Apical-Out Human Airway Organoids Modeling SARS-CoV-2 Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. Adult-Stem-Cell-Derived Human Lung Organoids
2.1.1. Establishment and Maintenance of Lung Organoids
2.1.2. Proximal Differentiation and Polarity Reversal of Airway Organoids
2.2. Viruses
2.2.1. Preparation of Virus Stock
2.2.2. Infection of Human Airway Organoids
2.3. Antibody Treatment
2.4. Dextran Permeability Test
2.5. RNA Extraction, RT and qPCR
2.6. Immunofluorescence Staining
2.7. Flow Cytometry Analysis
2.8. Statistical Analysis
3. Results
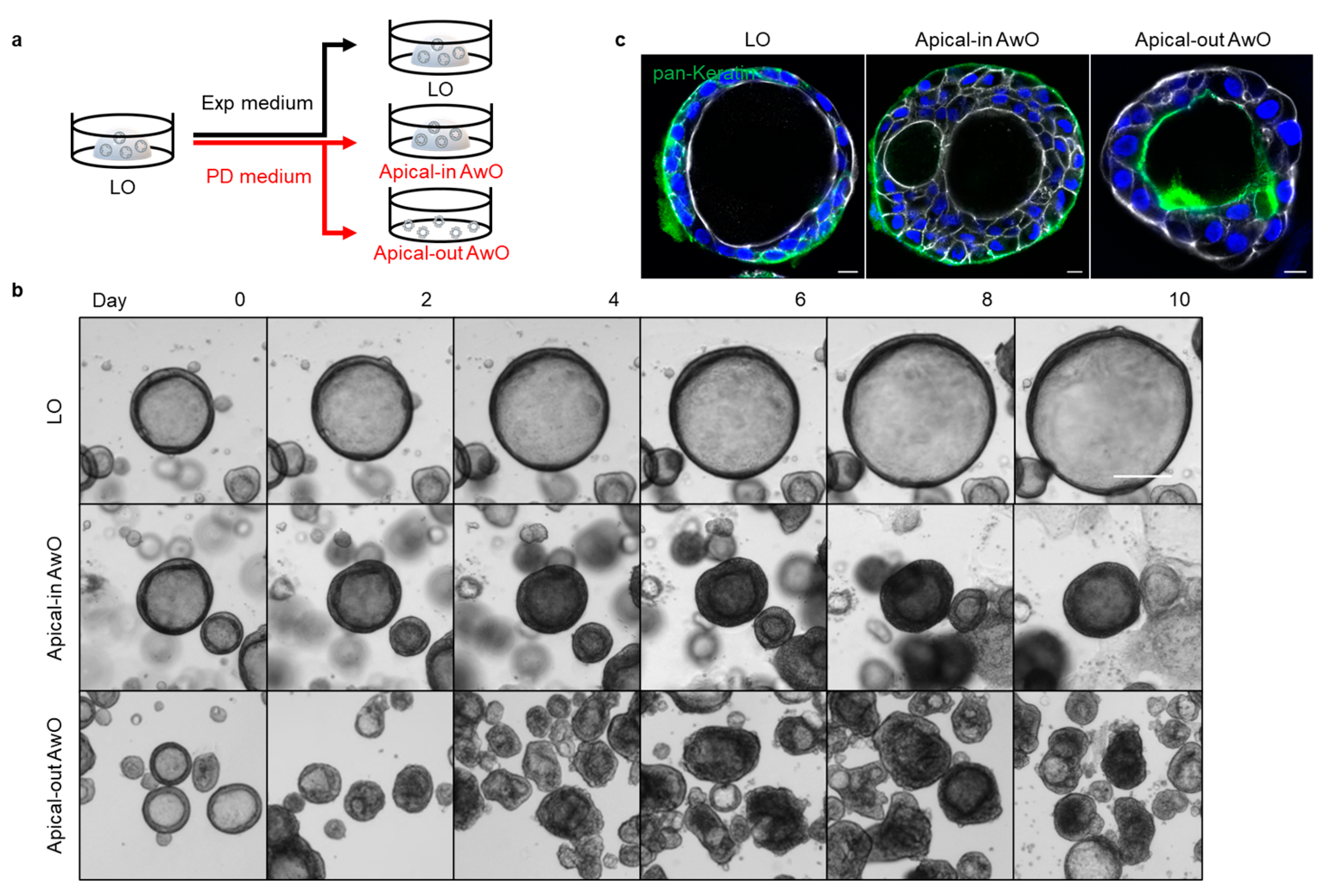
3.1. Establishment of Apical-Out Human Airway Organoid Model
3.1.1. Generation of Apical-Out Airway Organoids
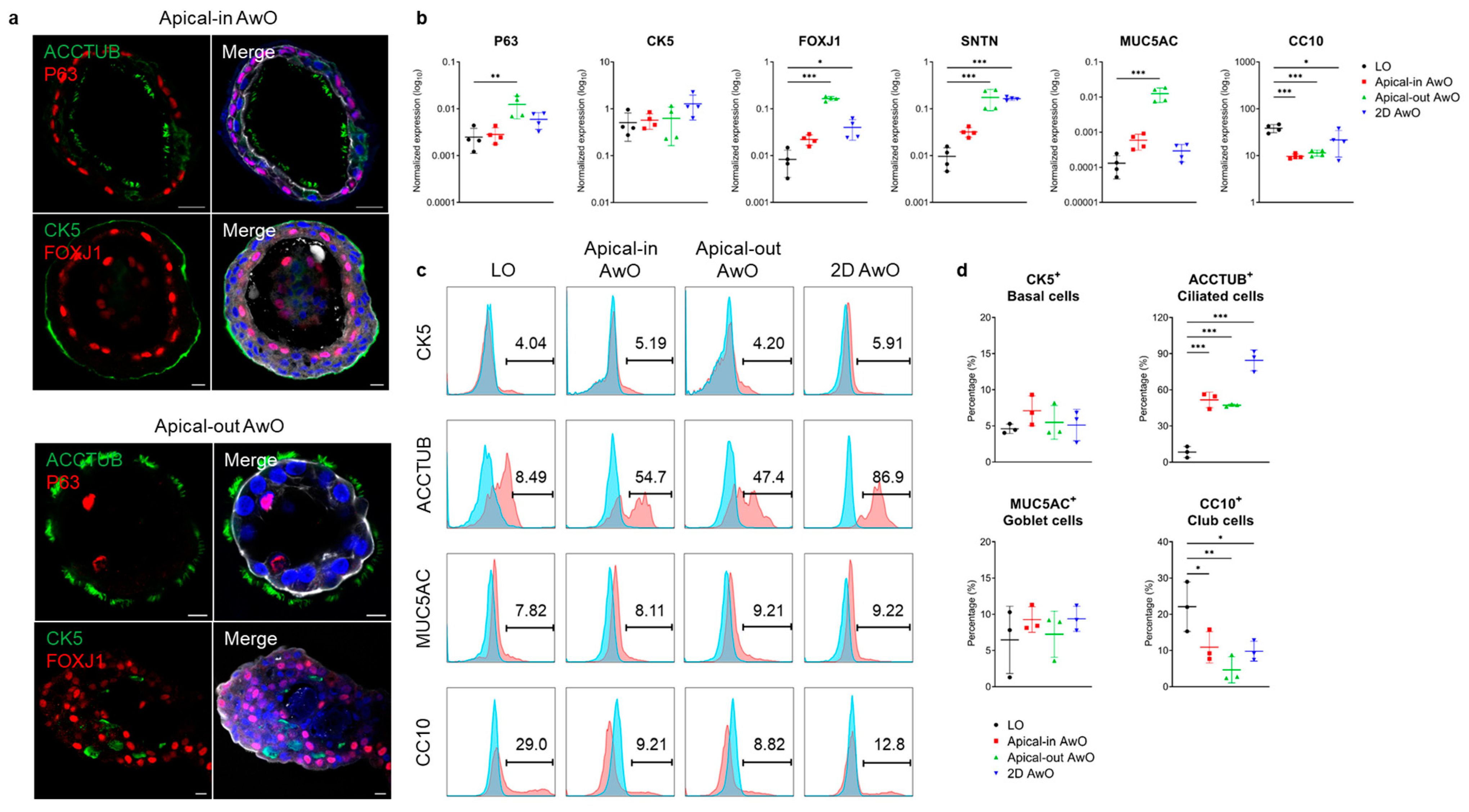
3.1.2. Characterization of Airway Organoids
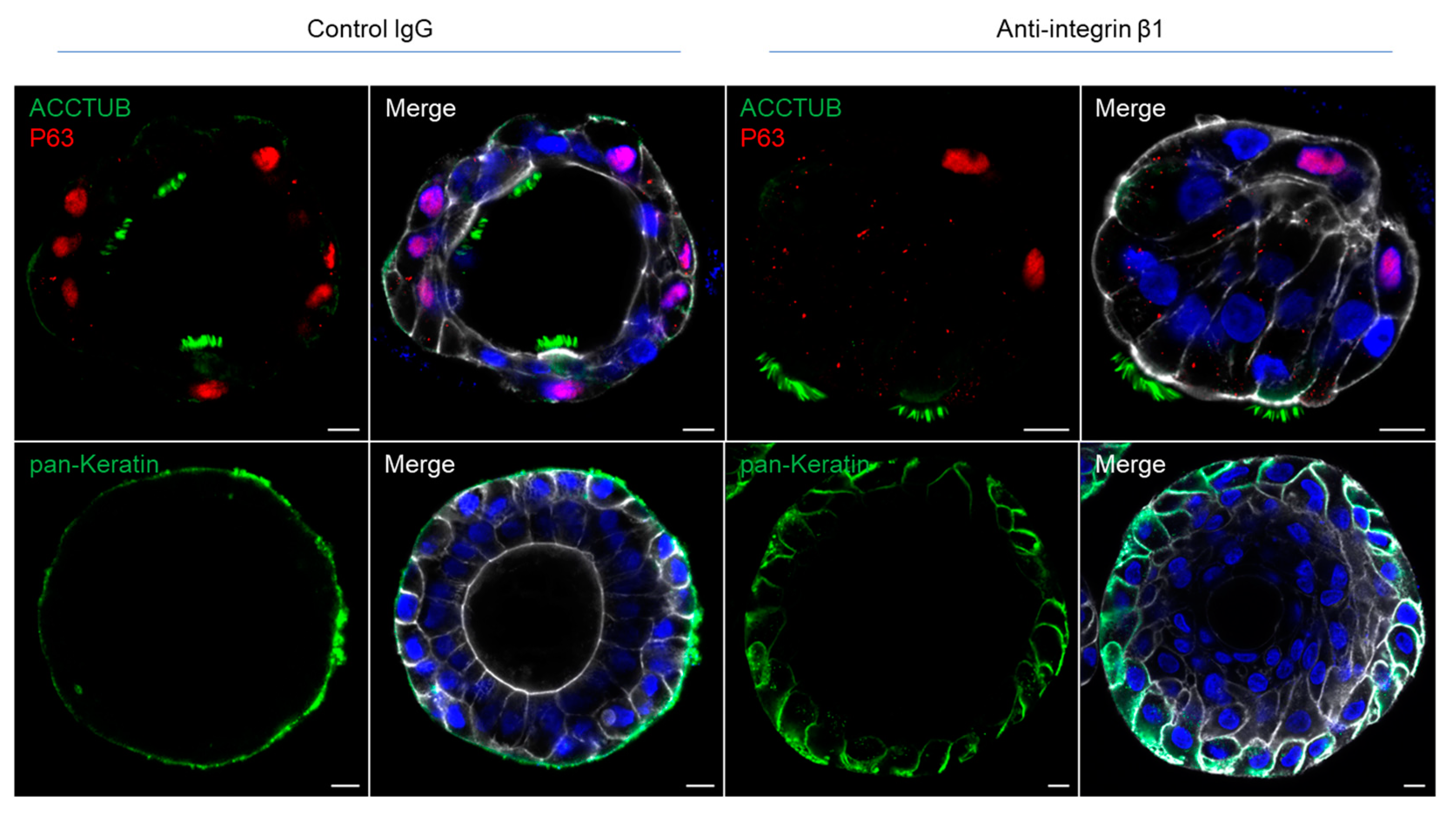
3.2. Integrin Beta 1 Controlling the Polarity of Airway Organoids
3.3. Differential Replicative Fitness of SARS-CoV-2 Variants in Apical-Out Airway Organoids
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Clevers, H. Modeling Development and Disease with Organoids. Cell 2016, 165, 1586–1597. [Google Scholar] [CrossRef]
- Dutta, D.; Heo, I.; Clevers, H. Disease Modeling in Stem Cell-Derived 3D Organoid Systems. Trends Mol. Med. 2017, 23, 393–410. [Google Scholar] [CrossRef]
- Zhou, J.; Li, C.; Sachs, N.; Chiu, M.C.; Wong, B.H.; Chu, H.; Poon, V.K.; Wang, D.; Zhao, X.; Wen, L.; et al. Differentiated human airway organoids to assess infectivity of emerging influenza virus. Proc. Natl. Acad. Sci. USA 2018, 115, 6822–6827. [Google Scholar] [CrossRef] [PubMed]
