A Metagenomic Investigation of the Viruses Associated with Shiraz Disease in Australia
Abstract
1. Introduction
2. Materials and Methods
2.1. Vineyard Locations and Grapevine Selection
2.2. Sample Collection
2.3. Nucleic Acid Extraction
2.3.1. Nucleic Acid Extraction for RT-PCR
2.3.2. Nucleic Acid Extraction for HTS
2.3.3. Nucleic Acid Quality Control
2.4. RT-PCR Reaction Conditions
2.5. Metagenomic High-Throughput Sequencing (Meta-HTS)
2.5.1. Nucleic Acid Pretreatments, Library Preparation and Sequencing
2.5.2. De Novo Assembly
2.5.3. Phylogenetic and Sequence Similarity Analysis
2.5.4. Phylogenetic Group Identification of the Grapevine Virus A Contigs
2.5.5. Multiple Sequence Alignment of the GVA RNA-Binding Protein
2.5.6. Recombination Analysis
3. Results
3.1. Virome Analysis by Endpoint RT-PCR and Meta-HTS
3.1.1. RT-PCR
3.1.2. Comparison of Virome Detection between Meta-HTS and Endpoint RT-PCR
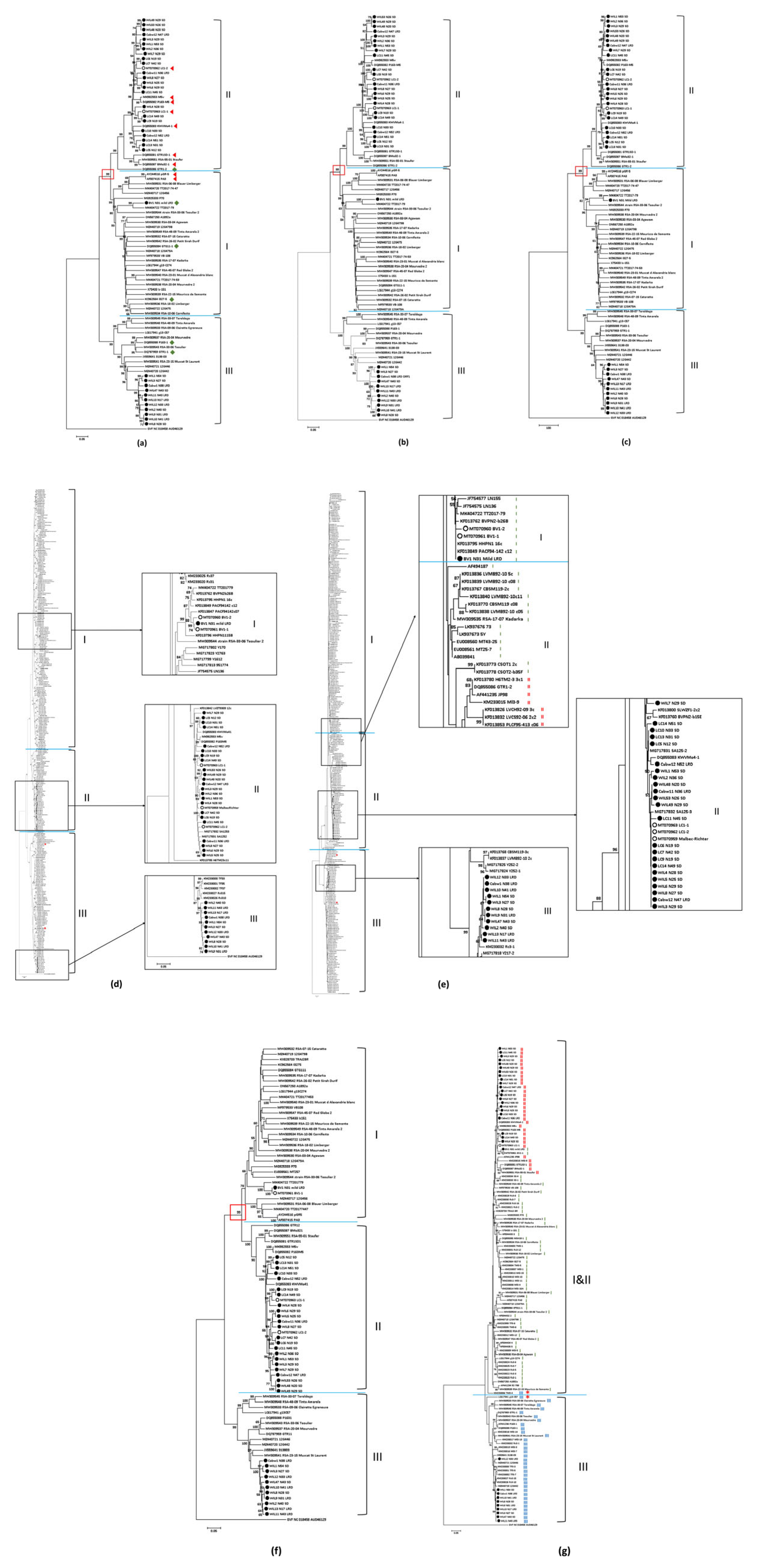
3.2. Phylogenetic Analysis of Grapevine Virus A (GVA)
3.2.1. Phylogenetic Groups (Phylogroups) of Grapevine Virus A (GVA)
3.2.2. Similarity within Phylogroups and between Australian GVA Isolates
3.3. Association between Amino acid Sequence of RNA-Binding Gene and Symptom Expression of Grapevine Virus A
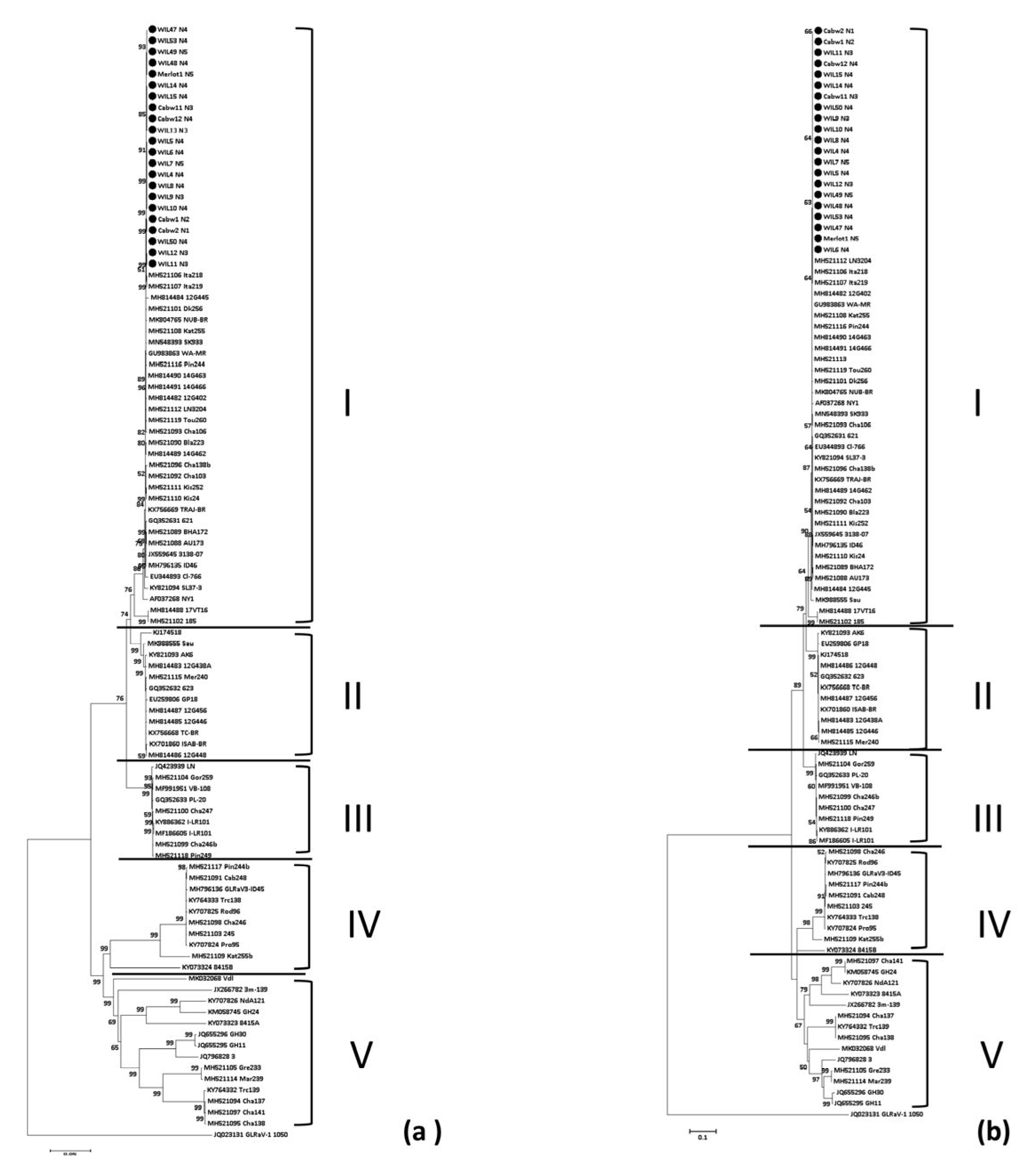
3.4. Phylogenetic Analysis of Grapevine Leafroll-Associated Virus 3
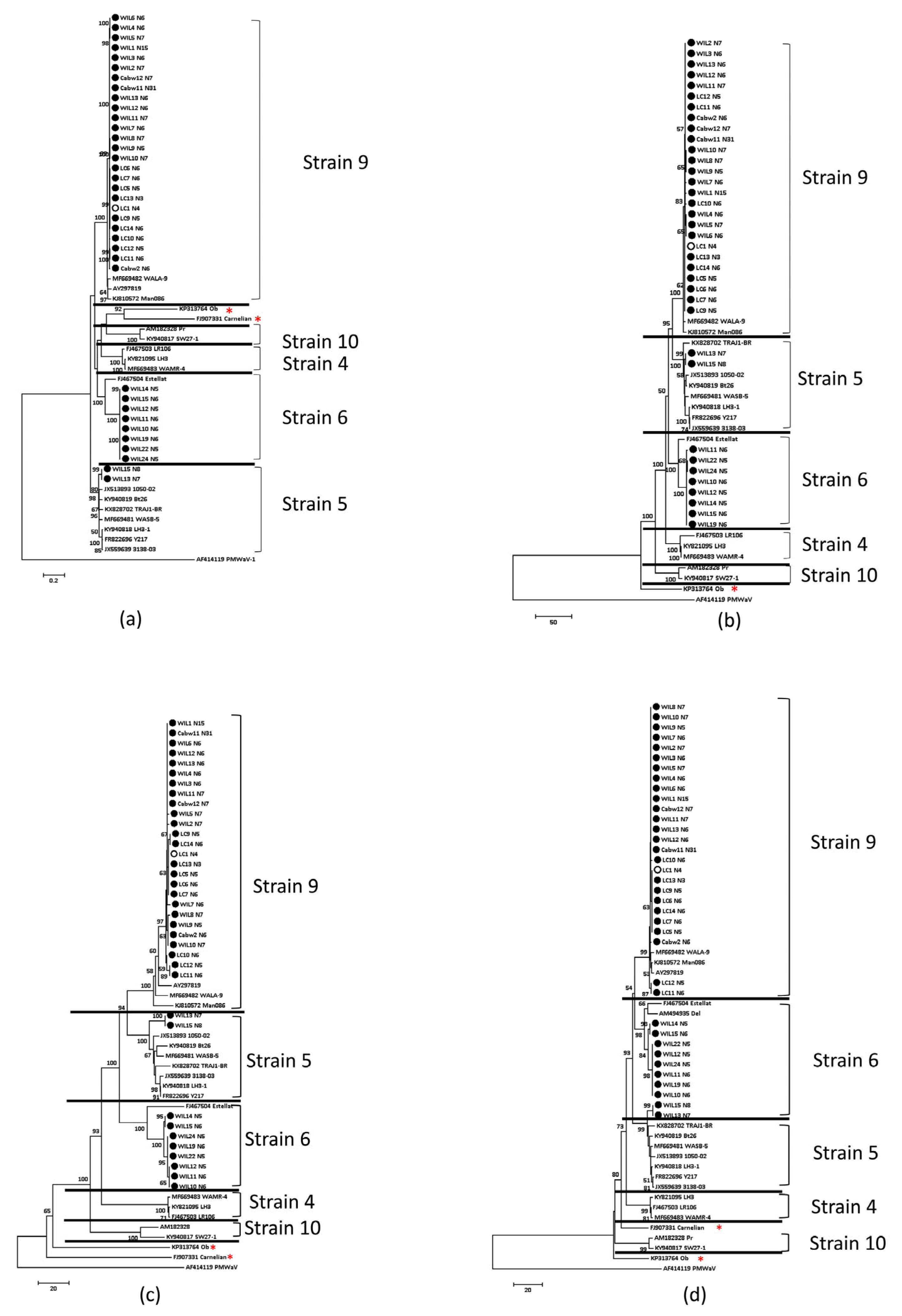
3.5. Phylogenetic Analysis of Grapevine Leafroll-Associated Virus 4, Strains 5, 6 and 9
3.6. Recombination Analysis
4. Discussion
4.1. Association between SD and Phylogenetic Groups of GVA
4.2. Amino Acid Sequence of RNA-Binding Gene of GVA and Symptom Expression
4.3. GVA Diversity
4.4. GLRaV-3
4.5. GLRaV-4
4.6. Other Viruses
4.7. Variability in Virus Detection
4.8. Association between SD and LRD Species
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wu, Q.; Habili, N.; Constable, F.; Al Rwahnih, M.; Goszczynski, D.E.; Wang, Y.; Pagay, V. Virus pathogens in Australian vineyards with an emphasis on Shiraz disease. Viruses 2020, 12, 818. [Google Scholar] [CrossRef]
- Habili, N.; Randles, J.W. Major yield loss in Shiraz vines infected with Australian Shiraz disease associated with grapevine virus A. In Proceedings of the 17th Meeting of the International Council for the Study of Viruses and Virus-like Diseases of the Grapevine, Davis, CA, USA, 7–14 October 2012; pp. 164–165. [Google Scholar]
- Australian Grape and Wine. Available online: https://www.agw.org.au/ (accessed on 6 February 2023).
