Web Resources for SARS-CoV-2 Genomic Database, Annotation, Analysis and Variant Tracking
Abstract
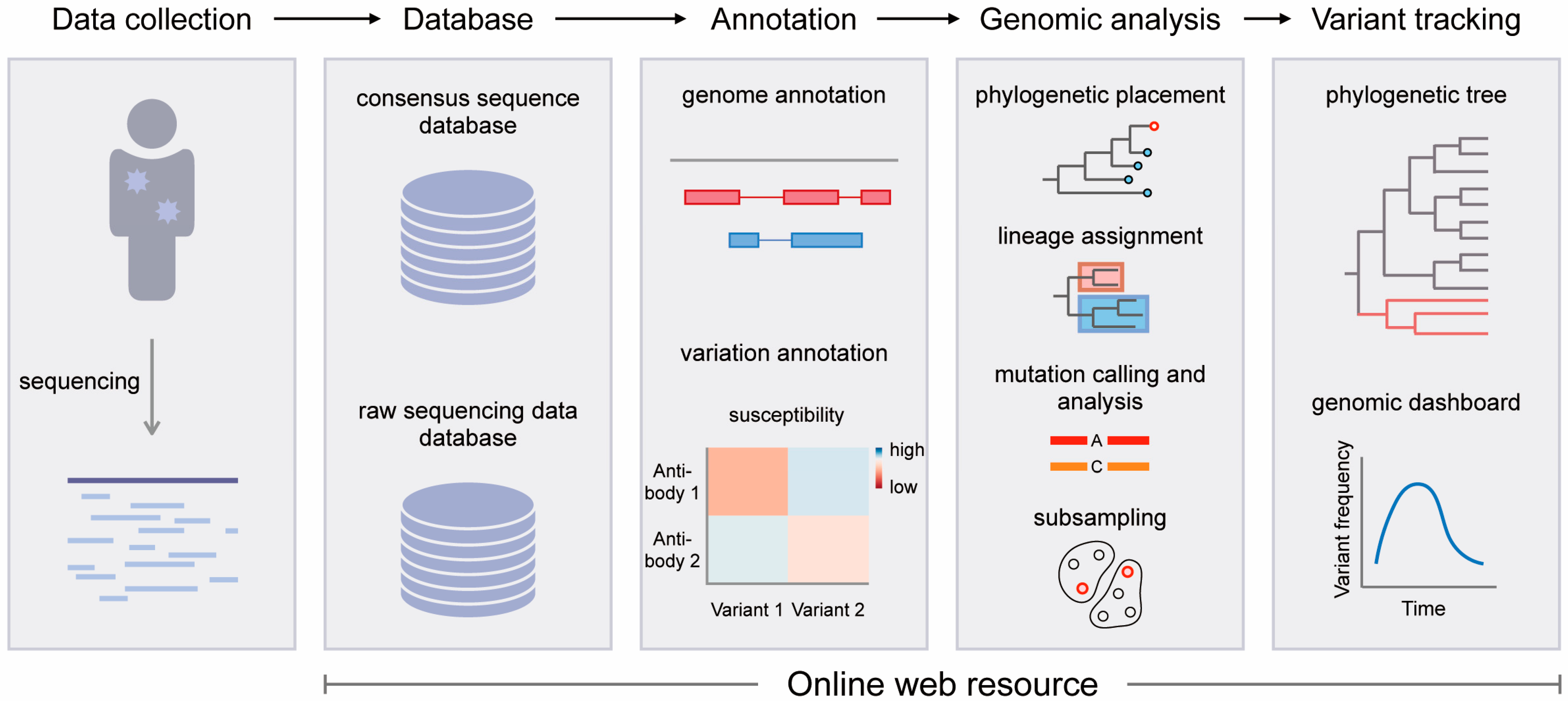
1. Introduction
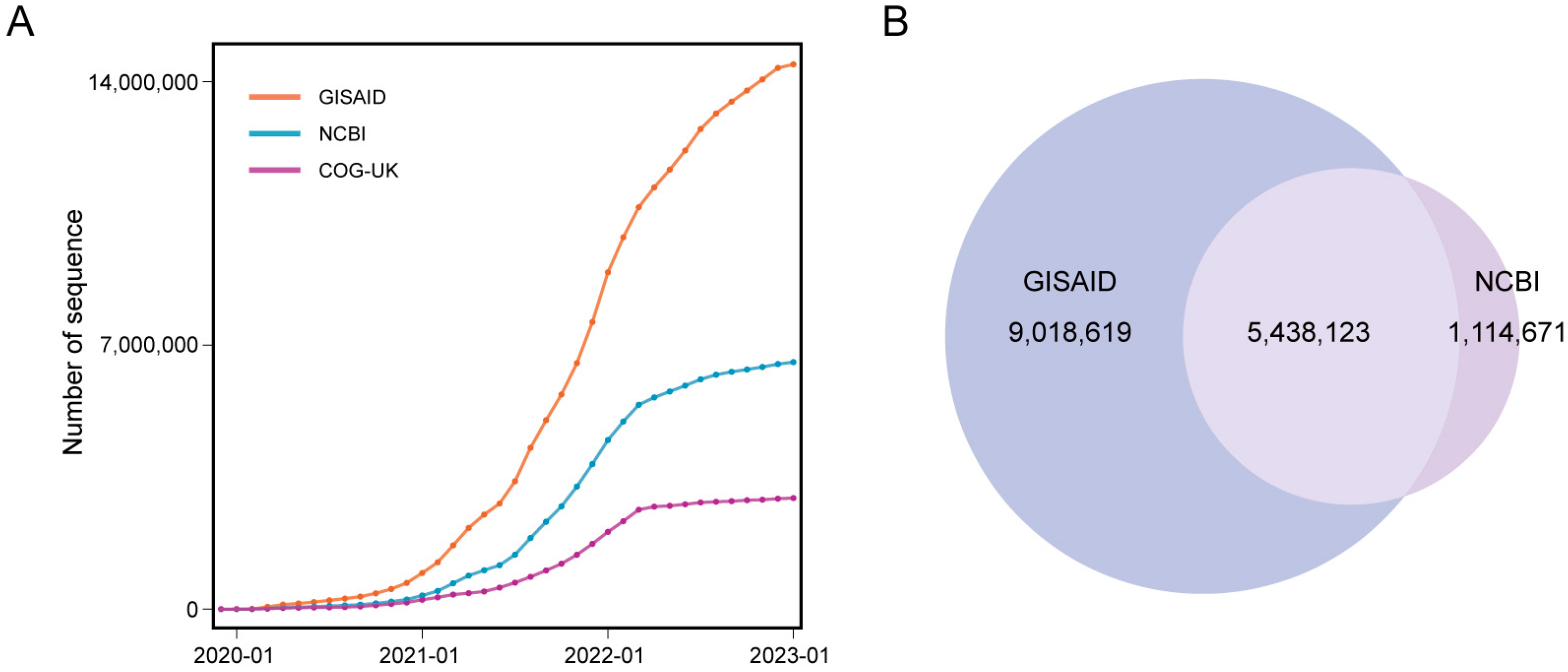
2. SARS-CoV-2 Genomic Databases
3. SARS-CoV-2 Genomic Annotation Web Resources
4. SARS-CoV-2 Genomic Analysis Web Tools
5. SARS-CoV-2 Variant Tracking Web Resources
6. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Gorbalenya, A.E.; Baker, S.C.; Baric, R.S.; de Groot, R.J.; Drosten, C.; Gulyaeva, A.A.; Haagmans, B.L.; Lauber, C.; Leontovich, A.M.; Neuman, B.W. The species severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar]
- Yang, H.; Rao, Z. Structural biology of SARS-CoV-2 and implications for therapeutic development. Nat. Rev. Microbiol 2021, 19, 685–700. [Google Scholar] [CrossRef] [PubMed]
- Shu, Y.; McCauley, J. GISAID: Global initiative on sharing all influenza data–from vision to reality. Eurosurveillance 2017, 22, 30494. [Google Scholar] [CrossRef] [PubMed]
- Elbe, S.; Buckland-Merrett, G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob. Chall. 2017, 1, 33–46. [Google Scholar] [CrossRef]
- Khare, S.; Gurry, C.; Freitas, L.; Schultz, M.B.; Bach, G.; Diallo, A.; Akite, N.; Ho, J.; Lee, R.T.; Yeo, W. GISAID’s role in pandemic response. China CDC Wkly. 2021, 3, 1049. [Google Scholar] [CrossRef]
- Hatcher, E.L.; Zhdanov, S.A.; Bao, Y.; Blinkova, O.; Nawrocki, E.P.; Ostapchuck, Y.; Schäffer, A.A.; Brister, J.R. Virus Variation Resource–improved response to emergent viral outbreaks. Nucleic Acids Res. 2017, 45, D482–D490. [Google Scholar] [CrossRef]
- Smith, D.; Bashton, M. An integrated national scale SARS-CoV-2 genomic surveillance network. Lancet Microbe 2020, 3, E99–E100. [Google Scholar]
- Song, S.; Ma, L.; Zou, D.; Tian, D.; Li, C.; Zhu, J.; Chen, M.; Wang, A.; Ma, Y.; Li, M.; et al. The Global Landscape of SARS-CoV-2 Genomes, Variants, and Haplotypes in 2019nCoVR. Genom. Proteom. Bioinform. 2020, 18, 749–759. [Google Scholar] [CrossRef]
- Gong, Z.; Zhu, J.W.; Li, C.P.; Jiang, S.; Ma, L.N.; Tang, B.X.; Zou, D.; Chen, M.L.; Sun, Y.B.; Song, S.H.; et al. An online coronavirus analysis platform from the National Genomics Data Center. Zool Res. 2020, 41, 705–708. [Google Scholar] [CrossRef]
- Bedford, T.; Greninger, A.L.; Roychoudhury, P.; Starita, L.M.; Famulare, M.; Huang, M.-L.; Nalla, A.; Pepper, G.; Reinhardt, A.; Xie, H. Cryptic transmission of SARS-CoV-2 in Washington state. Science 2020, 370, 571–575. [Google Scholar] [CrossRef]
