Natural History of DNA-Dependent DNA Polymerases: Multiple Pathways to the Origins of DNA
Abstract
1. Introduction
2. Materials and Methods
2.1. Obtaining the Analyzed Sequences
2.2. Alignment and Phylogeny
3. Results and Discussion
On the Origin of DNA-Dependent DNA Polymerase
4. Bacterial DNA-Dependent DNA Polymerase
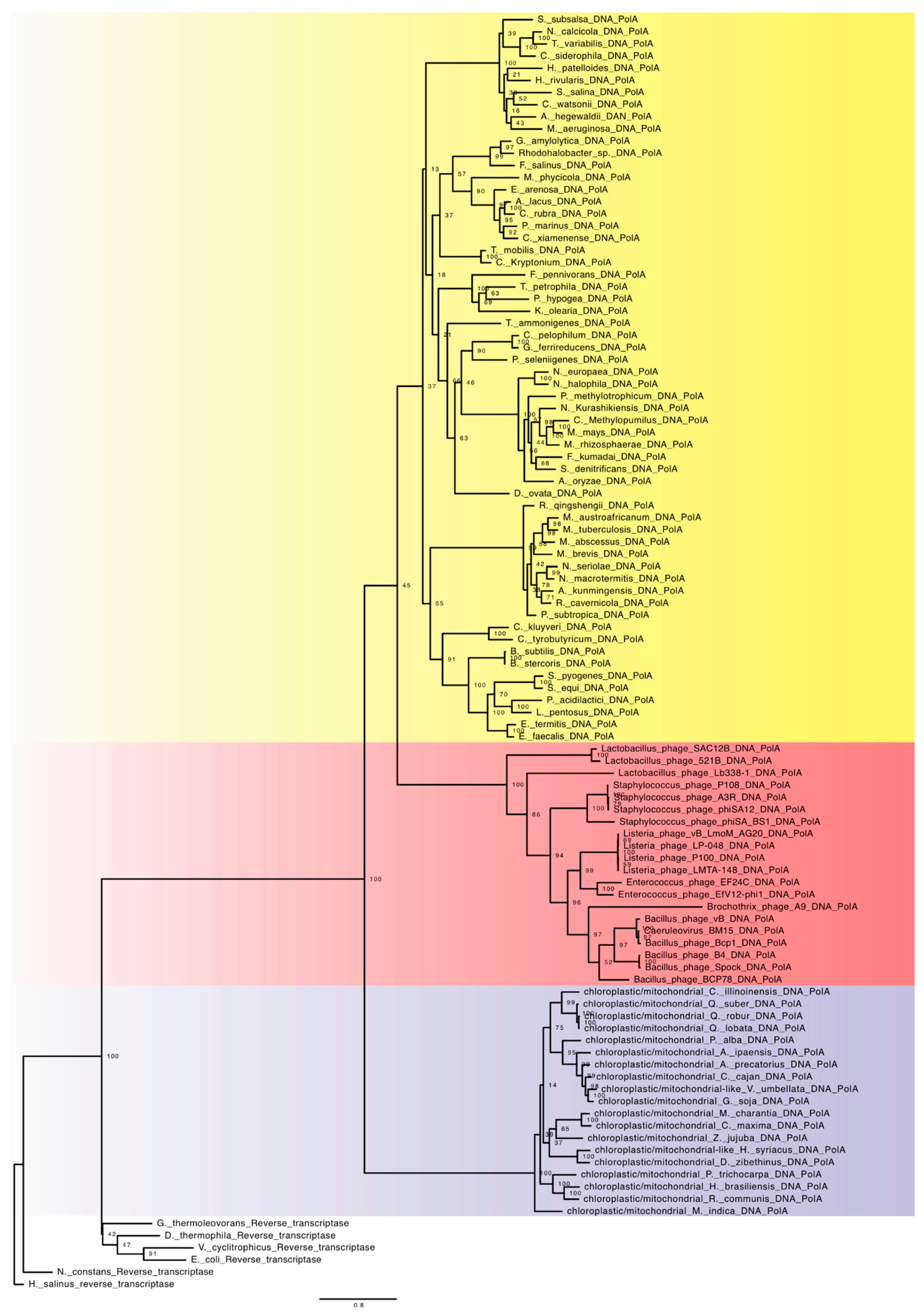
4.1. Family A
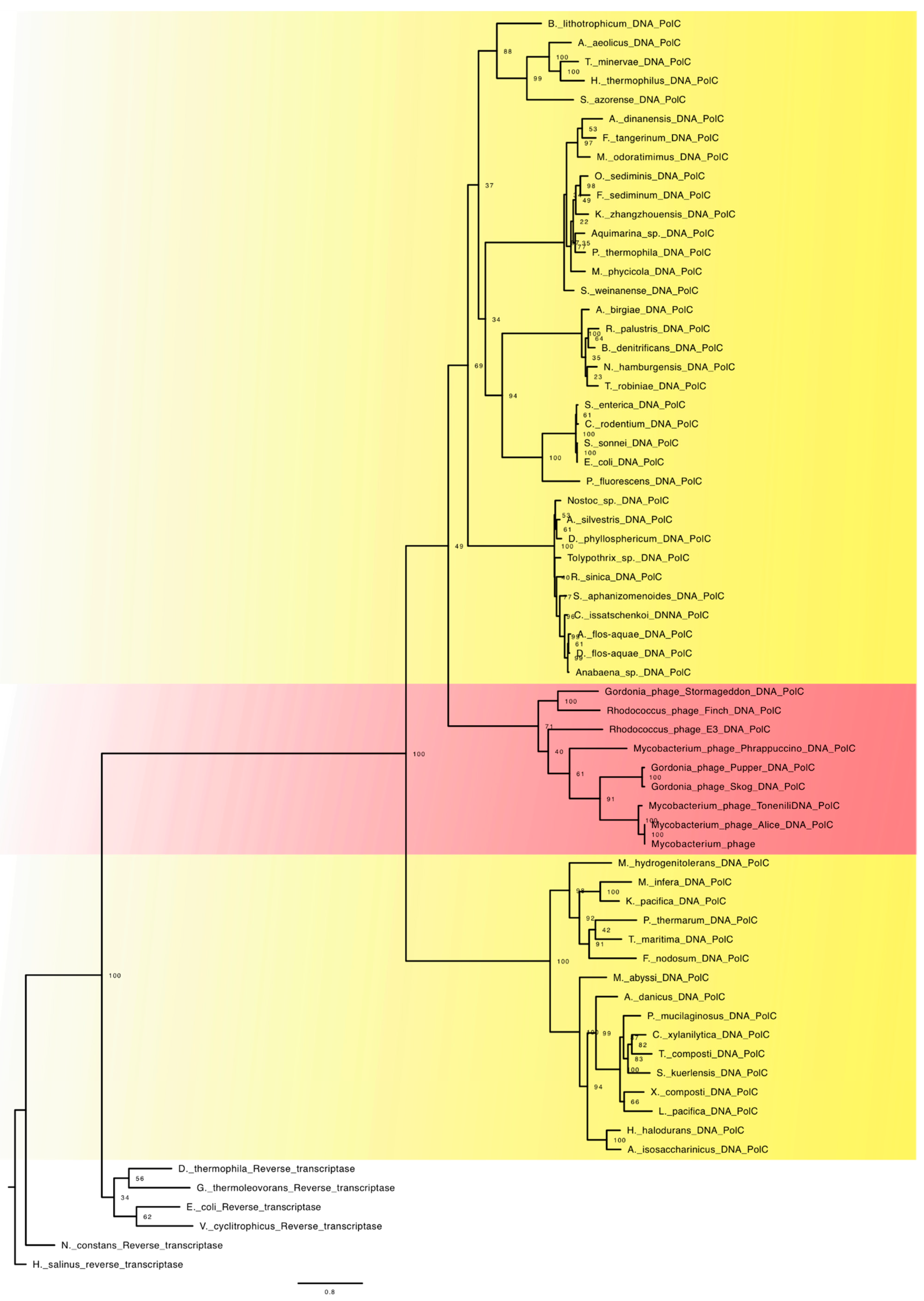
4.2. Family C
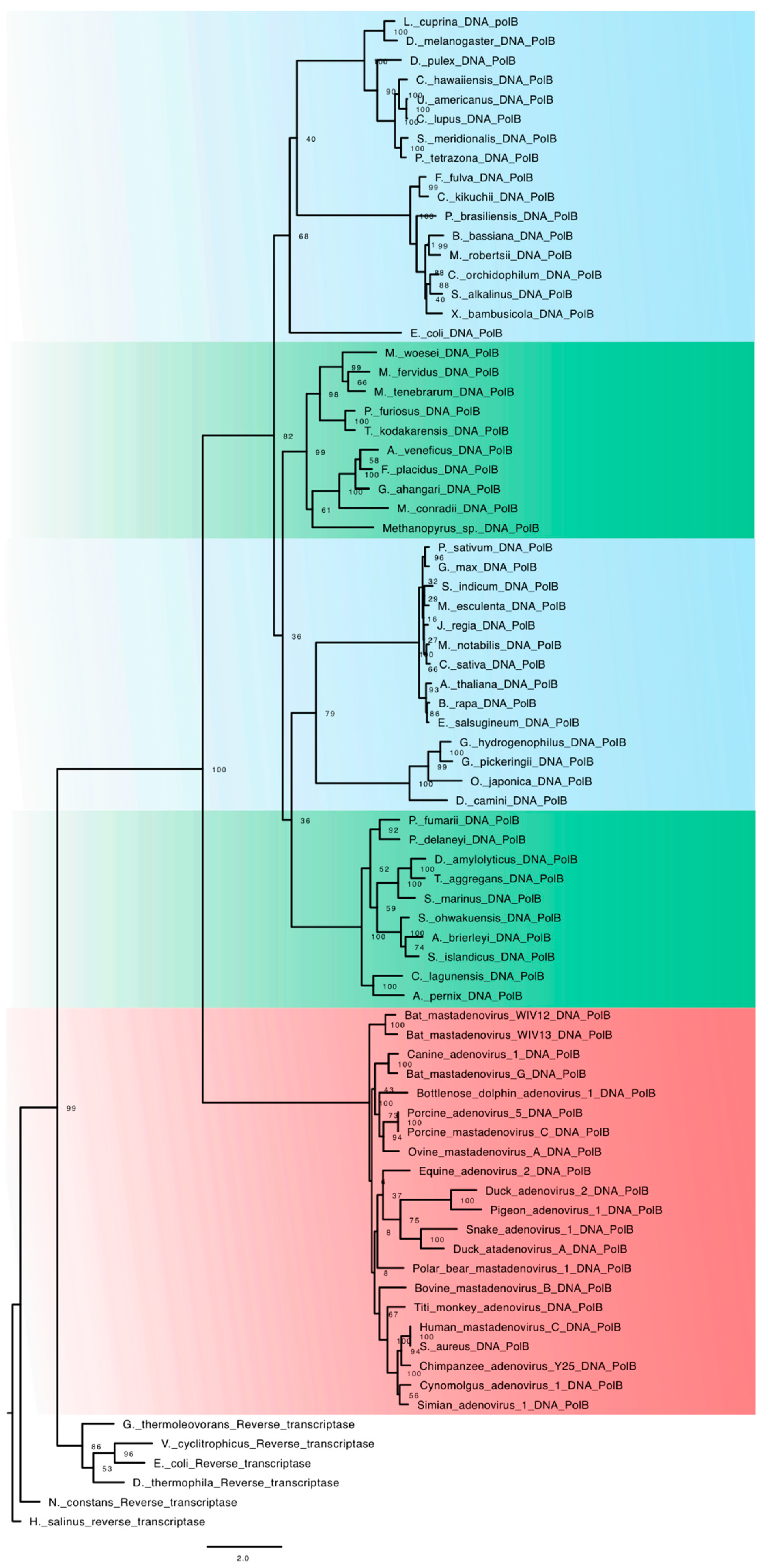
5. Archaeal and Eukaryotic DNA-Dependent DNA Polymerase
Family B
6. Viral DNA-Dependent DNA Polymerase
7. Last Considerations
Proposal of a Scenario for the Emergence of the DNA Genome in Cellular Lineages
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yarus, M. Boundaries for an RNA world. Curr. Opin. Chem. Biol. 1999, 3, 260–267. [Google Scholar] [CrossRef]
- Lazcano, A. The biochemical roots of the RNA world: From zymonucleic acid to ribozymes. Hist. Philos. Life Sci. 2012, 34, 407–423. [Google Scholar] [PubMed]
- Müller, U.F. Re-creating an RNA world. Cell. Mol. Life Sci. 2006, 63, 1278–1293. [Google Scholar] [CrossRef] [PubMed]
- Orgel, L.E. Prebiotic chemistry and the origin of the RNA world. Crit. Rev. Biochem. Mol. Biol. 2004, 39, 99–123. [Google Scholar] [CrossRef] [PubMed]
- Forterre, P. Genomics and early cellular evolution. The origin of the DNA world. Comptes Rendus Académie Sci. III 2001, 324, 1067–1076. [Google Scholar] [CrossRef]
- Forterre, P. The two ages of the RNA world, and the transition to the DNA world: A story of viruses and cells. Biochimie 2005, 87, 793–803. [Google Scholar] [CrossRef]
- Di Giulio, M. The late appearance of DNA, the nature of the LUCA and ancestors of the domains of life. Biosystems 2021, 202, 104330. [Google Scholar] [CrossRef]