- Martelli, G.P.; Ghanem-Sabanadzovic, N.A.; Agranovsky, A.A.; Al Rwahnih, M.; Dolja, V.V.; Dovas, C.I.; Fuchs, M.; Gugerli, P.; Hu, J.S.; Jelkmann, W.; et al. Taxonomic revision of the family Closteroviridae with special reference to the grapevine leafroll-associated members of the genus Ampelovirus and the putative species unassigned to the family. J. Plant Pathol. 2012, 94, 7–19. [Google Scholar]
- Maree, H.J.; Almeida, R.P.P.; Bester, R.; Chooi, K.M.; Cohen, D.; Dolja, V.V.; Fuchs, M.F.; Golino, D.A.; Jooste, A.E.C.; Martelli, G.P.; et al. Grapevine leafroll-associated virus 3. Front. Microbiol. 2013, 4, 82. [Google Scholar] [CrossRef] [PubMed]
- Cabaleiro, C.; Segura, A.; Garcia-Berrios, J.J. Effects of grapevine leafroll-associated virus 3 on the physiology and must of Vitis vinifera L. cv. Albarino following contamination in the field. Am. J. Enol. Vitic. 1999, 50, 40–44. [Google Scholar] [CrossRef]
- Mannini, F.; Mollo, A.; Credi, R. Field performance and wine quality modification in a clone of Vitis vinifera cv. Dolcetto after GLRaV-3 elimination. Am. J. Enol. Vitic. 2012, 63, 144–147. [Google Scholar] [CrossRef]
- Endeshaw, S.T.; Sabbatini, P.; Romanazzi, G.; Schilder, A.C.; Neri, D. Effects of grapevine leafroll associated virus 3 infection on growth, leaf gas exchange, yield and basic fruit chemistry of Vitis vinifera L. cv. Cabernet Franc. Sci. Hortic. 2014, 170, 228–236. [Google Scholar] [CrossRef]
- Almeida, R.; Daane, K.; Bell, V.; Blaisdell, G.K.; Cooper, M.; Herrbach, E.; Pietersen, G. Ecology and management of grapevine leafroll disease. Front. Microbiol. 2013, 4, 94. [Google Scholar] [CrossRef] [PubMed]
- Goszczynski, D.E.; Habili, N. Grapevine virus A variants of group II associated with Shiraz disease in South Africa are present in plants affected by Australian Shiraz disease, and have also been detected in the USA. Plant Pathol. 2012, 61, 205–214. [Google Scholar] [CrossRef]
- Goszczynski, D.E.; Du Preez, J.; Burger, J.T. Molecular divergence of grapevine virus A (GVA) variants associated with Shiraz disease in South Africa. Virus Res. 2008, 138, 105–110. [Google Scholar] [CrossRef]
- Corbett, M.K.; Wiid, J. Closterovirus-like particles in extracts from diseased grapevines. Phytopathol. Mediterr. 1985, 24, 91–100. [Google Scholar]
- Habili, N.; Scliefert, L. The increasing threat of grapevine virus A and its association with restricted spring growth in Australia. Aust. N. Z. Grapegrow. Winemak. 2001, 452, 22–26. [Google Scholar]
- Symons, B.; Habili, N. Grapevine virus A is associated with restricted growth in the spring. Aust. N. Z. Grapegrow. Winemak. 2000, 443, 17–19. [Google Scholar]
- Habili, N.; Randles, J.W. Descriptors for grapevine virus A-associated syndrome in Shiraz, Merlot and Ruby Cabernet in Australia, and its similarity to Shiraz disease in South Africa. Aust. N. Z. Grapegrow. Winemak. 2004, 488, 71–74. [Google Scholar]
- Habili, N. It’s not worth the gamble: Serious problems associated with top-working virus-infected vines. Aust. N. Z. Grapegrow. Winemak. 2003, 476, 84–89. [Google Scholar]
- Minafra, A.; Saldarelli, P.; Grieco, F.; Martelli, G.P. Nucleotide sequence of the 3′ terminal region of the RNA of two filamentous grapevine viruses. Arch. Virol. 1994, 137, 249–261. [Google Scholar] [CrossRef] [PubMed]