- Worobey, M.; Pekar, J.; Larsen, B.B.; Nelson, M.I.; Hill, V.; Joy, J.B.; Rambaut, A.; Suchard, M.A.; Wertheim, J.O.; Lemey, P. The emergence of SARS-CoV-2 in Europe and North America. Science 2020, 370, 564–570. [Google Scholar] [CrossRef]
- Kraemer, M.U.G.; Hill, V.; Ruis, C.; Dellicour, S.; Bajaj, S.; McCrone, J.T.; Baele, G.; Parag, K.V.; Battle, A.L.; Gutierrez, B.; et al. Spatiotemporal invasion dynamics of SARS-CoV-2 lineage B.1.1.7 emergence. Science 2021, 373, 889–895. [Google Scholar] [CrossRef]
- Washington, N.L.; Gangavarapu, K.; Zeller, M.; Bolze, A.; Cirulli, E.T.; Barrett, K.M.S.; Larsen, B.B.; Anderson, C.; White, S.; Cassens, T. Emergence and rapid transmission of SARS-CoV-2 B. 1.1. 7 in the United States. Cell 2021, 184, 2587–2594.e2587. [Google Scholar] [CrossRef]
- Alpert, T.; Brito, A.F.; Lasek-Nesselquist, E.; Rothman, J.; Valesano, A.L.; MacKay, M.J.; Petrone, M.E.; Breban, M.I.; Watkins, A.E.; Vogels, C.B. Early introductions and transmission of SARS-CoV-2 variant B. 1.1. 7 in the United States. Cell 2021, 184, 2595–2604.e2513. [Google Scholar] [CrossRef]
- Tegally, H.; Wilkinson, E.; Giovanetti, M.; Iranzadeh, A.; Fonseca, V.; Giandhari, J.; Doolabh, D.; Pillay, S.; San, E.J.; Msomi, N.; et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 2021, 592, 438–443. [Google Scholar] [CrossRef]
- Faria, N.R.; Mellan, T.A.; Whittaker, C.; Claro, I.M.; Candido, D.d.S.; Mishra, S.; Crispim, M.A.; Sales, F.C.; Hawryluk, I.; McCrone, J.T. Genomics and epidemiology of the P. 1 SARS-CoV-2 lineage in Manaus, Brazil. Science 2021, 372, 815–821. [Google Scholar] [CrossRef]
- McCrone, J.T.; Hill, V.; Bajaj, S.; Pena, R.E.; Lambert, B.C.; Inward, R.; Bhatt, S.; Volz, E.; Ruis, C.; Dellicour, S.; et al. Context-specific emergence and growth of the SARS-CoV-2 Delta variant. Nature 2022, 610, 154–160. [Google Scholar] [CrossRef]
- Viana, R.; Moyo, S.; Amoako, D.G.; Tegally, H.; Scheepers, C.; Althaus, C.L.; Anyaneji, U.J.; Bester, P.A.; Boni, M.F.; Chand, M.; et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature 2022, 603, 679–686. [Google Scholar] [CrossRef]
- Tegally, H.; Moir, M.; Everatt, J.; Giovanetti, M.; Scheepers, C.; Wilkinson, E.; Subramoney, K.; Makatini, Z.; Moyo, S.; Amoako, D.G.; et al. Emergence of SARS-CoV-2 Omicron lineages BA.4 and BA.5 in South Africa. Nat. Med. 2022, 28, 1785–1790. [Google Scholar] [CrossRef]
- Cheng, V.C.; Siu, G.K.; Wong, S.C.; Au, A.K.; Ng, C.S.; Chen, H.; Li, X.; Lee, L.K.; Leung, J.S.; Lu, K.K.; et al. Complementation of contact tracing by mass testing for successful containment of beta COVID-19 variant (SARS-CoV-2 VOC B.1.351) epidemic in Hong Kong. Lancet Reg Health West. Pac. 2021, 17, 100281. [Google Scholar] [CrossRef] [PubMed]
