Antagonisms of ASFV towards Host Defense Mechanisms: Knowledge Gaps in Viral Immune Evasion and Pathogenesis
Abstract
1. Introduction
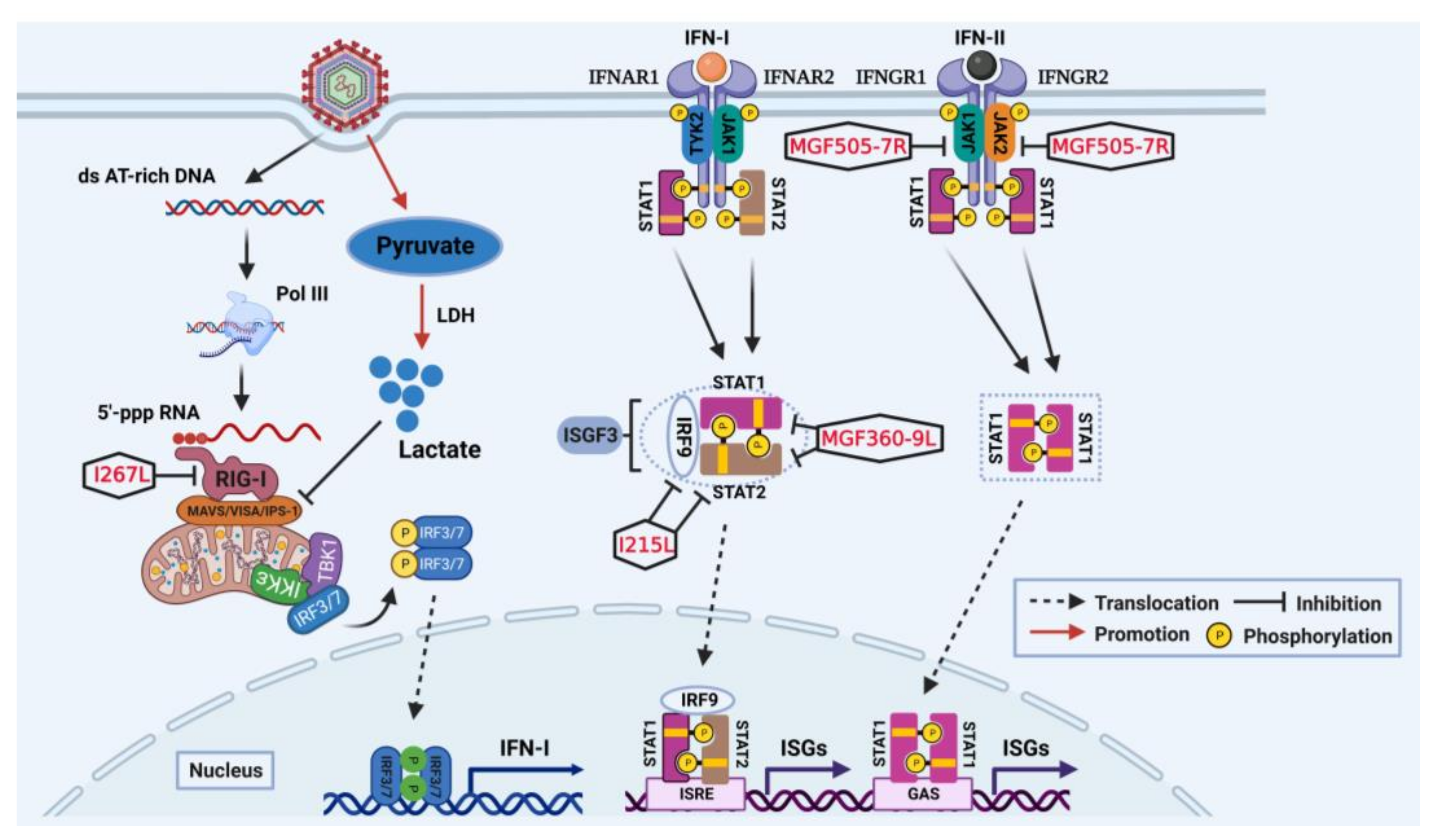
2. Suppression of IFN-I Production and IFN-Induced Antiviral Responses
2.1. Impairment on IFN-I Production Targeting cGAS-STING Axis
| ASFV Protein | Functional Site/Domain | Key Target | Underlying Mechanism | Reference |
|---|---|---|---|---|
| pEP364R, pC129R | Amino acids Y76 and N78 in pep364r | cGAMP | Selectively cleave cgamp | [41] |
| pE184L | Amino acids 1–20 in pe184l | STING | Impair STING oligomerization and dimerization | [43] |
| p17 (pD117L) | Amino acids 39–59 in p17 | STING | Interfere with the recruitment of TBK1 and Ikkε | [44] |
| pDP96R | Amino acids 30–96 in pdp96r | TBK1 | Suppress the phosphorylation of TBK1 | [49] |
| pI215L (UBCv1) | Unknown | TBK1 | Inhibit K63-linked polyubiquitination of TBK1 | [50] |
| pA137R | Unknown | TBK1 | Mediate the degradation of TBK1 | [52] |
| pM1249L | Unknown | TBK1; IRF3 | Suppress the phosphorylation of TBK1; mediate the degradation of IRF3 | [59] |
| pE120R | Amino acids 72–73 in pe120r | IRF3 | Suppress the phosphorylation of IRF3 | [58] |
| pE301R | Amino acids 1–200 in pe301r | IRF3 | Suppress the phosphorylation of IRF3 | [56] |
| pI226R | Unknown | IRF3 | Suppress the phosphorylation of IRF3 | [57] |
| pS273R | Amino acids 1–20 and 256–273 in ps273r | Ikkε | Affect the sumoylation of Ikkε | [53] |
| pMGF360-11L | Amino acids 167–353 in pmgf360-11L | TBK1; IRF7 | Mediate the degradation of TBK1 and IRF7 | [62] |
| pMGF505-11R | Amino acids 1–191 and 182–360 in pmgf505-11R | STING | Mediate the degradation of STING | [63] |
| pMGF360-14L | Unknown | IRF3 | Mediate the degradation of IRF3 | [64] |
| pMGF-505-7R | Unknown | TBK1; IRF7 | Mediate the degradation of TBK1 and IRF7 | [66] |
| STING | Mediate the degradation of STING | [67] |
2.2. Impairment on IFN-I Production Targeting RIG-I-MAVS Axis
2.3. Impairment on IFN-Induced Antiviral Responses Targeting JAK-STAT Pathway
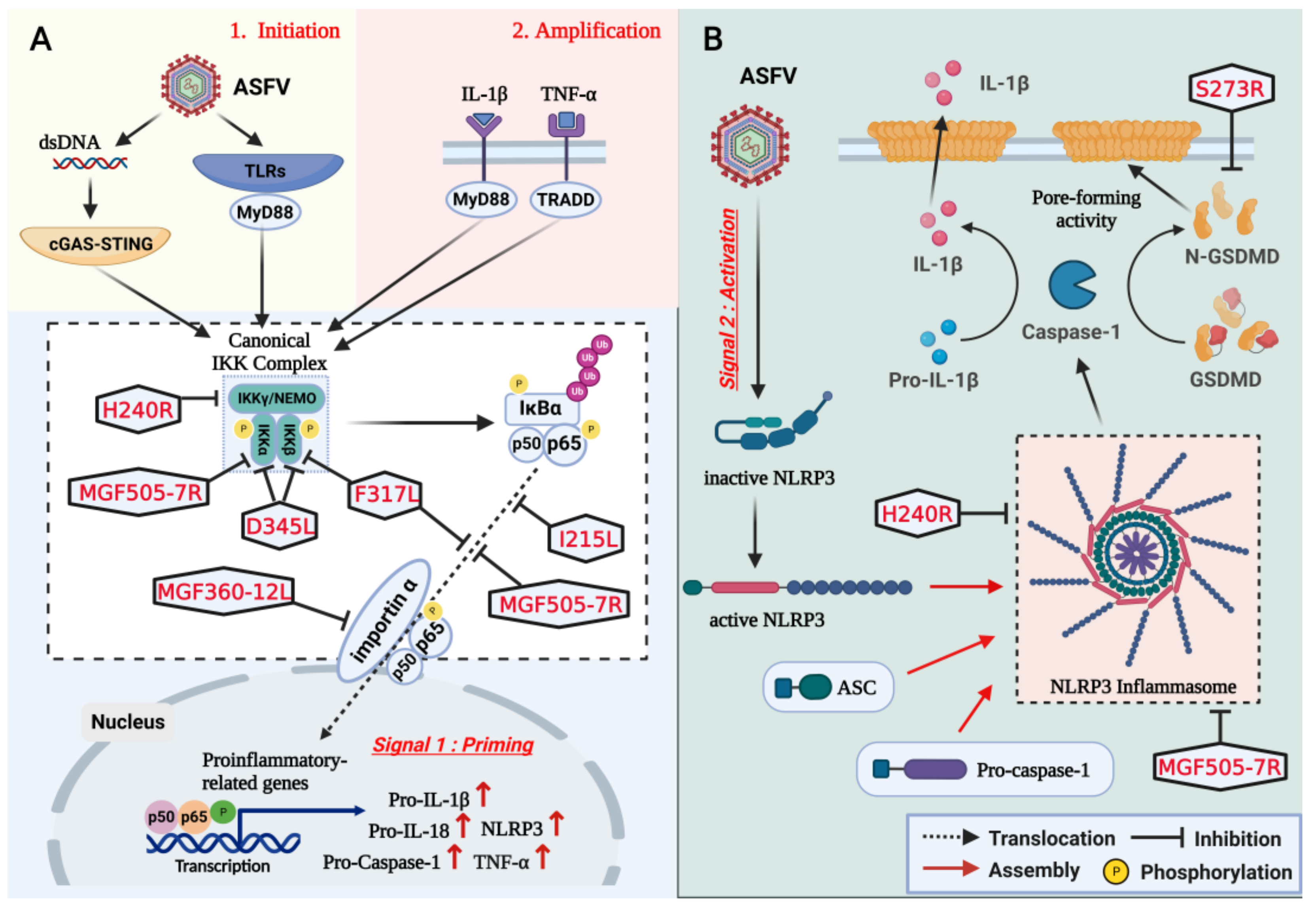
3. Inhibition of NLRP3 Inflammasome Activation and GSDMD-Mediated Pyroptosis
4. Effects of Immune Evasion on Viral Replication and Virulence
| LAV Candidates vs. Parental Strains | Viremia (Replication) | Clinical Signs (Pathogenicity) | Death (Virulence) | Reference |