- Sachs, N.; Papaspyropoulos, A.; Zomer-van Ommen, D.D.; Heo, I.; Bottinger, L.; Klay, D.; Weeber, F.; Huelsz-Prince, G.; Iakobachvili, N.; Amatngalim, G.D.; et al. Long-term expanding human airway organoids for disease modeling. EMBO J. 2019, 38, e100300. [Google Scholar] [CrossRef] [PubMed]
- Chiu, M.C.; Li, C.; Liu, X.; Yu, Y.; Huang, J.; Wan, Z.; Xiao, D.; Chu, H.; Cai, J.P.; Zhou, B.; et al. A bipotential organoid model of respiratory epithelium recapitulates high infectivity of SARS-CoV-2 Omicron variant. Cell Discov. 2022, 8, 57. [Google Scholar] [CrossRef] [PubMed]
- Dutta, D.; Clevers, H. Organoid culture systems to study host-pathogen interactions. Curr. Opin. Immunol. 2017, 48, 15–22. [Google Scholar] [CrossRef]
- Co, J.Y.; Margalef-Catala, M.; Li, X.; Mah, A.T.; Kuo, C.J.; Monack, D.M.; Amieva, M.R. Controlling Epithelial Polarity: A Human Enteroid Model for Host-Pathogen Interactions. Cell Rep. 2019, 26, 2509–2520.e4. [Google Scholar] [CrossRef]
- Li, C.; Chiu, M.C.; Yu, Y.; Liu, X.; Xiao, D.; Huang, J.; Wan, Z.; Zhou, J. Establishing Human Lung Organoids and Proximal Differentiation to Generate Mature Airway Organoids. J. Vis. Exp. 2022, 181, e63684. [Google Scholar]