- Jansen, L.; Tegomoh, B.; Lange, K.; Showalter, K.; Figliomeni, J.; Abdalhamid, B.; Iwen, P.C.; Fauver, J.; Buss, B.; Donahue, M. Investigation of a SARS-CoV-2 B. 1.1. 529 (omicron) variant cluster—Nebraska, November–December 2021. Morb. Mortal. Wkly. Rep. 2021, 70, 1782. [Google Scholar] [CrossRef] [PubMed]
- Chamie, G.; Marquez, C.; Crawford, E.; Peng, J.; Petersen, M.; Schwab, D.; Schwab, J.; Martinez, J.; Jones, D.; Black, D.; et al. Community Transmission of Severe Acute Respiratory Syndrome Coronavirus 2 Disproportionately Affects the Latinx Population During Shelter-in-Place in San Francisco. Clin. Infect. Dis 2021, 73, S127–S135. [Google Scholar] [CrossRef] [PubMed]
- Stoddard, G.; Black, A.; Ayscue, P.; Lu, D.; Kamm, J.; Bhatt, K.; Chan, L.; Kistler, A.L.; Batson, J.; Detweiler, A.; et al. Using genomic epidemiology of SARS-CoV-2 to support contact tracing and public health surveillance in rural Humboldt County, California. BMC Public Health 2022, 22, 456. [Google Scholar] [CrossRef]
- Lemieux, J.E.; Siddle, K.J.; Shaw, B.M.; Loreth, C.; Schaffner, S.F.; Gladden-Young, A.; Adams, G.; Fink, T.; Tomkins-Tinch, C.H.; Krasilnikova, L.A.; et al. Phylogenetic analysis of SARS-CoV-2 in Boston highlights the impact of superspreading events. Science 2021, 371, eabe3261. [Google Scholar] [CrossRef]
- Popa, A.; Genger, J.W.; Nicholson, M.D.; Penz, T.; Schmid, D.; Aberle, S.W.; Agerer, B.; Lercher, A.; Endler, L.; Colaco, H.; et al. Genomic epidemiology of superspreading events in Austria reveals mutational dynamics and transmission properties of SARS-CoV-2. Sci. Transl. Med. 2020, 12, eabe2555. [Google Scholar] [CrossRef]
- Chau, N.V.V.; Hong, N.T.T.; Ngoc, N.M.; Thanh, T.T.; Khanh, P.N.Q.; Nguyet, L.A.; Ny, N.T.H.; Man, D.N.H.; Hang, V.T.T.; Phong, N.T. Superspreading event of SARS-CoV-2 infection at a bar, Ho Chi Minh city, Vietnam. Emerg. Infect. Dis. 2021, 27, 310. [Google Scholar] [CrossRef]
- Chu, D.K.W.; Gu, H.; Chang, L.D.J.; Cheuk, S.S.Y.; Gurung, S.; Krishnan, P.; Ng, D.Y.M.; Liu, G.Y.Z.; Wan, C.K.C.; Tsang, D.N.C.; et al. SARS-CoV-2 Superspread in Fitness Center, Hong Kong, China, March 2021. Emerg Infect Dis 2021, 27, 2230–2232. [Google Scholar] [CrossRef]
- Rambaut, A. Phylodynamic Analysis. 176 Genomes. Virological. Available online: https://virological.org/t/phylodynamic-analysis-176-genomes-6-mar-2020/356 (accessed on 9 January 2023).
