat. Biotechnol. 2023, 41, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Lei, Y.; Ma, Y.; Liu, M.; Zheng, J.; Dan, D.; Gao, P. A Comprehensive Review of Fluorescence Correlation Spectroscopy. Front. Phys. 2021, 9, 1–21. [Google Scholar] [CrossRef]
- Chojnacki, J.; Eggeling, C. Super-Resolution STED Microscopy-Based Mobility Studies of the Viral Env Protein at HIV-1 Assembly Sites of Fully Infected T-Cells. Viruses 2021, 13, 608. [Google Scholar] [CrossRef]
- Müller, T.G.; Zila, V.; Peters, K.; Schifferdecker, S.; Stanic, M.; Lucic, B.; Laketa, V.; Lusic, M.; Müller, B.; Kräusslich, H.G. HIV-1 Uncoating by Release of Viral cDNA from Capsid-like Structures in the Nucleus of Infected Cells. Elife 2021, 10, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Large, E.E.; Silveria, M.A.; Zane, G.M.; Weerakoon, O.; Chapman, M.S. Adeno-Associated Virus (AAV) Gene Delivery: Dissecting Molecular Interactions upon Cell Entry. Viruses 2021, 13, 1336. [Google Scholar] [CrossRef] [PubMed]
- Wassie, A.T.; Zhao, Y.; Boyden, E.S. Expansion Microscopy: Principles and Uses in Biological Research. Nat. Methods 2019, 16, 33–41. [Google Scholar] [CrossRef]
- Truckenbrodt, S. Expansion Microscopy: Super-Resolution Imaging with Hydrogels. Anal. Chem. 2023, 95, 3–32. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Yu, C.C.; Gao, L.; Piatkevich, K.D.; Neve, R.L.; Munro, J.B.; Upadhyayula, S.; Boyden, E.S. A Highly Homogeneous Polymer Composed of Tetrahedron-like Monomers for High-Isotropy Expansion Microscopy. Nat. Nanotechnol. 2021, 16, 698–707. [Google Scholar] [CrossRef]
- Shaib, A.H.; Chouaib, A.A.; Imani, V.; Chowdhury, R.; Georgiev, S.V.; Mougios, N.; Monga, M.; Reshetniak, S.; Mihaylov, D.; Chen, H.; et al. Expansion Microscopy at One Nanometer Resolution. bioRxiv 2022, 17, 17. [Google Scholar] [CrossRef]
- Karsten, L.; Goett-Zink, L.; Schmitz, J.; Hoffrogge, R.; Grünberger, A.; Kottke, T.; Müller, K.M. Genetically Encoded Ratiometric PH Sensors for the Measurement of Intra- and Extracellular PH and Internalization Rates. Biosensors 2022, 12, 271. [Google Scholar] [CrossRef] [PubMed]
| Virus and Protein | Antibody Name | Binding Region | Amino Acids (VP1 Numbering) | Reference |
|---|---|---|---|---|
| AAV1 | 4E4 | protrusions across 2-fold axis | 456–459, 492–498 | [74] |
| AAV1 | ADK1a | 3-fold protrusions | 448, 450, 453–457, 500 | [70] |
| AAV1 | ADK1b | 2/5-fold wall; side of 3-fold | 256, 258, 259, 261, 263–266, 272, 385, 386, 547, 709, 710, 716–718, 720, 722 | [70] |
| AAV1, AAV6 | 5H7 | center of 3-fold symmetry axis | 494, 496–499, 582, 583, 588–591, 593–595, 597 | [74] |
| AAV1, AAV3, AAV5 | D3 | highly exposed on VP3 surface | 474–483 | [69] |
| AAV1,2,3,5,6,7,8,9, rh10, DJ VP1, VP2, VP3 | B1 | buried C-terminal aa | 726–734 | [69] |
| AAV2,3 | A20 | 2/5- fold wall and canyon | 253, 254, 258, 261, 262, 264, 384, 385, 548, 556, 658–660, 708, 717 | [75] |
| AAV2 | C37-B | 3-fold protrusions | 492–498, 585–589 | [74] |
| AAV2; VP1 | A1 | N-terminal VP1 domain | 122–131 | [69] |
| AAV2; VP1, VP2 | A69 | N-terminal VP2 domain | 171–182 | [69] |
| AAV5 | 3C5 | 2/5-fold wall | siteA: 254–261, 374, 375, 483, 485–492, 494, 496, 499, 500, 501 siteB: 246, 530, 532–538, 653, 654, 656, 657, 704–708 | [74] |
| AAV5 | ADK5a | 2/5- fold wall | 244, 246, 248–256, 263, 377, 378, 453, 456, 532, 533, 535–543, 546, 653, 654, 656, 697, 698, 704–710 | [70] |
| AAV5 | ADK5b | 2/5- fold wall to 5-fold symmetry axis | 248, 316–319, 443, 530–535, 540–543, 545, 546, 697, 704, 706, 708–710 | [70] |
| AAV5 | HL2476 | 3-fold protrusions | 481, 483, 484, 576 | [76] |
| AAV6 | ADK6 | 3-fold protrusions and 2/5-fold wall | K531 selectivity | [77] |
| AAV8 | ADK8 | 3-fold protrusions | 586–591 | [78] |
| AAV9 | PAV9.1 | center of 3-fold symmetry axis | 496–498, 588–592 | [79] |
| MAAP of AAV2 | Anti-MAAP GAL-KKI | polyclonal | 79–98 of MAAP | [11] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Golm, S.K.; Hübner, W.; Müller, K.M. Fluorescence Microscopy in Adeno-Associated Virus Research. Viruses 2023, 15, 1174. https://doi.org/10.3390/v15051174
Golm SK, Hübner W, Müller KM. Fluorescence Microscopy in Adeno-Associated Virus Research. Viruses. 2023; 15(5):1174. https://doi.org/10.3390/v15051174
Chicago/Turabian StyleGolm, Susanne K., Wolfgang Hübner, and Kristian M. Müller. 2023. "Fluorescence Microscopy in Adeno-Associated Virus Research" Viruses 15, no. 5: 1174. https://doi.org/10.3390/v15051174
APA StyleGolm, S. K., Hübner, W., & Müller, K. M. (2023). Fluorescence Microscopy in Adeno-Associated Virus Research. Viruses, 15(5), 1174. https://doi.org/10.3390/v15051174