|---|---|---|---|---|
| HLJ/18 | Unknown | Yes, 6/6 | Yes, 6/6 | [112] |
| HLJ/18-ΔH240R | Unknown | No, 0/6 | No, 0/6 | |
| HLJ/18 | Yes, high | Unknown | Yes, 5/5 | [100] |
| HLJ/18-Δ7R | Yes, medium | Unknown | Yes, 2/5 | |
| Georgia/2010 | Yes, high | Yes, 5/5 | Yes, 5/5 | [115] |
| Georgia/2010-ΔE184L | Yes, medium to high | Yes, 2/5 | Yes, 1/5 | |
| SY18 | Yes, high | Yes, 5/5 | Yes, 5/5 | [116] |
| SY18-ΔI226R | Yes, medium to high | No, 0/5 | No, 0/5 | |
| Georgia/2010 | Yes, high | Yes, 5/5 | Yes, 5/5 | [117] |
| Georgia/2010-ΔA137R | Yes, medium to high | No, 0/5 | No, 0/5 | |
| CN/GS/2018 | Yes, high | Yes, 6/6 | Yes, 6/6 | [114] |
| CN/GS/2018-Δ9L/Δ7R | Yes, low | No, 0/6 | No, 0/6 |
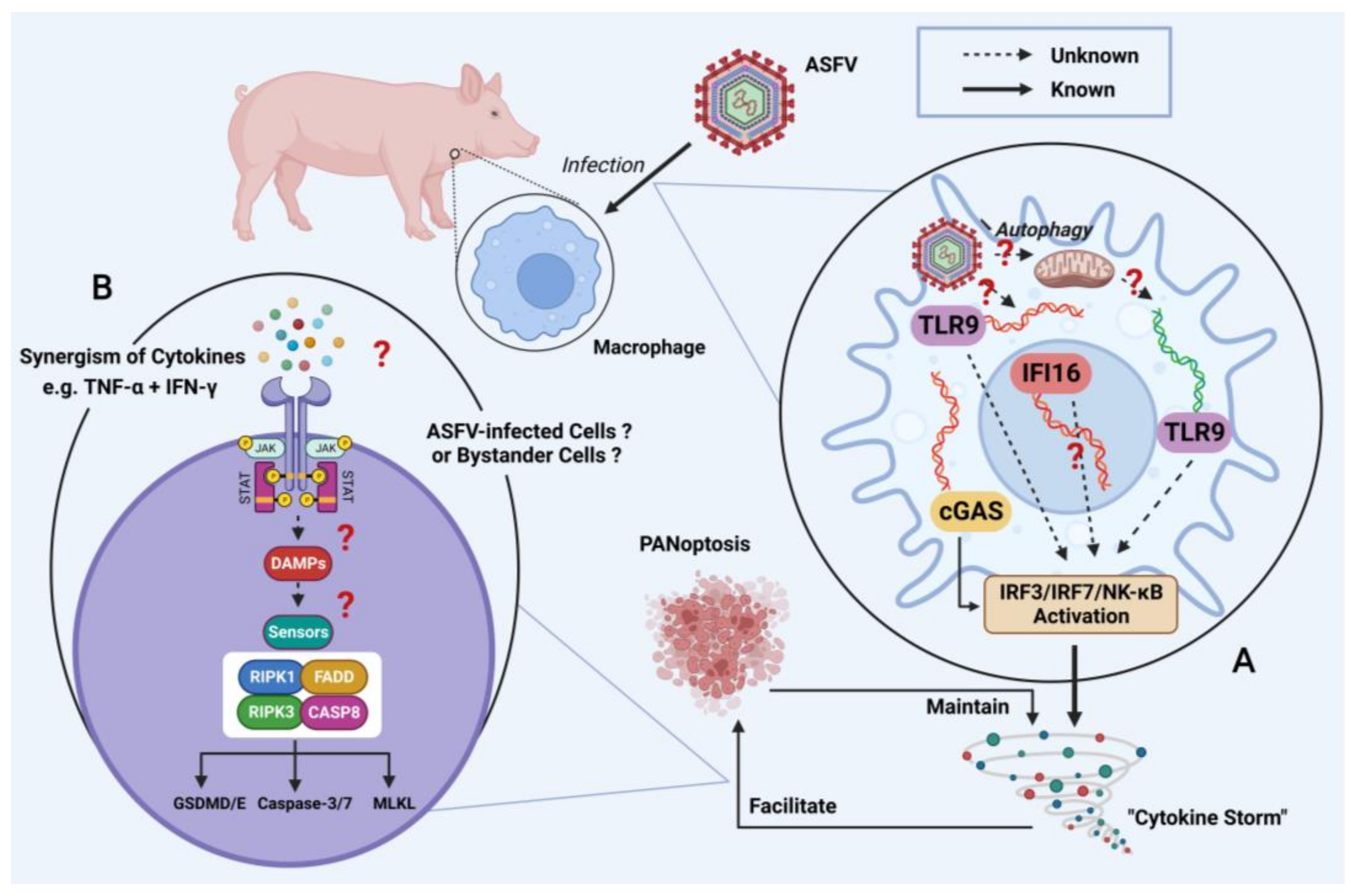
5. Future Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Qu, H.; Ge, S.; Zhang, Y.; Wu, X.; Wang, Z. A systematic review of genotypes and serogroups of African swine fever virus. Virus Genes 2022, 58, 77–87. [Google Scholar] [CrossRef]
- Zhou, X.; Li, N.; Luo, Y.; Liu, Y.; Miao, F.; Chen, T.; Zhang, S.; Cao, P.; Li, X.; Tian, K.; et al. Emergence of African Swine Fever in China, 2018. Transbound. Emerg. Dis. 2018, 65, 1482–1484. [Google Scholar] [CrossRef]
- Ge, S.; Li, J.; Fan, X.; Liu, F.; Li, L.; Wang, Q.; Ren, W.; Bao, J.; Liu, C.; Wang, H.; et al. Molecular Characterization of African Swine Fever Virus, China, 2018. Emerg. Infect. Dis. 2018, 24, 2131–2133. [Google Scholar] [CrossRef]
- Zhao, D.; Liu, R.; Zhang, X.; Li, F.; Wang, J.; Zhang, J.; Liu, X.; Wang, L.; Zhang, J.; Wu, X.; et al. Replication and virulence in pigs of the first African swine fever virus isolated in China. Emerg. Microbes. Infect. 2019, 8, 438–447. [Google Scholar] [CrossRef]
- Sun, E.; Zhang, Z.; Wang, Z.; He, X.; Zhang, X.; Wang, L.; Wang, W.; Huang, L.; Xi, F.; Huangfu, H.; et al. Emergence and prevalence of naturally occurring lower virulent African swine fever viruses in domestic pigs in China in 2020. Sci. China Life Sci. 2021, 64, 752–765. [Google Scholar] [CrossRef]
- Sun, E.; Huang, L.; Zhang, X.; Zhang, J.; Shen, D.; Zhang, Z.; Wang, Z.; Huo, H.; Wang, W.; Huangfu, H.; et al. Genotype I African swine fever viruses emerged in domestic pigs in China and caused chronic infection. Emerg. Microbes. Infect. 2021, 10, 2183–2193. [Google Scholar] [CrossRef]