- Tonkin-Hill, G.; Martincorena, I.; Amato, R.; Lawson, A.R.J.; Gerstung, M.; Johnston, I.; Jackson, D.K.; Park, N.; Lensing, S.V.; Quail, M.A.; et al. Patterns of within-host genetic diversity in SARS-CoV-2. Elife 2021, 10, e66857. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, D.; Zhang, L.; Sun, W.; Zhang, Z.; Chen, W.; Zhu, A.; Huang, Y.; Xiao, F.; Yao, J.; et al. Intra-host variation and evolutionary dynamics of SARS-CoV-2 populations in COVID-19 patients. Genome Med. 2021, 13, 30. [Google Scholar] [CrossRef]
- Braun, K.M.; Moreno, G.K.; Wagner, C.; Accola, M.A.; Rehrauer, W.M.; Baker, D.A.; Koelle, K.; O’Connor, D.H.; Bedford, T.; Friedrich, T.C.; et al. Acute SARS-CoV-2 infections harbor limited within-host diversity and transmit via tight transmission bottlenecks. PLoS Pathog. 2021, 17, e1009849. [Google Scholar] [CrossRef]
- Valesano, A.L.; Rumfelt, K.E.; Dimcheff, D.E.; Blair, C.N.; Fitzsimmons, W.J.; Petrie, J.G.; Martin, E.T.; Lauring, A.S. Temporal dynamics of SARS-CoV-2 mutation accumulation within and across infected hosts. PLoS Pathog. 2021, 17, e1009499. [Google Scholar] [CrossRef]
- Lythgoe, K.A.; Hall, M.; Ferretti, L.; de Cesare, M.; MacIntyre-Cockett, G.; Trebes, A.; Andersson, M.; Otecko, N.; Wise, E.L.; Moore, N.; et al. SARS-CoV-2 within-host diversity and transmission. Science 2021, 372, eabg0821. [Google Scholar] [CrossRef]
- San, J.E.; Ngcapu, S.; Kanzi, A.M.; Tegally, H.; Fonseca, V.; Giandhari, J.; Wilkinson, E.; Nelson, C.W.; Smidt, W.; Kiran, A.M.; et al. Transmission dynamics of SARS-CoV-2 within-host diversity in two major hospital outbreaks in South Africa. Virus Evol. 2021, 7, veab041. [Google Scholar] [CrossRef]
- Hannon, W.W.; Roychoudhury, P.; Xie, H.; Shrestha, L.; Addetia, A.; Jerome, K.R.; Greninger, A.L.; Bloom, J.D. Narrow transmission bottlenecks and limited within-host viral diversity during a SARS-CoV-2 outbreak on a fishing boat. Virus Evol. 2022, 8, veac052. [Google Scholar] [CrossRef]
- Phan, T. Genetic diversity and evolution of SARS-CoV-2. Infect. Genet. Evol. 2020, 81, 104260. [Google Scholar] [CrossRef]
- Telenti, A.; Hodcroft, E.B.; Robertson, D.L. The Evolution and Biology of SARS-CoV-2 Variants. Cold Spring Harb Perspect. Med. 2022, 12, a041390. [Google Scholar] [CrossRef]
- Simon-Loriere, E.; Schwartz, O. Towards SARS-CoV-2 serotypes? Nat. Rev. Microbiol 2022, 20, 187–188. [Google Scholar] [CrossRef]
- Lauring, A.S.; Hodcroft, E.B. Genetic variants of SARS-CoV-2—What do they mean? Jama 2021, 325, 529–531. [Google Scholar] [CrossRef]
- Peacock, T.P.; Penrice-Randal, R.; Hiscox, J.A.; Barclay, W.S. SARS-CoV-2 one year on: Evidence for ongoing viral adaptation. J. Gen. Virol. 2021, 102, 001584. [Google Scholar] [CrossRef]
- Wu, A.; Wang, L.; Zhou, H.Y.; Ji, C.Y.; Xia, S.Z.; Cao, Y.; Meng, J.; Ding, X.; Gold, S.; Jiang, T.; et al. One year of SARS-CoV-2 evolution. Cell Host Microbe 2021, 29, 503–507. [Google Scholar] [CrossRef]
- van Dorp, L.; Acman, M.; Richard, D.; Shaw, L.P.; Ford, C.E.; Ormond, L.; Owen, C.J.; Pang, J.; Tan, C.C.S.; Boshier, F.A.T.; et al. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect. Genet. Evol 2020, 83, 104351. [Google Scholar] [CrossRef]
- Rambaut, A.; Holmes, E.C.; O’Toole, A.; Hill, V.; McCrone, J.T.; Ruis, C.; du Plessis, L.; Pybus, O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020, 5, 1403–1407. [Google Scholar] [CrossRef] [PubMed]
- Hadfield, J.; Megill, C.; Bell, S.M.; Huddleston, J.; Potter, B.; Callender, C.; Sagulenko, P.; Bedford, T.; Neher, R.A. Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics 2018, 34, 4121–4123. [Google Scholar] [CrossRef] [PubMed]
- Konings, F.; Perkins, M.D.; Kuhn, J.H.; Pallen, M.J.; Alm, E.J.; Archer, B.N.; Barakat, A.; Bedford, T.; Bhiman, J.N.; Caly, L.; et al. SARS-CoV-2 Variants of Interest and Concern naming scheme conducive for global discourse. Nat. Microbiol. 2021, 6, 821–823. [Google Scholar] [CrossRef] [PubMed]
- Neher, R.A. Contributions of adaptation and purifying selection to SARS-CoV-2 evolution. Virus Evol. 2022, 8, veac113. [Google Scholar] [CrossRef]
- Tay, J.H.; Porter, A.F.; Wirth, W.; Duchene, S. The emergence of SARS-CoV-2 variants of concern is driven by acceleration of the substitution rate. Mol. Biol. Evol. 2022, 39, msac013. [Google Scholar] [CrossRef]
- Martin, D.P.; Lytras, S.; Lucaci, A.G.; Maier, W.; Grüning, B.; Shank, S.D.; Weaver, S.; MacLean, O.A.; Orton, R.J.; Lemey, P. Selection analysis identifies clusters of unusual mutational changes in Omicron lineage BA. 1 that likely impact Spike function. Mol. Biol. Evol. 2022, 39, msac061. [Google Scholar] [CrossRef]
- Hill, V.; Du Plessis, L.; Peacock, T.P.; Aggarwal, D.; Colquhoun, R.; Carabelli, A.M.; Ellaby, N.; Gallagher, E.; Groves, N.; Jackson, B.; et al. The origins and molecular evolution of SARS-CoV-2 lineage B.1.1.7 in the UK. Virus Evol. 2022, 8, veac080. [Google Scholar] [CrossRef]
- Ghafari, M.; Liu, Q.; Dhillon, A.; Katzourakis, A.; Weissman, D.B. Investigating the evolutionary origins of the first three SARS-CoV-2 variants of concern. Front. Virol. 2022, 76, 942555. [Google Scholar] [CrossRef]
- Mallapaty, S. Where did Omicron come from? Three key theories. Nature 2022, 602, 26–28. [Google Scholar] [CrossRef]
- Dennehy, J.J.; Gupta, R.K.; Hanage, W.P.; Johnson, M.C.; Peacock, T.P. Where is the next SARS-CoV-2 variant of concern? Lancet 2022, 399, 1938–1939. [Google Scholar] [CrossRef]
- Rambaut, A.; Loman, N.; Pybus, O.; Barclay, W.; Barrett, J.; Carabelli, A.; Connor, T.; Peacock, T.; Robertson, D.L.; Volz, E. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. Virological. Available online: https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563 (accessed on 8 May 2023).
