Novel Putative Tymoviridae-like Virus Isolated from Culex Mosquitoes in Colombia
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area and Entomological Collection
2.2. Virus Isolation and Preparation of Viral Stocks
2.3. RNA Purification
2.4. Metagenomic Sequencing, Assembly, and Virus Discovery
2.5. Primer Design for Virus Detection by RT-qPCR
2.6. 5′ and 3′ UTR Amplification Using RACE
2.7. Complete Genome Annotation
2.8. Evaluation of Virus Growth in Mosquito and Permissiveness of Mammalian Cells
2.9. Transmission Electron Microscopy (TEM)
2.10. Phylogenetic Analysis
3. Results
3.1. Cytopathic Effect Is Suggestive of Productive Viral Infection in C6/36 Cells
3.2. A Tymoviridae-like Virus Identified through Metagenomic Next-Generation Sequencing
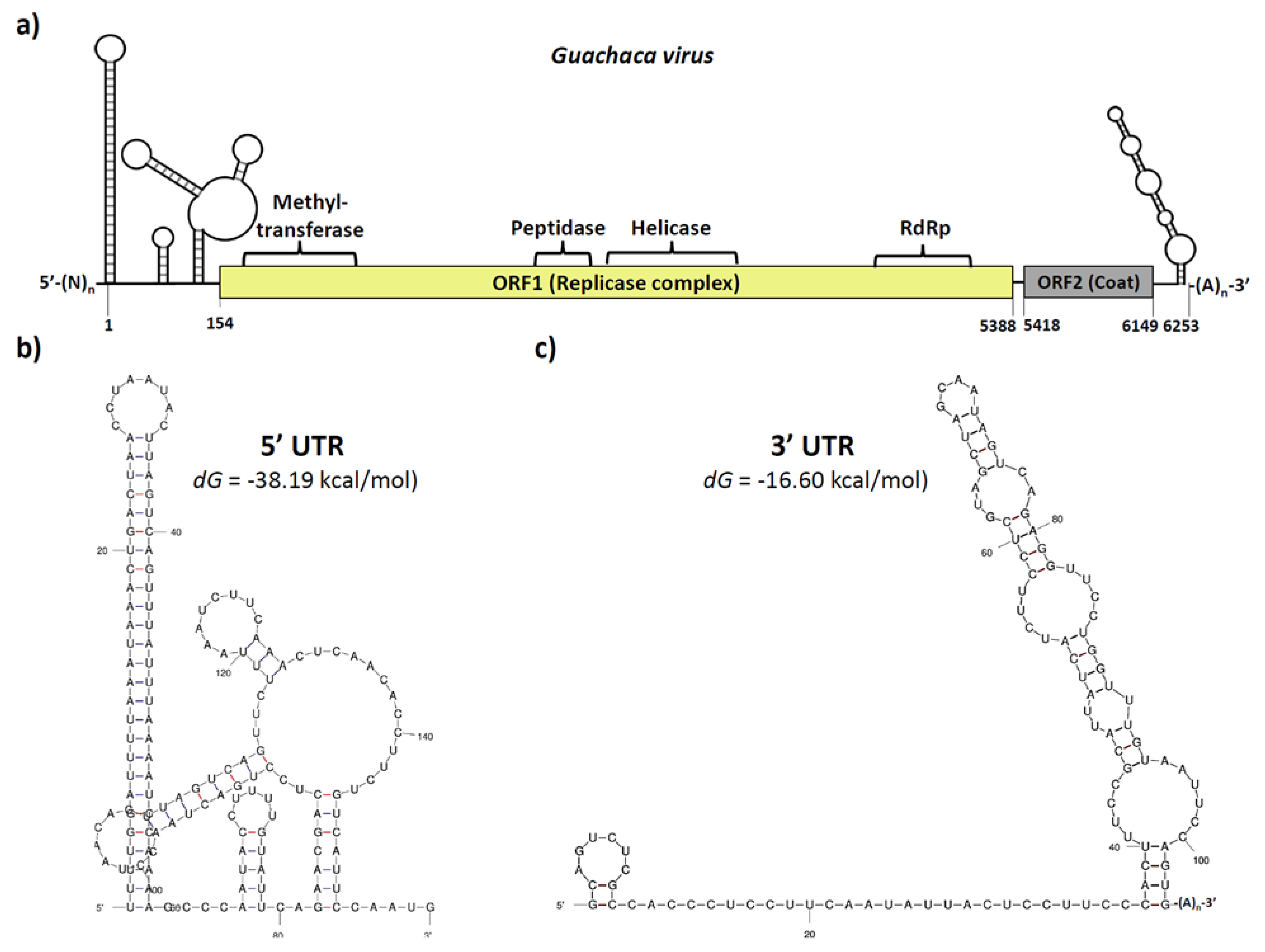
3.3. Putative Guachaca Virus Genome Organization
3.4. Phylogenetic Inference Supports the Proposal of a New Virus Species and Suggest the Need for New Genera for Insect-Specific Viruses from the Family Tymoviridae
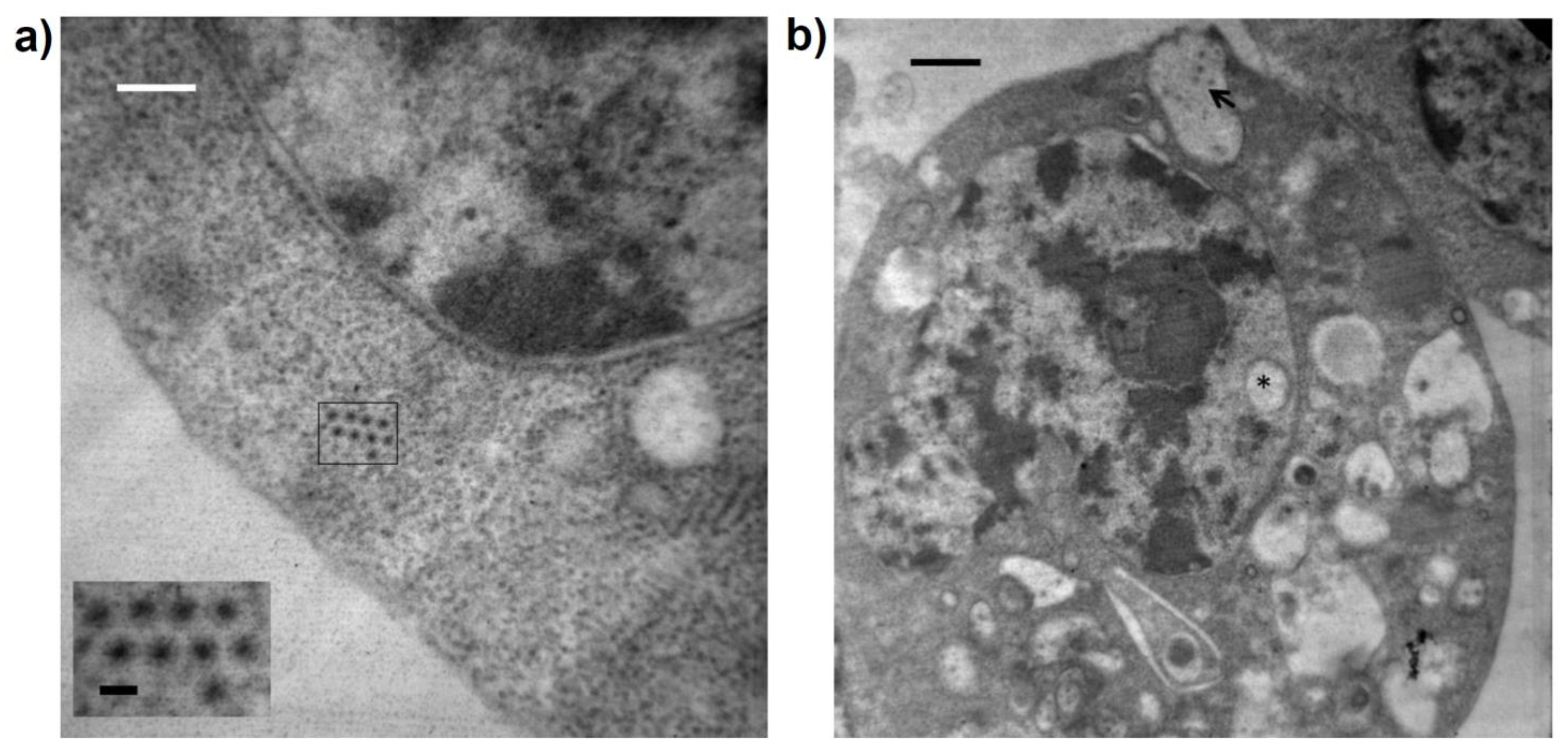
3.5. Ultrastructural Features of Guachaca Virus
3.6. Vertebrate Cells Were Not Permissible for GUAV Infection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Martelli, G.P.; Sabanadzovic, S.; Abou-Ghanem Sabanadzovic, N.; Edwards, M.C.; Dreher, T. The family Tymoviridae. Arch. Virol. 2002, 147, 1837–1846. [Google Scholar] [CrossRef] [PubMed]
- Ahlquist, P. Parallels among positive-strand RNA viruses, reverse-transcribing viruses and double-stranded RNA viruses. Nat. Rev. Microbiol. 2006, 4, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Elbeaino, T.; Digiaro, M.; Martelli, G.P. Complete sequence of Fig fleck-associated virus, a novel member of the family Tymoviridae. Virus Res. 2011, 161, 198–202. [Google Scholar] [CrossRef]
- Dreher, T.W. Turnip yellow mosaic virus: Transfer RNA mimicry, chloroplasts and a C-rich genome. Mol. Plant Pathol. 2004, 5, 367–375. [Google Scholar] [CrossRef]