- Wang, N.; Zhao, D.; Wang, J.; Zhang, Y.; Wang, M.; Gao, Y.; Li, F.; Wang, J.; Bu, Z.; Rao, Z.; et al. Architecture of African swine fever virus and implications for viral assembly. Science 2019, 366, 640–644. [Google Scholar] [CrossRef]
- Cackett, G.; Matelska, D.; Sykora, M.; Portugal, R.; Malecki, M.; Bahler, J.; Dixon, L.; Werner, F. The African Swine Fever Virus Transcriptome. J. Virol. 2020, 94, e00119-20. [Google Scholar] [CrossRef]
- Gaudreault, N.N.; Madden, D.W.; Wilson, W.C.; Trujillo, J.D.; Richt, J.A. African Swine Fever Virus: An Emerging DNA Arbovirus. Front. Vet. Sci. 2020, 7, 215. [Google Scholar] [CrossRef]
- Duan, X.; Ru, Y.; Yang, W.; Ren, J.; Hao, R.; Qin, X.; Li, D.; Zheng, H. Research progress on the proteins involved in African swine fever virus infection and replication. Front. Immunol. 2022, 13, 947180. [Google Scholar] [CrossRef]
- Wang, G.; Xie, M.; Wu, W.; Chen, Z. Structures and Functional Diversities of ASFV Proteins. Viruses 2021, 13, 2124. [Google Scholar] [CrossRef]
- Rappuoli, R. Vaccines: Science, health, longevity, and wealth. Proc. Natl. Acad. Sci. USA 2014, 111, 12282. [Google Scholar] [CrossRef]
- Urbano, A.C.; Ferreira, F. African swine fever control and prevention: An update on vaccine development. Emerg. Microbes. Infect. 2022, 11, 2021–2033. [Google Scholar] [CrossRef]
- Borca, M.V.; Ramirez-Medina, E.; Silva, E.; Vuono, E.; Rai, A.; Pruitt, S.; Holinka, L.G.; Velazquez-Salinas, L.; Zhu, J.; Gladue, D.P. Development of a Highly Effective African Swine Fever Virus Vaccine by Deletion of the I177L Gene Results in Sterile Immunity against the Current Epidemic Eurasia Strain. J. Virol. 2020, 94, e02017-19. [Google Scholar] [CrossRef]
- Tran, X.H.; Phuong, L.T.T.; Huy, N.Q.; Thuy, D.T.; Nguyen, V.D.; Quang, P.H.; Ngon, Q.V.; Rai, A.; Gay, C.G.; Gladue, D.P.; et al. Evaluation of the Safety Profile of the ASFV Vaccine Candidate ASFV-G-DeltaI177L. Viruses 2022, 14, 896. [Google Scholar] [CrossRef]
- Vietnam suspends African swine fever vaccine after pig deaths. Available online: https://www.reuters.com/world/asia-pacific/vietnam-suspends-african-swine-fever-vaccine-after-pig-deaths-2022-08-24/ (accessed on 27 December 2022).
- Wang, T.; Sun, Y.; Huang, S.; Qiu, H.J. Multifaceted Immune Responses to African Swine Fever Virus: Implications for Vaccine Development. Vet. Microbiol. 2020, 249, 108832. [Google Scholar] [CrossRef]