- Corey, L.; Beyrer, C.; Cohen, M.S.; Michael, N.L.; Bedford, T.; Rolland, M. SARS-CoV-2 variants in patients with immunosuppression. N. Engl. J. Med. 2021, 385, 562–566. [Google Scholar] [CrossRef]
- Chaguza, C.; Hahn, A.M.; Petrone, M.E.; Zhou, S.; Ferguson, D.; Breban, M.I.; Pham, K.; Pena-Hernandez, M.A.; Castaldi, C.; Hill, V.; et al. Accelerated SARS-CoV-2 intrahost evolution leading to distinct genotypes during chronic infection. Cell Rep. Med. 2023, 4, 100943. [Google Scholar] [CrossRef]
- Wilkinson, S.A.J.; Richter, A.; Casey, A.; Osman, H.; Mirza, J.D.; Stockton, J.; Quick, J.; Ratcliffe, L.; Sparks, N.; Cumley, N.; et al. Recurrent SARS-CoV-2 mutations in immunodeficient patients. Virus Evol. 2022, 8, veac050. [Google Scholar] [CrossRef]
- Caccuri, F.; Messali, S.; Bortolotti, D.; Di Silvestre, D.; De Palma, A.; Cattaneo, C.; Bertelli, A.; Zani, A.; Milanesi, M.; Giovanetti, M. Competition for dominance within replicating quasispecies during prolonged SARS-CoV-2 infection in an immunocompromised host. Virus Evol. 2022, 8, veac042. [Google Scholar] [CrossRef]
- Harari, S.; Tahor, M.; Rutsinsky, N.; Meijer, S.; Miller, D.; Henig, O.; Halutz, O.; Levytskyi, K.; Ben-Ami, R.; Adler, A.; et al. Drivers of adaptive evolution during chronic SARS-CoV-2 infections. Nat. Med. 2022, 28, 1501–1508. [Google Scholar] [CrossRef]
- Sonnleitner, S.T.; Prelog, M.; Sonnleitner, S.; Hinterbichler, E.; Halbfurter, H.; Kopecky, D.B.; Almanzar, G.; Koblmüller, S.; Sturmbauer, C.; Feist, L. Cumulative SARS-CoV-2 mutations and corresponding changes in immunity in an immunocompromised patient indicate viral evolution within the host. Nat. Commun. 2022, 13, 2560. [Google Scholar] [CrossRef]
- Munnink, B.B.O.; Nijhuis, R.H.T.; Worp, N.; Boter, M.; Weller, B.; Verstrepen, B.E.; GeurtsvanKessel, C.; Corsten, M.F.; Russcher, A.; Koopmans, M. Highly Divergent SARS-CoV-2 Alpha Variant in Chronically Infected Immunocompromised Person. Emerg Infect. Dis 2022, 28, 1920–1923. [Google Scholar] [CrossRef]
- Weigang, S.; Fuchs, J.; Zimmer, G.; Schnepf, D.; Kern, L.; Beer, J.; Luxenburger, H.; Ankerhold, J.; Falcone, V.; Kemming, J. Within-host evolution of SARS-CoV-2 in an immunosuppressed COVID-19 patient as a source of immune escape variants. Nat. Commun. 2021, 12, 6405. [Google Scholar] [CrossRef]
- Elena, S.F.; Sanjuan, R. Adaptive value of high mutation rates of RNA viruses: Separating causes from consequences. J. Virol. 2005, 79, 11555–11558. [Google Scholar] [CrossRef] [PubMed]
- Rochman, N.D.; Wolf, Y.I.; Faure, G.; Mutz, P.; Zhang, F.; Koonin, E.V. Ongoing global and regional adaptive evolution of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2021, 118, e2104241118. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.P.; Weaver, S.; Tegally, H.; San, J.E.; Shank, S.D.; Wilkinson, E.; Lucaci, A.G.; Giandhari, J.; Naidoo, S.; Pillay, Y. The emergence and ongoing convergent evolution of the SARS-CoV-2 N501Y lineages. Cell 2021, 184, 5189–5200.e5187. [Google Scholar] [CrossRef] [PubMed]
- Ramazzotti, D.; Angaroni, F.; Maspero, D.; Mauri, M.; D’Aliberti, D.; Fontana, D.; Antoniotti, M.; Elli, E.M.; Graudenzi, A.; Piazza, R. Large-scale analysis of SARS-CoV-2 synonymous mutations reveals the adaptation to the human codon usage during the virus evolution. Virus Evol. 2022, 8, veac026. [Google Scholar] [CrossRef]
- Ji, C.Y.; Han, N.; Cheng, Y.X.; Shang, J.; Weng, S.; Yang, R.; Zhou, H.Y.; Wu, A. Detecting Potentially Adaptive Mutations from the Parallel and Fixed Patterns in SARS-CoV-2 Evolution. Viruses 2022, 14, 1087. [Google Scholar] [CrossRef]
- Kistler, K.E.; Huddleston, J.; Bedford, T. Rapid and parallel adaptive mutations in spike S1 drive clade success in SARS-CoV-2. Cell Host Microbe 2022, 30, 545–555.e544. [Google Scholar] [CrossRef]
- Cao, Y.; Jian, F.; Wang, J.; Yu, Y.; Song, W.; Yisimayi, A.; Wang, J.; An, R.; Chen, X.; Zhang, N.; et al. Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. Nature 2022, 614, 521–529. [Google Scholar] [CrossRef]
- Hufsky, F.; Lamkiewicz, K.; Almeida, A.; Aouacheria, A.; Arighi, C.; Bateman, A.; Baumbach, J.; Beerenwinkel, N.; Brandt, C.; Cacciabue, M.; et al. Computational strategies to combat COVID-19: Useful tools to accelerate SARS-CoV-2 and coronavirus research. Brief. Bioinform. 2021, 22, 642–663. [Google Scholar] [CrossRef]
- Mercatelli, D.; Holding, A.N.; Giorgi, F.M. Web tools to fight pandemics: The COVID-19 experience. Brief. Bioinform. 2021, 22, 690–700. [Google Scholar] [CrossRef]
- Hu, T.; Li, J.; Zhou, H.; Li, C.; Holmes, E.C.; Shi, W. Bioinformatics resources for SARS-CoV-2 discovery and surveillance. Brief. Bioinform. 2021, 22, 631–641. [Google Scholar] [CrossRef]
- Fernandes, J.D.; Hinrichs, A.S.; Clawson, H.; Gonzalez, J.N.; Lee, B.T.; Nassar, L.R.; Raney, B.J.; Rosenbloom, K.R.; Nerli, S.; Rao, A.A.; et al. The UCSC SARS-CoV-2 Genome Browser. Nat. Genet. 2020, 52, 991–998. [Google Scholar] [CrossRef]