- Minafra, A.; Saldarelli, P.; Martelli, G.P. Grapevine virus A: Nucleotide sequence, genome organization, and relationship in the Trichovirus genus. Arch. Virol. 1997, 142, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Haviv, S.; Iddan, Y.; Goszczynski, D.E.; Mawassi, M. The ORF5 of grapevine virus A is involved in symptoms expression in Nicotiana benthamiana plants. Ann. Appl. Biol. 2012, 160, 181–190. [Google Scholar] [CrossRef]
- Galiakparov, N.; Tanne, E.; Mawassi, M.; Gafny, R.; Sela, I. ORF 5 of grapevine virus A encodes a nucleic acid-binding protein and affects pathogenesis. Virus Genes 2003, 27, 257–262. [Google Scholar] [CrossRef]
- Dry, P.R.; Coombe, B.G.; Anderson, C.J. Viticulture: Volume 2. Practices, 2nd ed.; Winetitles: Adelaide, Australia, 2004. [Google Scholar]
- Balijja, A.; Kvarnheden, A.; Turchetti, T. A non-phenol–chloroform extraction of double-stranded RNA from plant and fungal tissues. J. Virol. Methods 2008, 152, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Nassuth, A.; Pollari, E.; Helmeczy, K.; Stewart, S.; Kofalvi, S.A. Improved RNA extraction and one-tube RT-PCR assay for simultaneous detection of control plant RNA plus several viruses in plant extracts. J. Virol. Methods 2000, 90, 37–49. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J. Precipitation of RNA with Ethanol. Cold Spring Harb. Protoc. 2020, 101717, 89–91. [Google Scholar] [CrossRef] [PubMed]
- Krueger, F. Trim Galore, v. 0.4.2. 2012. Available online: https://github.com/FelixKrueger/TrimGalore (accessed on 20 February 2021).
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Bushnell, B. BBMap: A Fast, Accurate, Splice-Aware Aligner; Lawrence Berkeley National Lab (LBNL): Berkeley, CA, USA, 2014. [Google Scholar]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Muhire, B.M.; Varsani, A.; Martin, D.P. SDT: A virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE 2014, 9, e108277. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Goszczynski, D.E. Single-strand conformation polymorphism (SSCP), cloning and sequencing reveal a close association between related molecular variants of Grapevine virus A (GVA) and Shiraz disease in South Africa. Plant Pathol. 2007, 56, 755–762. [Google Scholar] [CrossRef]
- Goszczynski, D.E.; Jooste, A.E.C. Identification of grapevines infected with divergent variants of grapevine virus A using variant-specific RT-PCR. J. Virol. Methods 2003, 112, 157–164. [Google Scholar] [CrossRef]
- Martin, D.P.; Varsani, A.; Roumagnac, P.; Botha, G.; Maslamoney, S.; Schwab, T.; Kelz, Z.; Kumar, V.; Murrell, B. RDP5: A computer program for analyzing recombination in, and removing signals of recombination from, nucleotide sequence datasets. Virus Evol. 2021, 7, veaa087. [Google Scholar] [CrossRef]
- Martin, D.; Rybicki, E. RDP: Detection of recombination amongst aligned sequences. Bioinformatics 2000, 16, 562–563. [Google Scholar] [CrossRef]
- Sawyer, S.A. GENECONV: A Computer Package for the Statistical Detection of Gene Conversion; Department of Mathematics, Washington University in Louis: Washington, WA, USA, 1999. [Google Scholar]