- Leipe, D.D.; Aravind, L.; Koonin, E.V. Did DNA replication evolve twice independently? Nucleic Acids Res. 1999, 27, 3389–3401. [Google Scholar] [CrossRef]
- Davis, B.K. Molecular evolution before the origin of species. Prog. Biophys. Mol. Biol. 2002, 79, 77–133. [Google Scholar] [CrossRef]
- Filée, J.; Forterre, P.; Sen-Lin, T.; Laurent, J. Evolution of DNA polymerase families: Evidences for multiple gene exchange between cellular and viral proteins. J. Mol. Evol. 2002, 54, 763–773. [Google Scholar] [CrossRef]
- Mushegian, A. Gene content of LUCA, the last universal common ancestor. Front. Biosci. 2008, 13, 4657–4666. [Google Scholar] [CrossRef]
- Goldman, A.D.; Bernhard, T.M.; Dolzhenko, E.; Landweber, L.F. LUCApedia: A database for the study of ancient life. Nucleic Acids Res. 2013, 41, D1079–D1082. [Google Scholar] [CrossRef]
- Glansdorff, N.; Xu, Y.; Labedan, B. The last universal common ancestor: Emergence, constitution and genetic legacy of an elusive forerunner. Biol. Direct. 2008, 3, 29. [Google Scholar] [CrossRef] [PubMed]
- Di Giulio, M. The last universal common ancestor (LUCA) and the ancestors of archaea and bacteria were progenotes. J. Mol. Evol. 2011, 72, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Di Giulio, M. LUCA as well as the ancestors of archaea, bacteria and eukaryotes were progenotes: Inference from the distribution and diversity of the reading mechanism of the AUA and AUG codons in the domains of life. Biosystems 2020, 198, 104239. [Google Scholar] [CrossRef] [PubMed]
- Di Giulio, M. The RNase P, LUCA, the ancestors of the life domains, the progenote, and the tree of life. Biosystems 2022, 212, 104604. [Google Scholar] [CrossRef]
- Di Giulio, M. The origins of the cell membrane, the progenote, and the universal ancestor (LUCA). Biosystems 2022, 222, 104799. [Google Scholar] [CrossRef]