- Lau, S.Y.; Wang, P.; Mok, B.W.; Zhang, A.J.; Chu, H.; Lee, A.C.; Deng, S.; Chen, P.; Chan, K.H.; Song, W.; et al. Attenuated SARS-CoV-2 variants with deletions at the S1/S2 junction. Emerg. Microbes Infect. 2020, 9, 837–842. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Mok, B.W.Y.; Chen, L.L.; Chan, J.M.C.; Tsang, O.T.Y.; Lam, B.H.S.; Chuang, V.W.M.; Chu, A.W.H.; Chan, W.M.; Ip, J.D.; et al. Neutralization of Severe Acute Respiratory Syndrome Coronavirus 2 Omicron Variant by Sera From BNT162b2 or CoronaVac Vaccine Recipients. Clin. Infect. Dis. 2022, 75, e822–e826. [Google Scholar] [CrossRef]
- Ong, C.P.; Ye, Z.W.; Tang, K.; Liang, R.; Xie, Y.; Zhang, H.; Qin, Z.; Sun, H.; Wang, T.Y.; Cheng, Y.; et al. Comparative analysis of SARS-CoV-2 Omicron BA.2.12.1 and BA.5.2 variants. J. Med. Virol. 2023, 95, e28326. [Google Scholar] [CrossRef] [PubMed]
- Chiu, M.C.; Li, C.; Liu, X.; Song, W.; Wan, Z.; Yu, Y.; Huang, J.; Xiao, D.; Chu, H.; Cai, J.P.; et al. Human Nasal Organoids Model SARS-CoV-2 Upper Respiratory Infection and Recapitulate the Differential Infectivity of Emerging Variants. mBio 2022, 13, e0194422. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Li, C.; Liu, X.; Chiu, M.C.; Zhao, X.; Wang, D.; Wei, Y.; Lee, A.; Zhang, A.J.; Chu, H.; et al. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat. Med. 2020, 26, 1077–1083. [Google Scholar] [CrossRef] [PubMed]
- Sheppard, D. Functions of pulmonary epithelial integrins: From development to disease. Physiol. Rev. 2003, 83, 673–686. [Google Scholar] [CrossRef]