- Flynn, J.A.; Purushotham, D.; Choudhary, M.N.; Zhuo, X.; Fan, C.; Matt, G.; Li, D.; Wang, T. Exploring the coronavirus pandemic with the WashU Virus Genome Browser. Nat. Genet. 2020, 52, 986–991. [Google Scholar] [CrossRef]
- De Silva, N.H.; Bhai, J.; Chakiachvili, M.; Contreras-Moreira, B.; Cummins, C.; Frankish, A.; Gall, A.; Genez, T.; Howe, K.L.; Hunt, S.E.; et al. The Ensembl COVID-19 resource: Ongoing integration of public SARS-CoV-2 data. Nucleic Acids Res. 2022, 50, D765–D770. [Google Scholar] [CrossRef]
- Starr, T.N.; Greaney, A.J.; Hilton, S.K.; Ellis, D.; Crawford, K.H.; Dingens, A.S.; Navarro, M.J.; Bowen, J.E.; Tortorici, M.A.; Walls, A.C. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell 2020, 182, 1295–1310.e1220. [Google Scholar] [CrossRef]
- Starr, T.N.; Greaney, A.J.; Hannon, W.W.; Loes, A.N.; Hauser, K.; Dillen, J.R.; Ferri, E.; Farrell, A.G.; Dadonaite, B.; McCallum, M.; et al. Shifting mutational constraints in the SARS-CoV-2 receptor-binding domain during viral evolution. Science 2022, 377, 420–424. [Google Scholar] [CrossRef]
- Starr, T.N.; Greaney, A.J.; Stewart, C.M.; Walls, A.C.; Hannon, W.W.; Veesler, D.; Bloom, J.D. Deep mutational scans for ACE2 binding, RBD expression, and antibody escape in the SARS-CoV-2 Omicron BA.1 and BA.2 receptor-binding domains. PLoS Pathog. 2022, 18, e1010951. [Google Scholar] [CrossRef]
- Greaney, A.J.; Starr, T.N.; Bloom, J.D. An antibody-escape estimator for mutations to the SARS-CoV-2 receptor-binding domain. Virus Evol. 2022, 8, veac021. [Google Scholar] [CrossRef]
- Chen, J.; Gao, K.; Wang, R.; Wei, G.W. Prediction and mitigation of mutation threats to COVID-19 vaccines and antibody therapies. Chem. Sci. 2021, 12, 6929–6948. [Google Scholar] [CrossRef]
- Wang, R.; Chen, J.; Gao, K.; Wei, G.W. Vaccine-escape and fast-growing mutations in the United Kingdom, the United States, Singapore, Spain, India, and other COVID-19-devastated countries. Genomics 2021, 113, 2158–2170. [Google Scholar] [CrossRef]
- Tzou, P.L.; Tao, K.; Pond, S.L.K.; Shafer, R.W. Coronavirus Resistance Database (CoV-RDB): SARS-CoV-2 susceptibility to monoclonal antibodies, convalescent plasma, and plasma from vaccinated persons. PLoS ONE 2022, 17, e0261045. [Google Scholar] [CrossRef]
- Sun, Q.; Shu, C.; Shi, W.; Luo, Y.; Fan, G.; Nie, J.; Bi, Y.; Wang, Q.; Qi, J.; Lu, J.; et al. VarEPS: An evaluation and prewarning system of known and virtual variations of SARS-CoV-2 genomes. Nucleic Acids Res. 2022, 50, D888–D897. [Google Scholar] [CrossRef] [PubMed]
- Turakhia, Y.; Thornlow, B.; Hinrichs, A.S.; De Maio, N.; Gozashti, L.; Lanfear, R.; Haussler, D.; Corbett-Detig, R. Ultrafast Sample placement on Existing tRees (UShER) enables real-time phylogenetics for the SARS-CoV-2 pandemic. Nat. Genet. 2021, 53, 809–816. [Google Scholar] [CrossRef]
- O’Toole, A.; Scher, E.; Underwood, A.; Jackson, B.; Hill, V.; McCrone, J.T.; Colquhoun, R.; Ruis, C.; Abu-Dahab, K.; Taylor, B.; et al. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol. 2021, 7, veab064. [Google Scholar] [CrossRef] [PubMed]
- Aksamentov, I.; Roemer, C.; Hodcroft, E.B.; Neher, R.A. Nextclade: Clade assignment, mutation calling and quality control for viral genomes. J. Open Source Softw. 2021, 6, 3773. [Google Scholar] [CrossRef]
- Cheng, Y.; Ji, C.; Han, N.; Li, J.; Xu, L.; Chen, Z.; Yang, R.; Zhou, H.Y.; Wu, A. covSampler: A subsampling method with balanced genetic diversity for large-scale SARS-CoV-2 genome data sets. Virus Evol. 2022, 8, veac071. [Google Scholar] [CrossRef]
- McBroome, J.; Thornlow, B.; Hinrichs, A.S.; Kramer, A.; De Maio, N.; Goldman, N.; Haussler, D.; Corbett-Detig, R.; Turakhia, Y. A daily-updated database and tools for comprehensive SARS-CoV-2 mutation-annotated trees. Mol. Biol. Evol. 2021, 38, 5819–5824. [Google Scholar] [CrossRef]
- Sanderson, T. Taxonium, a web-based tool for exploring large phylogenetic trees. eLife 2022, 11, e82392. [Google Scholar] [CrossRef]
- McBroome, J.; Martin, J.; de Bernardi Schneider, A.; Turakhia, Y.; Corbett-Detig, R. Identifying SARS-CoV-2 regional introductions and transmission clusters in real time. Virus Evol. 2022, 8, veac048. [Google Scholar] [CrossRef]
- Ferreira, R.C.; Wong, E.; Gugan, G.; Wade, K.; Liu, M.; Baena, L.M.; Chato, C.; Lu, B.; Olabode, A.S.; Poon, A.F.Y. CoVizu: Rapid analysis and visualization of the global diversity of SARS-CoV-2 genomes. Virus Evol. 2021, 7, veab092. [Google Scholar] [CrossRef]
- Chen, C.; Nadeau, S.; Yared, M.; Voinov, P.; Xie, N.; Roemer, C.; Stadler, T. CoV-Spectrum: Analysis of globally shared SARS-CoV-2 data to identify and characterize new variants. Bioinformatics 2022, 38, 1735–1737. [Google Scholar] [CrossRef]