- Zhu, J.J. African Swine Fever Vaccinology: The Biological Challenges from Immunological Perspectives. Viruses 2022, 14, 2021. [Google Scholar] [CrossRef]
- Wu, L.; Yang, B.; Yuan, X.; Hong, J.; Peng, M.; Chen, J.L.; Song, Z. Regulation and Evasion of Host Immune Response by African Swine Fever Virus. Front. Microbiol. 2021, 12, 698001. [Google Scholar] [CrossRef]
- Zhu, J.J.; Ramanathan, P.; Bishop, E.A.; O’Donnell, V.; Gladue, D.P.; Borca, M.V. Mechanisms of African swine fever virus pathogenesis and immune evasion inferred from gene expression changes in infected swine macrophages. PLoS ONE 2019, 14, e0223955. [Google Scholar] [CrossRef]
- Chathuranga, K.; Lee, J.-S. African Swine Fever Virus (ASFV): Immunity and Vaccine Development. Vaccines 2023, 11, 199. [Google Scholar] [CrossRef]
- Franzoni, G.; Dei Giudici, S.; Oggiano, A. Infection, modulation and responses of antigen-presenting cells to African swine fever viruses. Virus Res. 2018, 258, 73–80. [Google Scholar] [CrossRef]
- Dixon, L.K.; Islam, M.; Nash, R.; Reis, A.L. African swine fever virus evasion of host defences. Virus Res. 2019, 266, 25–33. [Google Scholar] [CrossRef]
- Reis, A.L.; Netherton, C.; Dixon, L.K. Unraveling the Armor of a Killer: Evasion of Host Defenses by African Swine Fever Virus. J. Virol. 2017, 91, e02338-16. [Google Scholar] [CrossRef]
- He, W.R.; Yuan, J.; Ma, Y.H.; Zhao, C.Y.; Yang, Z.Y.; Zhang, Y.; Han, S.; Wan, B.; Zhang, G.P. Modulation of Host Antiviral Innate Immunity by African Swine Fever Virus: A Review. Animals 2022, 12, 2935. [Google Scholar] [CrossRef]
- Zheng, X.; Nie, S.; Feng, W.H. Regulation of antiviral immune response by African swine fever virus (ASFV). Virol. Sin. 2022, 37, 157–167. [Google Scholar] [CrossRef]
- Ayanwale, A.; Trapp, S.; Guabiraba, R.; Caballero, I.; Roesch, F. New Insights in the Interplay Between African Swine Fever Virus and Innate Immunity and Its Impact on Viral Pathogenicity. Front. Microbiol. 2022, 13, 958307. [Google Scholar] [CrossRef]
- Seo, Y.J.; Hahm, B. Type I interferon modulates the battle of host immune system against viruses. Adv. Appl. Microbiol. 2010, 73, 83–101. [Google Scholar] [CrossRef]
- Fan, W.; Jiao, P.; Zhang, H.; Chen, T.; Zhou, X.; Qi, Y.; Sun, L.; Shang, Y.; Zhu, H.; Hu, R.; et al. Inhibition of African Swine Fever Virus Replication by Porcine Type I and Type II Interferons. Front. Microbiol. 2020, 11, 1203. [Google Scholar] [CrossRef]