- Yu, W.; Datta, A.; Leroy, P.; O’Brien, L.E.; Mak, G.; Jou, T.S.; Matlin, K.S.; Mostov, K.E.; Zegers, M.M. Beta1-integrin orients epithelial polarity via Rac1 and laminin. Mol. Biol. Cell 2005, 16, 433–445. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef]
- Ahn, J.H.; Kim, J.; Hong, S.P.; Choi, S.Y.; Yang, M.J.; Ju, Y.S.; Kim, Y.T.; Kim, H.M.; Rahman, M.D.T.; Chung, M.K.; et al. Nasal ciliated cells are primary targets for SARS-CoV-2 replication in the early stage of COVID-19. J. Clin. Invest. 2021, 131, e148517. [Google Scholar] [CrossRef]
- Viana, R.; Moyo, S.; Amoako, D.G.; Tegally, H.; Scheepers, C.; Althaus, C.L.; Anyaneji, U.J.; Bester, P.A.; Boni, M.F.; Chand, M.; et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature 2022, 603, 679–686. [Google Scholar] [CrossRef] [PubMed]
- Tegally, H.; Moir, M.; Everatt, J.; Giovanetti, M.; Scheepers, C.; Wilkinson, E.; Subramoney, K.; Makatini, Z.; Moyo, S.; Amoako, D.G.; et al. Emergence of SARS-CoV-2 Omicron lineages BA.4 and BA.5 in South Africa. Nat. Med. 2022, 28, 1785–1790. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Guo, Y.; Iketani, S.; Nair, M.S.; Li, Z.; Mohri, H.; Wang, M.; Yu, J.; Bowen, A.D.; Chang, J.Y.; et al. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4 and BA.5. Nature 2022, 608, 603–608. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Gao, Y.; Li, S.; Wu, C.; Wang, J.; Zheng, N. Modulation of Mucin (MUC2, MUC5AC and MUC5B) mRNA Expression and Protein Production and Secretion in Caco-2/HT29-MTX Co-Cultures Following Exposure to Individual and Combined Aflatoxin M1 and Ochratoxin A. Toxins 2019, 11, 132. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Yang, L.; Lacko, L.A.; Chen, S. Human organoid models to study SARS-CoV-2 infection. Nat. Methods 2022, 19, 418–428. [Google Scholar] [CrossRef] [PubMed]
- Stroulios, G.; Brown, T.; Moreni, G.; Kondro, D.; Dei, A.; Eaves, A.; Louis, S.; Hou, J.; Chang, W.; Pajkrt, D.; et al. Apical-out airway organoids as a platform for studying viral infections and screening for antiviral drugs. Sci. Rep. 2022, 12, 7673. [Google Scholar] [CrossRef] [PubMed]
- Ravindra, N.G.; Alfajaro, M.M.; Gasque, V.; Huston, N.C.; Wan, H.; Szigeti-Buck, K.; Yasumoto, Y.; Greaney, A.M.; Habet, V.; Chow, R.D.; et al. Single-cell longitudinal analysis of SARS-CoV-2 infection in human airway epithelium identifies target cells, alterations in gene expression, and cell state changes. PLoS Biol. 2021, 19, e3001143. [Google Scholar] [CrossRef]
- Liu, J.; Li, Y.; Liu, Q.; Yao, Q.; Wang, X.; Zhang, H.; Chen, R.; Ren, L.; Min, J.; Deng, F.; et al. SARS-CoV-2 cell tropism and multiorgan infection. Cell Discov. 2021, 7, 17. [Google Scholar] [CrossRef]

| Antibodies/Stains | Suppliers | Catalog No. |
|---|---|---|
| Integrin β1 | abcam | ab24693 |
| pan-Keratin | abcam | ab8068 |
| CK5 | abcam | ab128190 |
| P63 | abcam | ab124762 |
| TUBULIN | Sigma-Aldrich | T7941 |
| FOXJ1 | Sigma-Aldrich | HPA005714 |
| MUC5AC | abcam | ab3649 |
| CC10 | R&D systems | MAB4218 |