- Farias, S.T.; Jose, M.V.; Prosdocimi, F. Is it possible that cells have had more than one origin? Biosystems 2021, 202, 104371. [Google Scholar] [CrossRef]
- Di Giulio, M. The universal ancestor, the deeper nodes of the tree of life, and the fundamental types of primary cells (cellular domains). J. Theor. Biol. 2019, 460, 142–143. [Google Scholar] [CrossRef]
- Forterre, P. The origin of viruses and their possible roles in major evolutionary transitions. Virus Res. 2006, 117, 5–16. [Google Scholar] [CrossRef]
- Dos Santos Junior, P.A.; José, M.V.; Farias, S.T. From RNA to DNA: Insights about the transition of informational molecule in the biological systems based on the structural proximity between the polymerases. Biosystems 2021, 206, 104442. [Google Scholar] [CrossRef] [PubMed]
- Raia, P.; Delarue, M.; Sauguet, L. An updated structural classification of replicative DNA polymerases. Biochem. Soc. Trans. 2019, 47, 239–249. [Google Scholar] [CrossRef] [PubMed]
- Mönttinen, H.A.; Ravantti, J.J.; Stuart, D.I.; Poranen, M.M. Automated structural comparisons clarify the phylogeny of the right-hand-shaped polymerases. Mol. Biol. Evol. 2014, 31, 2741–2752. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Miller, M.A.; Schwartz, T.; Pickett, B.E.; He, S.; Klem, E.B.; Scheuermann, R.H.; Passarotti, M.; Kaufman, S.; O’Leary, M.A. A RESTful API for Access to Phylogenetic Tools via the CIPRES Science Gateway. Evol. Bioinform. Online 2015, 16, 43–48. [Google Scholar] [CrossRef]
- Farias, S.T.; Dos Santos Junior, A.P.; Rêgo, T.G.; José, M.V. Origin and Evolution of RNA-Dependent RNA Polymerase. Front. Genet. 2017, 20, 125. [Google Scholar] [CrossRef]