- Hogenhout, S.A.; Ammar, E.-D.; Whitfield, A.E.; Redinbaugh, M.G. Insect Vector Interactions with Persistently Transmitted Viruses. Annu. Rev. Phytopathol. 2008, 46, 327–359. [Google Scholar] [CrossRef] [PubMed]
- Dietzgen, R.; Mann, K.; Johnson, K. Plant Virus–Insect Vector Interactions: Current and Potential Future Research Directions. Viruses 2016, 8, 303. [Google Scholar] [CrossRef]
- Foster, W.A. Mosquito Sugar Feeding and Reproductive Energetics. Annu. Rev. Entomol. 1995, 40, 443–474. [Google Scholar] [CrossRef]
- Batson, J.; Dudas, G.; Haas-Stapleton, E.; Kistler, A.L.; Li, L.M.; Logan, P.; Ratnasiri, K.; Retallack, H. Single mosquito metatranscriptomics identifies vectors, emerging pathogens and reservoirs in one assay. Elife 2021, 10, e68353. [Google Scholar] [CrossRef]
- Faizah, A.N.; Kobayashi, D.; Isawa, H.; Amoa-Bosompem, M.; Murota, K.; Higa, Y.; Futami, K.; Shimada, S.; Kim, K.S.; Itokawa, K.; et al. Deciphering the Virome of Culex vishnui Subgroup Mosquitoes, the Major Vectors of Japanese Encephalitis, in Japan. Viruses 2020, 12, 264. [Google Scholar] [CrossRef]
- Frey, K.G.; Biser, T.; Hamilton, T.; Santos, C.J.; Pimentel, G.; Mokashi, V.P.; Bishop-Lilly, K.A. Bioinformatic Characterization of Mosquito Viromes within the Eastern United States and Puerto Rico: Discovery of Novel Viruses. Evol. Bioinform. Online 2016, 12, 1–12. [Google Scholar] [CrossRef]
- Öhlund, P.; Hayer, J.; Lundén, H.; Hesson, J.C.; Blomström, A.-L. Viromics Reveal a Number of Novel RNA Viruses in Swedish Mosquitoes. Viruses 2019, 11, 1027. [Google Scholar] [CrossRef] [PubMed]
- Pettersson, J.H.O.; Shi, M.; Eden, J.S.; Holmes, E.C.; Hesson, J.C. Meta-Transcriptomic Comparison of the RNA Viromes of the Mosquito Vectors Culex pipiens and Culex torrentium in Northern Europe. Viruses 2019, 11, 1033. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi, M.; Altan, E.; Deng, X.; Barker, C.M.; Fang, Y.; Coffey, L.L.; Delwart, E. Virome of >12 thousand Culex mosquitoes from throughout California. Virology 2018, 523, 74–88. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Neville, P.; Nicholson, J.; Eden, J.-S.; Imrie, A.; Holmes, E.C. High-Resolution Metatranscriptomics Reveals the Ecological Dynamics of Mosquito-Associated RNA Viruses in Western Australia. J. Virol. 2017, 91, e00680-17. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Wang, Y.; Shi, C.; Atoni, E.; Zhao, L.; Yuan, Z. Comparative Metagenomic Profiling of Viromes Associated with Four Common Mosquito Species in China. Virol. Sin. 2018, 33, 59–66. [Google Scholar] [CrossRef]
- Xiao, P.; Li, C.; Zhang, Y.; Han, J.; Guo, X.; Xie, L.; Tian, M.; Li, Y.; Wang, M.; Liu, H.; et al. Metagenomic Sequencing from Mosquitoes in China Reveals a Variety of Insect and Human Viruses. Front. Cell. Infect. Microbiol. 2018, 8, 364. [Google Scholar] [CrossRef]
- Charles, J.; Tangudu, C.S.; Hurt, S.L.; Tumescheit, C.; Firth, A.E.; Garcia-Rejon, J.E.; Machain-Williams, C.; Blitvich, B.J. Discovery of a novel Tymoviridae-like virus in mosquitoes from Mexico. Arch. Virol. 2019, 164, 649–652. [Google Scholar] [CrossRef]
- Miranda, K.K.P.; Galvão, G.J.P.; da Silva Araújo, P.A.; da Silva Ribeiro, A.C.; da Silva, S.P.; da Silva Lemos, P.; Martins, L.C.; Nunes, M.R.T.; da Costa Vasconcelos, P.F.; da Costa Ferreira, V.; et al. Discovery and genome sequencing of a new virus related to members of the family Tymoviridae, isolated from mosquitoes of the genus Mansonia in Brazil. Arch. Virol. 2022, 167, 1889–1892. [Google Scholar] [CrossRef]
- Wang, L.; Lv, X.; Zhai, Y.; Fu, S.; Wang, D.; Rayner, S.; Tang, Q.; Liang, G. Genomic Characterization of a Novel Virus of the Family Tymoviridae Isolated from Mosquitoes. PLoS ONE 2012, 7, e39845. [Google Scholar] [CrossRef]
- Conceição-Neto, N.; Yinda, K.C.; Van Ranst, M.; Matthijnssens, J. NetoVIR: Modular approach to customize sample preparation procedures for viral Metagenomics. Methods Mol. Biol. 2018, 1838, 85–95. [Google Scholar] [CrossRef]