- Garcia-Belmonte, R.; Perez-Nunez, D.; Pittau, M.; Richt, J.A.; Revilla, Y. African Swine Fever Virus Armenia/07 Virulent Strain Controls Interferon Beta Production through the cGAS-STING Pathway. J. Virol. 2019, 93, e02298-18. [Google Scholar] [CrossRef]
- Ran, Y.; Li, D.; Xiong, M.G.; Liu, H.N.; Feng, T.; Shi, Z.W.; Li, Y.H.; Wu, H.N.; Wang, S.Y.; Zheng, H.X.; et al. African swine fever virus I267L acts as an important virulence factor by inhibiting RNA polymerase III-RIG-I-mediated innate immunity. PLoS Pathog. 2022, 18, e1010270. [Google Scholar] [CrossRef]
- Ezeonwumelu, I.J.; Garcia-Vidal, E.; Ballana, E. JAK-STAT Pathway: A Novel Target to Tackle Viral Infections. Viruses 2021, 13, 2379. [Google Scholar] [CrossRef]
- Cai, S.; Zheng, Z.; Cheng, J.; Zhong, L.; Shao, R.; Zheng, F.; Lai, Z.; Ou, J.; Xu, L.; Zhou, P.; et al. Swine Interferon-Inducible Transmembrane Proteins Potently Inhibit African Swine Fever Virus Replication. Front. Immunol. 2022, 13, 827709. [Google Scholar] [CrossRef]
- Munoz-Moreno, R.; Cuesta-Geijo, M.A.; Martinez-Romero, C.; Barrado-Gil, L.; Galindo, I.; Garcia-Sastre, A.; Alonso, C. Antiviral Role of IFITM Proteins in African Swine Fever Virus Infection. PLoS ONE 2016, 11, e0154366. [Google Scholar] [CrossRef]
- Razzuoli, E.; Franzoni, G.; Carta, T.; Zinellu, S.; Amadori, M.; Modesto, P.; Oggiano, A. Modulation of Type I Interferon System by African Swine Fever Virus. Pathogens 2020, 9, 361. [Google Scholar] [CrossRef]
- Portugal, R.; Leitao, A.; Martins, C. Modulation of type I interferon signaling by African swine fever virus (ASFV) of different virulence L60 and NHV in macrophage host cells. Vet. Microbiol. 2018, 216, 132–141. [Google Scholar] [CrossRef]
- Sun, L.; Wu, J.; Du, F.; Chen, X.; Chen, Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 2013, 339, 786–791. [Google Scholar] [CrossRef]
- Shang, G.; Zhang, C.; Chen, Z.J.; Bai, X.C.; Zhang, X. Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP. Nature 2019, 567, 389–393. [Google Scholar] [CrossRef]
- Eaglesham, J.B.; Pan, Y.; Kupper, T.S.; Kranzusch, P.J. Viral and metazoan poxins are cGAMP-specific nucleases that restrict cGAS-STING signalling. Nature 2019, 566, 259–263. [Google Scholar] [CrossRef]