- Menéndez-Arias, L. Mutation rates and intrinsic fidelity of retroviral reverse transcriptases. Viruses 2009, 1, 1137–1165. [Google Scholar] [CrossRef]
- Bao, K.; Cohen, S.N. Reverse transcriptase activity innate to DNA polymerase I and DNA topoisomerase I proteins of Streptomyces telomere complex. Proc. Natl. Acad. Sci. USA 2004, 101, 14361–14366. [Google Scholar] [CrossRef]
- Choi, W.S.; He, P.; Pothukuchy, A.; Gollihar, J.; Ellington, A.D.; Yang, W. How a B family DNA polymerase has been evolved to copy RNA. Proc. Natl. Acad. Sci. USA 2020, 117, 21274–21280. [Google Scholar] [CrossRef]
- Ricchetti, M.; Buc, H.E. coli DNA polymerase I as a reverse transcriptase. EMBO J. 1993, 12, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Albà, M. Replicative DNA polymerases. Genome Biol. 2001, 2, reviews3002.1. [Google Scholar] [CrossRef] [PubMed]
- Taanman, J.W. The mitochondrial genome: Structure, transcription, translation and replication. Biochim. Biophys. Acta 1999, 1410, 103–123. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.P.; Ito, J. DNA polymerase C of the thermophilic bacterium Thermus aquaticus: Classification and phylogenetic analysis of the family C DNA polymerases. J. Mol. Evol. 1999, 48, 756–769. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, T. Days weaving the lagging strand synthesis of DNA-A personal recollection of the discovery of Okazaki fragments and studies on discontinuous replication mechanism. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2017, 93, 322–338. [Google Scholar] [CrossRef]
- Kazlauskas, D.; Krupovic, M.; Guglielmini, J.; Forterre, P.; Venclovas, Č. Diversity and evolution of B-family DNA polymerases. Nucleic Acids Res. 2020, 48, 10142–10156. [Google Scholar] [CrossRef]