- Gangavarapu, K.; Latif, A.A.; Mullen, J.L.; Alkuzweny, M.; Hufbauer, E.; Tsueng, G.; Haag, E.; Zeller, M.; Aceves, C.M.; Zaiets, K.; et al. Outbreak.info genomic reports: Scalable and dynamic surveillance of SARS-CoV-2 variants and mutations. Nat. Methods 2023, 20, 512–522. [Google Scholar] [CrossRef]
- Tsueng, G.; Mullen, J.L.; Alkuzweny, M.; Cano, M.; Rush, B.; Haag, E.; Lin, J.; Welzel, D.J.; Zhou, X.; Qian, Z.; et al. Outbreak.info Research Library: A standardized, searchable platform to discover and explore COVID-19 resources. Nat. Methods 2023, 20, 536–540. [Google Scholar] [CrossRef]
- Chen, A.T.; Altschuler, K.; Zhan, S.H.; Chan, Y.A.; Deverman, B.E. COVID-19 CG enables SARS-CoV-2 mutation and lineage tracking by locations and dates of interest. Elife 2021, 10, e63409. [Google Scholar] [CrossRef]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B. Tracking changes in SARS-CoV-2 spike: Evidence that D614G increases infectivity of the COVID-19 virus. Cell 2020, 182, 812–827.e819. [Google Scholar] [CrossRef]
- Alam, I.; Radovanovic, A.; Incitti, R.; Kamau, A.A.; Alarawi, M.; Azhar, E.I.; Gojobori, T. CovMT: An interactive SARS-CoV-2 mutation tracker, with a focus on critical variants. Lancet Infect. Dis 2021, 21, 602. [Google Scholar] [CrossRef]
- Áine, T.; Verity, H.; Oliver G, P.; Alexander, W.; Issac I, B.; Kamran, K.; Jane P, M.; Houriiyah, T.; Richard R, L.; Jennifer, G.; et al. Tracking the international spread of SARS-CoV-2 lineages B.1.1.7 and B.1.351/501Y-V2 with grinch. Wellcome Open Res. 2021, 6, 121. [Google Scholar]
- Xavier, J.S.; Moir, M.; Tegally, H.; Sitharam, N.; Abdool Karim, W.; San, J.E.; Linhares, J.; Wilkinson, E.; Ascher, D.B.; Baxter, C. SARS-CoV-2 Africa dashboard for real-time COVID-19 information. Nat. Microbiol. 2022, 8, 1–4. [Google Scholar] [CrossRef]
- Bernasconi, A.; Canakoglu, A.; Masseroli, M.; Pinoli, P.; Ceri, S. A review on viral data sources and search systems for perspective mitigation of COVID-19. Brief. Bioinform 2021, 22, 664–675. [Google Scholar] [CrossRef]
- Lenharo, M. GISAID in crisis: Can the controversial COVID genome database survive? Nature 2023. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280.e278. [Google Scholar] [CrossRef]
- Ju, B.; Zhang, Q.; Ge, J.; Wang, R.; Sun, J.; Ge, X.; Yu, J.; Shan, S.; Zhou, B.; Song, S.; et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 2020, 584, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Shan, C.; Duan, X.; Chen, Z.; Liu, P.; Song, J.; Song, T.; Bi, X.; Han, C.; Wu, L.; et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 2020, 584, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Zost, S.J.; Gilchuk, P.; Case, J.B.; Binshtein, E.; Chen, R.E.; Nkolola, J.P.; Schafer, A.; Reidy, J.X.; Trivette, A.; Nargi, R.S.; et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature 2020, 584, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Greaney, A.J.; Starr, T.N.; Gilchuk, P.; Zost, S.J.; Binshtein, E.; Loes, A.N.; Hilton, S.K.; Huddleston, J.; Eguia, R.; Crawford, K.H. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe 2021, 29, 44–57.e49. [Google Scholar] [CrossRef]
- Greaney, A.J.; Loes, A.N.; Crawford, K.H.; Starr, T.N.; Malone, K.D.; Chu, H.Y.; Bloom, J.D. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe 2021, 29, 463–476.e466. [Google Scholar] [CrossRef]
- Frost, S.D.; Pybus, O.G.; Gog, J.R.; Viboud, C.; Bonhoeffer, S.; Bedford, T. Eight challenges in phylodynamic inference. Epidemics 2015, 10, 88–92. [Google Scholar] [CrossRef]
- Morel, B.; Barbera, P.; Czech, L.; Bettisworth, B.; Hübner, L.; Lutteropp, S.; Serdari, D.; Kostaki, E.-G.; Mamais, I.; Kozlov, A.M. Phylogenetic analysis of SARS-CoV-2 data is difficult. Mol. Biol. Evol. 2021, 38, 1777–1791. [Google Scholar] [CrossRef]
- Hodcroft, E.B.; De Maio, N.; Lanfear, R.; MacCannell, D.R.; Minh, B.Q.; Schmidt, H.A.; Stamatakis, A.; Goldman, N.; Dessimoz, C. Want to track pandemic variants faster? Fix the bioinformatics bottleneck. Nature 2021, 591, 30–33. [Google Scholar] [CrossRef]
- Shneider, A.; Su, M.; Hinrichs, A.; Wang, J.; Amin, H.; Bell, J.; Wadford, D.; O’toole, A.; Scher, E.; Perry, M. SARS-CoV-2 lineage assignment is more stable with UShER. Virological. Available online: https://virological.org/t/sars-cov-2-lineage-assignment-is-more-stable-with-usher/781 (accessed on 8 May 2023).
- Bolyen, E.; Dillon, M.R.; Bokulich, N.A.; Ladner, J.T.; Larsen, B.B.; Hepp, C.M.; Lemmer, D.; Sahl, J.W.; Sanchez, A.; Holdgraf, C.; et al. Reproducibly sampling SARS-CoV-2 genomes across time, geography, and viral diversity. F1000Res 2020, 9, 657. [Google Scholar] [CrossRef]
- De Maio, N.; Walker, C.; Borges, R.; Weilguny, L.; Slodkowicz, G.; Goldman, N. Issues with SARS-CoV-2 sequencing data. Virological. Available online: https://virological.org/t/issues-with-sars-cov-2-sequencing-data/473 (accessed on 8 May 2023).