- Hernaez, B.; Alonso, G.; Georgana, I.; El-Jesr, M.; Martin, R.; Shair, K.H.Y.; Fischer, C.; Sauer, S.; Maluquer de Motes, C.; Alcami, A. Viral cGAMP nuclease reveals the essential role of DNA sensing in protection against acute lethal virus infection. Sci. Adv. 2020, 6, eabb4565. [Google Scholar] [CrossRef]
- Dodantenna, N.; Ranathunga, L.; Chathuranga, W.A.G.; Weerawardhana, A.; Cha, J.W.; Subasinghe, A.; Gamage, N.; Haluwana, D.K.; Kim, Y.; Jheong, W.; et al. African Swine Fever Virus EP364R and C129R Target Cyclic GMP-AMP To Inhibit the cGAS-STING Signaling Pathway. J. Virol. 2022, 96, e0102222. [Google Scholar] [CrossRef]
- Ghosh, M.; Saha, S.; Bettke, J.; Nagar, R.; Parrales, A.; Iwakuma, T.; van der Velden, A.W.M.; Martinez, L.A. Mutant p53 suppresses innate immune signaling to promote tumorigenesis. Cancer Cell 2021, 39, 494–508.e5. [Google Scholar] [CrossRef]
- Zhu, Z.; Li, S.; Ma, C.; Yang, F.; Cao, W.; Liu, H.; Chen, X.; Feng, T.; Shi, Z.; Tian, H.; et al. African Swine Fever Virus E184L Protein Interacts with Innate Immune Adaptor STING to Block IFN Production for Viral Replication and Pathogenesis. J. Immunol. 2023, 210, 442–458. [Google Scholar] [CrossRef]
- Zheng, W.; Xia, N.; Zhang, J.; Cao, Q.; Jiang, S.; Luo, J.; Wang, H.; Chen, N.; Zhang, Q.; Meurens, F.; et al. African Swine Fever Virus Structural Protein p17 Inhibits cGAS-STING Signaling Pathway Through Interacting With STING. Front. Immunol. 2022, 13, 941579. [Google Scholar] [CrossRef]
- Liu, S.; Cai, X.; Wu, J.; Cong, Q.; Chen, X.; Li, T.; Du, F.; Ren, J.; Wu, Y.T.; Grishin, N.V.; et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 2015, 347, aaa2630. [Google Scholar] [CrossRef]
- Zhou, R.; Zhang, Q.; Xu, P. TBK1, a central kinase in innate immune sensing of nucleic acids and beyond. Acta. Biochim. Biophys. Sin. 2020, 52, 757–767. [Google Scholar] [CrossRef]
- Larabi, A.; Devos, J.M.; Ng, S.L.; Nanao, M.H.; Round, A.; Maniatis, T.; Panne, D. Crystal structure and mechanism of activation of TANK-binding kinase 1. Cell Rep. 2013, 3, 734–746. [Google Scholar] [CrossRef]
- Tu, D.; Zhu, Z.; Zhou, A.Y.; Yun, C.H.; Lee, K.E.; Toms, A.V.; Li, Y.; Dunn, G.P.; Chan, E.; Thai, T.; et al. Structure and ubiquitination-dependent activation of TANK-binding kinase 1. Cell Rep. 2013, 3, 747–758. [Google Scholar] [CrossRef]