- Ajami, N.J.; Wong, M.C.; Ross, M.C.; Lloyd, R.E.; Petrosino, J.F. Maximal viral information recovery from sequence data using VirMAP. Nat. Commun. 2018, 9, 3205. [Google Scholar] [CrossRef] [PubMed]
- Blum, M.; Chang, H.-Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R.; Teeling, E. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Minh, B.Q.; Nguyen, M.A.T.; von Haeseler, A. Ultrafast Approximation for Phylogenetic Bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Thurner, C.; Witwer, C.; Hofacker, I.L.; Stadler, P.F. Conserved RNA secondary structures in Flaviviridae genomes. J. Gen. Virol. 2004, 85, 1113–1124. [Google Scholar] [CrossRef]
- Martinez-Salas, E.; Francisco-Velilla, R.; Fernandez-Chamorro, J.; Embarek, A.M. Insights into Structural and Mechanistic Features of Viral IRES Elements. Front. Microbiol. 2018, 8, 2629. [Google Scholar] [CrossRef]
- Fernández-Sanlés, A.; Ríos-Marco, P.; Romero-López, C.; Berzal-Herranz, A. Functional Information Stored in the Conserved Structural RNA Domains of Flavivirus Genomes. Front. Microbiol. 2017, 8, 546. [Google Scholar] [CrossRef]
- Charles, J.; Tangudu, C.S.; Hurt, S.L.; Tumescheit, C.; Firth, A.E.; Garcia-Rejon, J.E.; Machain-Williams, C.; Blitvich, B.J. Detection of novel and recognized RNA viruses in mosquitoes from the Yucatan Peninsula of Mexico using metagenomics and characterization of their in vitro host ranges. J. Gen. Virol. 2018, 99, 1729–1738. [Google Scholar] [CrossRef]
- French, R.K.; Holmes, E.C. An Ecosystems Perspective on Virus Evolution and Emergence. Trends Microbiol. 2020, 28, 165–175. [Google Scholar] [CrossRef] [PubMed]


| Primer Name | Target Gene | Genomic Position a | Sequence (5′–3′) | Annealing T (°C) |
|---|---|---|---|---|
| Guachaca-F | Viral helicase | 3014–3035 | tgacgcccgagacaacttacct | 57.5 |
| Guachaca-P | Viral helicase | 3067–3090 | FAM-acttacaggtctccagccaacatc-BHQ1 | 55 |
| Guachaca-R | Viral helicase | 3164–3142 | gagtgacggaaagagcgggagta | 58 |
| Group | Amino Acid Differences (d(S.E)) a | |||||