| Web resource | Link | Reference |
|---|---|---|
| Database | ||
| GISAID | https://gisaid.org/, accessed on 20 February 2023 | [4,5,6] |
| NCBI | https://www.ncbi.nlm.nih.gov/sars-cov-2/, accessed on 20 February 2023 | [7] |
| COG-UK | https://www.cogconsortium.uk/priority-areas/data-linkage-analysis/public-data-analysis/, accessed on 20 February 2023 | [8] |
| CNCB RCoV19 | https://ngdc.cncb.ac.cn/ncov/release_genome, accessed on 20 February 2023 | [9,10] |
| Annotation | ||
| UCSC SARS-CoV-2 Genome Browser | https://genome.ucsc.edu/covid19.html, accessed on 12 January 2023 | [73] |
| WashU SARS-CoV-2 Genome Browser | https://virusgateway.wustl.edu/, accessed on 12 January 2023 | [74] |
| Ensembl COVID-19 Browser | https://covid-19.ensembl.org, accessed on 12 January 2023 | [75] |
| NCBI SARS-CoV-2 Annotation | https://www.ncbi.nlm.nih.gov/nuccore/NC_045512.2?report=graph, accessed on 11 January 2023 | [7] |
| CNCB RCoV19 Annotation | https://ngdc.cncb.ac.cn/ncov/knowledge/gene, accessed on 13 January 2023 | [9,10] |
| SARS-CoV-2 RBD DMS | https://jbloomlab.github.io/SARS-CoV-2-RBD_DMS/, accessed on 13 January 2023 https://jbloomlab.github.io/SARS-CoV-2-RBD_DMS_variants/, accessed on 13 January 2023 https://jbloomlab.github.io/SARS-CoV-2-RBD_DMS_Omicron/, accessed on 13 January 2023 | [76,77,78] |
| Antibody-escape estimator | https://jbloomlab.github.io/SARS2_RBD_Ab_escape_maps/escape-calc/, accessed on 13 January 2023 | [79] |
| Mutation analyzer | https://weilab.math.msu.edu/MutationAnalyzer/, accessed on 13 January 2023 | [80,81] |
| CoV-RDB | https://covdb.stanford.edu/, accessed on 14 January 2023 | [82] |
| VarEPS | https://nmdc.cn/ncovn/, accessed on 18 January 2023 | [83] |
| Analysis | ||
| UShER | https://genome.ucsc.edu/cgi-bin/hgPhyloPlace, accessed on 14 January 2023 | [84] |
| Pangolin | https://pangolin.cog-uk.io/, accessed on 14 January 2023 | [85] |
| CoVsurver | https://corona.bii.a-star.edu.sg/, accessed on 14 January 2023 | [6] |
| Nextclade | https://clades.nextstrain.org/, accessed on 14 January 2023 | [86] |
| covSampler | https://www.covsampler.net/, accessed on 14 January 2023 | [87] |
| Variant tracking | ||
| Cov2Tree | https://cov2tree.org/, accessed on 15 January 2023 | [88,89] |
| Cluster-Tracker | https://clustertracker.gi.ucsc.edu/, accessed on 15 January 2023 | [90] |
| CoVizu | https://filogeneti.ca/CoVizu/, accessed on 15 January 2023 | [91] |
| Nextstrain | https://nextstrain.org/, accessed on 15 January 2023 | [45] |
| CoVariants | https://covariants.org/, accessed on 15 January 2023 | / |
| covSpectrum | https://cov-spectrum.org/, accessed on 15 January 2023 | [92] |
| Outbreak.info | https://outbreak.info/, accessed on 15 January 2023 | [93,94] |
| COVID CG | https://covidcg.org/, accessed on 15 January 2023 | [95] |
| CoVerage | https://sarscoverage.org/, accessed on 15 January 2023 | / |
| CovGlobe | https://covglobe.org/, accessed on 15 January 2023 | / |
| REGENERON COVID-19 Dashboard | https://covid19dashboard.regeneron.com/, accessed on 15 January 2023 | / |
| COVID-19 Viral Genome Analysis Pipeline | https://cov.lanl.gov/, accessed on 15 January 2023 | [96] |
| CovMT | https://www.cbrc.kaust.edu.sa/covmt/, accessed on 15 January 2023 https://www.cbrc.kaust.edu.sa/covmtdev/, accessed on 15 January 2023 | [97] |
| cov-lineages.org | https://cov-lineages.org/, accessed on 15 January 2023 | [98] |
| SARS-CoV-2 Africa dashboard | https://climade.health/dashboard/covid-africa/, accessed on 15 January 2023 | [99] |
| Wellcome Sanger Institute COVID-19 Genomic surveillance dashboard | https://covid19.sanger.ac.uk/, accessed on 15 January 2023 | / |
| covidtag | http://covidtag.paseq.org/, accessed on 15 January 2023 | / |
| Web Resource | Data Source | Region |
|---|---|---|
| CoVariants | GISAID | Global |
| covSpectrum | GISIAD and NCBI | Global |
| Outbreak.info | GISAID | Global |
| COVID CG | GISAID | Global |
| CoVerage | GISAID | Global |
| CovGlobe | GISAID | Global |
| Regeneron COVID-19 Dashboard | GISAID | Global |
| COVID-19 Viral Genome Analysis Pipeline | GISAID | Global |
| CovMT | GISAID | Global |
| cov-lineages.org | GISAID | Global |
| SARS-CoV-2 Africa Dashboard | GISAID | Africa |
| Wellcome Sanger Institute COVID-19 Genomic Surveillance Dashboard | COG-UK | England |
| covidtag | GISAID | Spain |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, Y.; Ji, C.; Zhou, H.-Y.; Zheng, H.; Wu, A. Web Resources for SARS-CoV-2 Genomic Database, Annotation, Analysis and Variant Tracking. Viruses 2023, 15, 1158. https://doi.org/10.3390/v15051158
Cheng Y, Ji C, Zhou H-Y, Zheng H, Wu A. Web Resources for SARS-CoV-2 Genomic Database, Annotation, Analysis and Variant Tracking. Viruses. 2023; 15(5):1158. https://doi.org/10.3390/v15051158
Chicago/Turabian StyleCheng, Yexiao, Chengyang Ji, Hang-Yu Zhou, Heng Zheng, and Aiping Wu. 2023. "Web Resources for SARS-CoV-2 Genomic Database, Annotation, Analysis and Variant Tracking" Viruses 15, no. 5: 1158. https://doi.org/10.3390/v15051158
APA StyleCheng, Y., Ji, C., Zhou, H.-Y., Zheng, H., & Wu, A. (2023). Web Resources for SARS-CoV-2 Genomic Database, Annotation, Analysis and Variant Tracking. Viruses, 15(5), 1158. https://doi.org/10.3390/v15051158