- Wang, X.; Wu, J.; Wu, Y.; Chen, H.; Zhang, S.; Li, J.; Xin, T.; Jia, H.; Hou, S.; Jiang, Y.; et al. Inhibition of cGAS-STING-TBK1 signaling pathway by DP96R of ASFV China 2018/1. Biochem. Biophys. Res. Commun. 2018, 506, 437–443. [Google Scholar] [CrossRef]
- Huang, L.; Xu, W.; Liu, H.; Xue, M.; Liu, X.; Zhang, K.; Hu, L.; Li, J.; Liu, X.; Xiang, Z.; et al. African Swine Fever Virus pI215L Negatively Regulates cGAS-STING Signaling Pathway through Recruiting RNF138 to Inhibit K63-Linked Ubiquitination of TBK1. J. Immunol. 2021, 207, 2754–2769. [Google Scholar] [CrossRef]
- Song, G.; Liu, B.; Li, Z.; Wu, H.; Wang, P.; Zhao, K.; Jiang, G.; Zhang, L.; Gao, C. E3 ubiquitin ligase RNF128 promotes innate antiviral immunity through K63-linked ubiquitination of TBK1. Nat. Immunol. 2016, 17, 1342–1351. [Google Scholar] [CrossRef]
- Sun, M.; Yu, S.; Ge, H.; Wang, T.; Li, Y.; Zhou, P.; Pan, L.; Han, Y.; Yang, Y.; Sun, Y.; et al. The A137R Protein of African Swine Fever Virus Inhibits Type I Interferon Production via the Autophagy-Mediated Lysosomal Degradation of TBK1. J. Virol. 2022, 96, e0195721. [Google Scholar] [CrossRef]
- Luo, J.; Zhang, J.; Ni, J.; Jiang, S.; Xia, N.; Guo, Y.; Shao, Q.; Cao, Q.; Zheng, W.; Chen, N.; et al. The African swine fever virus protease pS273R inhibits DNA sensing cGAS-STING pathway by targeting IKKepsilon. Virulence 2022, 13, 740–756. [Google Scholar] [CrossRef]
- Balka, K.R.; Louis, C.; Saunders, T.L.; Smith, A.M.; Calleja, D.J.; D’Silva, D.B.; Moghaddas, F.; Tailler, M.; Lawlor, K.E.; Zhan, Y.; et al. TBK1 and IKKepsilon Act Redundantly to Mediate STING-Induced NF-kappaB Responses in Myeloid Cells. Cell Rep. 2020, 31, 107492. [Google Scholar] [CrossRef]
- Luo, W.W.; Tong, Z.; Cao, P.; Wang, F.B.; Liu, Y.; Zheng, Z.Q.; Wang, S.Y.; Li, S.; Wang, Y.Y. Transcription-independent regulation of STING activation and innate immune responses by IRF8 in monocytes. Nat. Commun. 2022, 13, 4822. [Google Scholar] [CrossRef]
- Liu, X.; Liu, H.; Ye, G.; Xue, M.; Yu, H.; Feng, C.; Zhou, Q.; Liu, X.; Zhang, L.; Jiao, S.; et al. African swine fever virus pE301R negatively regulates cGAS-STING signaling pathway by inhibiting the nuclear translocation of IRF3. Vet. Microbiol. 2022, 274, 109556. [Google Scholar] [CrossRef]
- Hong, J.; Chi, X.; Yuan, X.; Wen, F.; Rai, K.R.; Wu, L.; Song, Z.; Wang, S.; Guo, G.; Chen, J.L. I226R Protein of African Swine Fever Virus Is a Suppressor of Innate Antiviral Responses. Viruses 2022, 14, 575. [Google Scholar] [CrossRef]