- Nasir, A.; Sun, F.J.; Kim, K.M.; Caetano-Anollés, G. Untangling the origin of viruses and their impact on cellular evolution. Ann. N. Y. Acad. Sci. 2015, 1341, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Krupovic, M.; Koonin, E.V. Multiple origins of viral capsid proteins from cellular ancestors. Proc. Natl. Acad. Sci. USA 2017, 114, E2401–E2410. [Google Scholar] [CrossRef]
- Sinkovics, J.; Horvath, J.; Horak, A. The origin and evolution of viruses (a review). Acta Microbiol. Immunol. Hung. 1998, 45, 349–390. [Google Scholar]
- Farias, S.T.; Jheeta, S.; Prosdocimi, F. Viruses as a survival strategy in the armory of life. Hist. Philos. Life Sci. 2019, 41, 45. [Google Scholar] [CrossRef]
- Edgell, D.R.; Doolittle, W.F. Archaea and the origin(s) of DNA replication proteins. Cell 1997, 89, 995–998. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Farias, S.T.; Furtado, A.N.M.; dos Santos Junior, A.P.; José, M.V. Natural History of DNA-Dependent DNA Polymerases: Multiple Pathways to the Origins of DNA. Viruses 2023, 15, 749. https://doi.org/10.3390/v15030749
de Farias ST, Furtado ANM, dos Santos Junior AP, José MV. Natural History of DNA-Dependent DNA Polymerases: Multiple Pathways to the Origins of DNA. Viruses. 2023; 15(3):749. https://doi.org/10.3390/v15030749
Chicago/Turabian Stylede Farias, Sávio Torres, Ariadne Nobrega Marinho Furtado, Ariosvaldo Pereira dos Santos Junior, and Marco V. José. 2023. "Natural History of DNA-Dependent DNA Polymerases: Multiple Pathways to the Origins of DNA" Viruses 15, no. 3: 749. https://doi.org/10.3390/v15030749
APA Stylede Farias, S. T., Furtado, A. N. M., dos Santos Junior, A. P., & José, M. V. (2023). Natural History of DNA-Dependent DNA Polymerases: Multiple Pathways to the Origins of DNA. Viruses, 15(3), 749. https://doi.org/10.3390/v15030749