- Posada, D.; Crandall, K.A. Evaluation of methods for detecting recombination from DNA sequences: Computer simulations. Proc. Natl. Acad. Sci. USA 2001, 98, 13757–13762. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.M. Analyzing the mosaic structure of genes. J. Mol. Evol. 1992, 34, 126–129. [Google Scholar] [CrossRef] [PubMed]
- Salminen, M.O.; Carr, J.K.; Burke, D.S.; McCutchan, F.E. Identification of breakpoints in intergenotypic recombinants of HIV type 1 by bootscanning. AIDS Res. Hum. Retrovir. 1995, 11, 1423–1425. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, M.J.; Armstrong, J.S.; Gibbs, A.J. Sister-scanning: A Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics 2000, 16, 573–582. [Google Scholar] [CrossRef]
- Boni, M.F.; Posada, D.; Feldman, M.W. An exact nonparametric method for inferring mosaic structure in sequence triplets. Genetics 2007, 176, 1035–1047. [Google Scholar] [CrossRef]
- Rast, H.E.; James, D.; Habili, N.; Masri, S.A. Genome organisation and characterization of a novel variant of grapevine leafroll-associated virus 3. In Proceedings of the 17th Meeting of the International Council for the Study of Virus and Virus-like Diseases of the Grapevine, Davis, CA, USA, 7–14 October 2012; pp. 61–63. [Google Scholar]
- Habili, N.; Cameron, I.; Randles, J. A mild strain of grapevine leafroll-associated virus 3 is present in desirable clones of Crimson seedless table grapes in Western Australia. In Proceedings of the 16th Meeting of the International Council for the Study of Virus and Virus-like Diseases of the Grapevine, Dijon, France, 31 August–4 September 2009; pp. 237–238. [Google Scholar]
- Diaz-Lara, A.; Klaassen, V.; Stevens, K.; Sudarshana, M.R.; Rowhani, A.; Maree, H.J.; Chooi, K.M.; Blouin, A.G.; Habili, N.; Song, Y.; et al. Characterization of grapevine leafroll-associated virus 3 genetic variants and application towards RT-qPCR assay design. PLoS ONE 2018, 13, e0208862. [Google Scholar] [CrossRef]
- Gouveia, P.; Santos, M.T.; Eiras-Dias, J.E.; Nolasco, G. Five phylogenetic groups identified in the coat protein gene of grapevine leafroll-associated virus 3 obtained from Portuguese grapevine varieties. Arch. Virol. 2011, 156, 413–420. [Google Scholar] [CrossRef]
- Ghanem-Sabanadzovic, A.; Sabanadzovic, S.; Uyemoto, J.K.; Golino, D.; Rowhani, A. A putative new ampelovirus associated with grapevine leafroll disease. Arch. Virol. 2010, 155, 1871–1876. [Google Scholar] [CrossRef]
- Reynard, J.-S.; Schneeberger, P.H.H.; Frey, J.E.; Schaerer, S. Biological, serological, and molecular characterization of a highly divergent strain of grapevine leafroll-associated virus 4 causing grapevine leafroll disease. Phytopathology 2015, 105, 1262–1269. [Google Scholar] [CrossRef]
- Osman, F.; Leutenegger, C.; Golino, D.; Rowhani, A. Comparison of low-density arrays, RT-PCR and real-time TaqMan® RT-PCR in detection of grapevine viruses. J. Virol. Methods 2008, 149, 292–299. [Google Scholar] [CrossRef]
- Čepin, U.; Gutiérrez-Aguirre, I.; Balažic, L.; Pompe-Novak, M.; Gruden, K.; Ravnikar, M. A one-step reverse transcription real-time PCR assay for the detection and quantitation of grapevine fanleaf virus. J. Virol. Methods 2010, 170, 47–56. [Google Scholar] [CrossRef]
- Le Maguet, J.; Beuve, M.; Herrbach, E.; Lemaire, O. Transmission of six ampeloviruses and two vitiviruses to grapevine by Phenacoccus aceris. Phytopathology 2012, 102, 717–723. [Google Scholar] [CrossRef]