| J2 (dsRNA) | Scicons | 10010500 |
| NP | in-house [5,12] | n/a |
| Target Genes | Forward Primer | Reverse Primer |
|---|---|---|
| P63 | CAGACTCAATTTAGTGAGCC | CTGCTGGTCCATGCTGTT |
| CK5 | GAGGAATGCAGACTCAGTGGA | TAGCTTCCACTGCTACCTCCG |
| FOXJ1 | TCGTATGCCACGCTCATCTG | CGGATTGAATTCTGCCAGGT |
| SNTN | GCTGCAAACCCAATTTAGGA | TGCTCATCAAGTTCAGAAAGGA |
| MUC5AC | CCTACAAAGCTGAGGCCTGT | GACCCTCCTCTCAATGGTGC |
| CC10 | AGCATCATTAAGCTCATGGAAAAA | GTGGACTCAAAGCATGGCAG |
| GAPDH | GGAGCGAGATCCCTCCAAAAT | GGCTGTTGTCATACTTCTCATGG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chiu, M.C.; Zhang, S.; Li, C.; Liu, X.; Yu, Y.; Huang, J.; Wan, Z.; Zhu, X.; Zhou, J. Apical-Out Human Airway Organoids Modeling SARS-CoV-2 Infection. Viruses 2023, 15, 1166. https://doi.org/10.3390/v15051166
Chiu MC, Zhang S, Li C, Liu X, Yu Y, Huang J, Wan Z, Zhu X, Zhou J. Apical-Out Human Airway Organoids Modeling SARS-CoV-2 Infection. Viruses. 2023; 15(5):1166. https://doi.org/10.3390/v15051166
Chicago/Turabian StyleChiu, Man Chun, Shuxin Zhang, Cun Li, Xiaojuan Liu, Yifei Yu, Jingjing Huang, Zhixin Wan, Xiaoxin Zhu, and Jie Zhou. 2023. "Apical-Out Human Airway Organoids Modeling SARS-CoV-2 Infection" Viruses 15, no. 5: 1166. https://doi.org/10.3390/v15051166
APA StyleChiu, M. C., Zhang, S., Li, C., Liu, X., Yu, Y., Huang, J., Wan, Z., Zhu, X., & Zhou, J. (2023). Apical-Out Human Airway Organoids Modeling SARS-CoV-2 Infection. Viruses, 15(5), 1166. https://doi.org/10.3390/v15051166