|---|---|---|---|---|---|---|
| Marafivirus | Maculavirus | Tymovirus | IAT_Lineage_1 | IAT_Lineage_2 | IAT_Lineage_3 | |
| Marafivirus | 0.344(0.010) | |||||
| Maculavirus | 0.456(0.013) | 0.024(0.005) | ||||
| Tymovirus | 0.500(0.012) | 0.515(0.013) | 0.381(0.010) | |||
| IST_Lineage_1 | 0.581(0.013) | 0.573(0.014) | 0.622(0.013) | 0.083(0.006) | ||
| IST_Lineage_2 | 0.505(0.012) | 0.520(0.013) | 0.536(0.012) | 0.556(0.014) | 0.358(0.011) | |
| IST_Lineage_3 | 0.536(0.013) | 0.534(0.015) | 0.555(0.013) | 0.602(0.014) | 0.540(0.014) | 0.254(0.010) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laiton-Donato, K.; Guzmán, C.; Perdomo-Balaguera, E.; Sarmiento, L.; Torres-Fernandez, O.; Ruiz, H.A.; Rosales-Munar, A.; Peláez-Carvajal, D.; Navas, M.-C.; Wong, M.C.; et al. Novel Putative Tymoviridae-like Virus Isolated from Culex Mosquitoes in Colombia. Viruses 2023, 15, 953. https://doi.org/10.3390/v15040953
Laiton-Donato K, Guzmán C, Perdomo-Balaguera E, Sarmiento L, Torres-Fernandez O, Ruiz HA, Rosales-Munar A, Peláez-Carvajal D, Navas M-C, Wong MC, et al. Novel Putative Tymoviridae-like Virus Isolated from Culex Mosquitoes in Colombia. Viruses. 2023; 15(4):953. https://doi.org/10.3390/v15040953
Chicago/Turabian StyleLaiton-Donato, Katherine, Camila Guzmán, Erik Perdomo-Balaguera, Ladys Sarmiento, Orlando Torres-Fernandez, Héctor Alejandro Ruiz, Alicia Rosales-Munar, Dioselina Peláez-Carvajal, Maria-Cristina Navas, Matthew C. Wong, and et al. 2023. "Novel Putative Tymoviridae-like Virus Isolated from Culex Mosquitoes in Colombia" Viruses 15, no. 4: 953. https://doi.org/10.3390/v15040953
APA StyleLaiton-Donato, K., Guzmán, C., Perdomo-Balaguera, E., Sarmiento, L., Torres-Fernandez, O., Ruiz, H. A., Rosales-Munar, A., Peláez-Carvajal, D., Navas, M.-C., Wong, M. C., Junglen, S., Ajami, N. J., Parra-Henao, G., & Usme-Ciro, J. A. (2023). Novel Putative Tymoviridae-like Virus Isolated from Culex Mosquitoes in Colombia. Viruses, 15(4), 953. https://doi.org/10.3390/v15040953