- Habili, N.; Wu, Q.; Pagay, V. Virus-associated Shiraz disease may lead Shiraz to become an endangered variety in Australia. Wine Vitic. J. 2016, 31, 47–50. [Google Scholar]
- Minafra, A.; Hadidi, A. Sensitive detection of grapevine virus A, B, or leafroll-associated III from viruliferous mealybugs and infected tissue by cDNA amplification. J. Virol. Methods 1994, 47, 175–187. [Google Scholar] [CrossRef]
- Fazeli, C.F.; Rezaian, M.A. Nucleotide sequence and organization of ten open reading frames in the genome of grapevine leafroll-associated virus 1 and identification of three subgenomic RNAs. J. Gen. Virol. 2000, 81, 605–615. [Google Scholar] [CrossRef] [PubMed]
- Bertazzon, N.; Angelini, E. Advances in the detection of grapevine leafroll-associated virus 2 variants. J. Plant Pathol. 2004, 86, 283–290. [Google Scholar]
- Habili, N.; Fazeli, C.F.; Ewart, A.; Hamilton, R.; Cirami, R.; Saldarelli, P.; Minafra, A.; Rezaian, M.A. Natural spread and molecular analysis of grapevine leafroll-associated virus 3 in Australia. Phytopathology 1995, 85, 1418–1422. [Google Scholar] [CrossRef]
- Good, X.; Monis, J. Partial genome organization, identification of the coat protein gene, and detection of Grapevine leafroll-associated virus-5. Phytopathology 2001, 91, 274–281. [Google Scholar] [CrossRef]
- Mahfoudhi, N.; Habili, N.; Masri, S.A.; Dhouibi, M.H. First report on the occurrence of grapevine leafroll-associated viruses 5 and 9 in Tunisian grapevines. Plant Dis. 2007, 91, 1359. [Google Scholar] [CrossRef]
- Wu, Q.; Kehoe, M.; Kinoti, W.M.; Wang, C.; Rinaldo, A.; Tyerman, S.; Habili, N.; Constable, F.E. First report of grapevine rupestris vein feathering virus in grapevine in Australia. Plant Dis. 2020, 105, 515. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-P.; Uyemoto, J.K.; Golino, D.A.; Rowhani, A. Nucleotide sequence and RT-PCR detection of a virus associated with grapevine rupestris stem-pitting disease. Phytopathology 1998, 88, 1231–1237. [Google Scholar] [CrossRef] [PubMed]
- MacKenzie, D.J.; McLean, M.A.; Mukerji, S.; Green, M. Improved RNA extraction from woody plants for the detection of viral pathogens by reverse transcription-polymerase chain reaction. Plant Dis. 1997, 81, 222–226. [Google Scholar] [CrossRef] [PubMed]
- National Center of Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 13 February 2023).


| Location | Year Selected | Variety and Clone | Year Established | Grafted on Rootstock? | Total Grapevines | No. of SD Grapevines | No. of LRD Grapevines | No. of Asymptomatic Grapevines | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RT-PCR | HTS | RT-PCR | HTS | RT-PCR | HTS | RT-PCR | HTS | |||||
| Langhorne Creek 1 (LC) | 2018 | Shiraz BVRC12 | 2004 | Chardonnay | 30 | 14 | 15 | 9 | N/A | 0 | 15 | 5 |
| Willunga (WIL) | 2018&2020 | Shiraz BVRC12 | 2004 | No | 80 4 | 24 | 8 | 13 | 7 | 7 | 15 | 4 |
| 2020 | Cabernet Sauvignon SA125 2 | 2004 | No | 4 | 4 | N/A | N/A | 4 | 4 | N/A | N/A | |
| 2020 | Merlot unknown clone 2 | Unknown | Unknown | 1 | 1 | 1 | 1 | N/A | N/A | N/A | N/A | |
| Barossa Valley 3 (BV) | 2018 | heritage Shiraz, unknown clone | ≈1900σ | No | 6 | 3 | N/A | N/A | 3 | 2 | 3 | 1 |
| Coombe vineyard (CV) | 2020 | Shiraz 2 BVRC12 | 1993 | No | 2 | 2 | N/A | N/A | N/A | N/A | 2 | 2 |
| 2020 | Cabernet Sauvignon SA125 2 | 1993 | No | 2 | 2 | N/A | N/A | 2 | 2 | N/A | N/A | |
| Sample ID 1 | Total Number of Grapevines with Symptom and Virus Status Combination | Symptoms 2 | GVA (I,II,II), GLRaV-1 (1), GLRaV-3 (3) and GLRaV-4 (4) Status by RT-PCR 3 | GVA (I,II,III), GLRaV-1 (1), GLRaV-3 (3) and GLRaV-4 (4) Status by Meta-HTS 4 |
|---|---|---|---|---|
| WIL19, WIL22 | 2 | Asymptomatic | 4 | 4 |
| WIL17, LC16, LC18, LC20, LC24, LC27, BV6, CVP5, CVP6 | 9 | None | None | |
| WIL24 | 1 | None | 4 | |
| WIL14, 15 | 2 | LRD | 3, 4 | 3, 4 |
| WIL9, WIL10, WIL11, WIL12 | 4 | III, 3, 4 | III, 3, 4 | |
| Cabw1, Cabw2, Cabw12 | 3 | I/II, III, 3, 4 | II and III, 3, 4 | |
| Cabw11 | 1 | I/II, 3, 4 | II, 3, 4 | |
| WIL13 * | 1 | 3, 4 | III, 3, 4 | |
| BV1, 3 | 2 | Mild LRD | I/II, 1 | I, 1 |
| CabSA125_R3V30, CabSA125_R3V44 | 2 | I/II, III, 4 | II and III, 4 | |
| WIL8 | 1 | SD | I/II, III, 3, 4 | II and III, 3, 4 |
| LC10, LC11, LC12, LC13, LC14 | 5 | I/II, 4 | II, 4 | |
| WIL48, WIL49, WIL50, WIL53 | 4 | 1/II, 3 | II, 3 | |
| WIL4, WIL5, WIL6, WIL7 | 4 | I/II, 3, 4 | II, 3, 4 | |
| LC5, LC6, LC7, LC9 * | 4 | I/II | II, 4 | |
| WIL47 * | 1 | I/II, 3 | III, 3 | |
| Melort1 * | 1 | I/II, III, 3 | II and III, 3, 4 | |
| WIL1, WIL2, WIL3 * | 3 | I/II, III, 3, 4 | II andIII, 4 |
| Genes Compared 1 | I 2 | II 2 | III 2 | ||||
|---|---|---|---|---|---|---|---|
| Australian Isolates Only | All Isolates | Australian Isolates Only | All Isolates | Australian Isolates Only | All Isolates | ||
| Whole genome | N/A 3 | 76.33–99.80 | 91.45–99.90 | 79.92–99.90 | 94.85–99.94 | 76.52–99.94 | |
| RdRp | RdRp nt | N/A | 74.96–99.88 | 90.65–99.90 | 79.53–99.90 | 94.12–99.96 | 74.76–99.96 |
| RdRp aa | N/A | 84.72–99.53 | 96.60–100 | 90.28–100 | 97.25–100 | 86.59–100 | |
| MP | MP nt | 99.88 4 | 77.30–100 | 91.07–100 | 83.03–100 | 96.31–100 | 81.67–100 |
| MP aa | 100 4 | 78.85–100 | 92.47–100 | 89.25–100 | 95.70–100 | 87.46–100 | |
| CP | CP nt | 98.49–99.83 | 80.23–99.66 | 94.30–100 | 84.09–100 | 97.82–100 | 77.39–100 |
| CP aa | 98.99–100 | 79.90–100 | 96.98–100 | 88.94–100 | 98.99–100 | 82.41–100 | |
| RB | RB nt | 95.24–100 5 | 86.08–100 5 | See GVAI | 97.43–100 | 91.58–100 | |
| RB aa | 94.51–100 5 | 84.62–100 5 | 97.80–100 | 92.31–100 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Q.; Habili, N.; Kinoti, W.M.; Tyerman, S.D.; Rinaldo, A.; Zheng, L.; Constable, F.E. A Metagenomic Investigation of the Viruses Associated with Shiraz Disease in Australia. Viruses 2023, 15, 774. https://doi.org/10.3390/v15030774
Wu Q, Habili N, Kinoti WM, Tyerman SD, Rinaldo A, Zheng L, Constable FE. A Metagenomic Investigation of the Viruses Associated with Shiraz Disease in Australia. Viruses. 2023; 15(3):774. https://doi.org/10.3390/v15030774
Chicago/Turabian StyleWu, Qi, Nuredin Habili, Wycliff M. Kinoti, Stephen D. Tyerman, Amy Rinaldo, Linda Zheng, and Fiona E. Constable. 2023. "A Metagenomic Investigation of the Viruses Associated with Shiraz Disease in Australia" Viruses 15, no. 3: 774. https://doi.org/10.3390/v15030774
APA StyleWu, Q., Habili, N., Kinoti, W. M., Tyerman, S. D., Rinaldo, A., Zheng, L., & Constable, F. E. (2023). A Metagenomic Investigation of the Viruses Associated with Shiraz Disease in Australia. Viruses, 15(3), 774. https://doi.org/10.3390/v15030774