- Liu, H.; Zhu, Z.; Feng, T.; Ma, Z.; Xue, Q.; Wu, P.; Li, P.; Li, S.; Yang, F.; Cao, W.; et al. African Swine Fever Virus E120R Protein Inhibits Interferon Beta Production by Interacting with IRF3 To Block Its Activation. J. Virol. 2021, 95, e0082421. [Google Scholar] [CrossRef]
- Cui, S.; Wang, Y.; Gao, X.; Xin, T.; Wang, X.; Yu, H.; Chen, S.; Jiang, Y.; Chen, Q.; Jiang, F.; et al. African swine fever virus M1249L protein antagonizes type I interferon production via suppressing phosphorylation of TBK1 and degrading IRF3. Virus Res. 2022, 319, 198872. [Google Scholar] [CrossRef]
- Zhu, Z.; Chen, H.; Liu, L.; Cao, Y.; Jiang, T.; Zou, Y.; Peng, Y. Classification and characterization of multigene family proteins of African swine fever viruses. Brief. Bioinform. 2021, 22, bbaa380. [Google Scholar] [CrossRef]
- Cackett, G.; Sykora, M.; Werner, F. Transcriptome view of a killer: African swine fever virus. Biochem. Soc. Trans. 2020, 48, 1569–1581. [Google Scholar] [CrossRef]
- Yang, K.; Xue, Y.; Niu, H.; Shi, C.; Cheng, M.; Wang, J.; Zou, B.; Wang, J.; Niu, T.; Bao, M.; et al. African swine fever virus MGF360–11L negatively regulates cGAS-STING-mediated inhibition of type I interferon production. Vet. Res. 2022, 53, 7. [Google Scholar] [CrossRef]
- Yang, K.; Huang, Q.; Wang, R.; Zeng, Y.; Cheng, M.; Xue, Y.; Shi, C.; Ye, L.; Yang, W.; Jiang, Y.; et al. African swine fever virus MGF505–11R inhibits type I interferon production by negatively regulating the cGAS-STING-mediated signaling pathway. Vet. Microbiol. 2021, 263, 109265. [Google Scholar] [CrossRef]
- Wang, Y.; Cui, S.; Xin, T.; Wang, X.; Yu, H.; Chen, S.; Jiang, Y.; Gao, X.; Jiang, Y.; Guo, X.; et al. African Swine Fever Virus MGF360–14L Negatively Regulates Type I Interferon Signaling by Targeting IRF3. Front. Cell Infect. Microbiol. 2021, 11, 818969. [Google Scholar] [CrossRef]
- Huang, L.; Li, J.; Zheng, J.; Li, D.; Weng, C. Multifunctional pMGF505–7R Is a Key Virulence-Related Factor of African Swine Fever Virus. Front. Microbiol. 2022, 13, 852431. [Google Scholar] [CrossRef]
- Yang, K.; Xue, Y.; Niu, T.; Li, X.; Cheng, M.; Bao, M.; Zou, B.; Shi, C.; Wang, J.; Yang, W.; et al. African swine fever virus MGF505–7R protein interacted with IRF7and TBK1 to inhibit type I interferon production. Virus Res. 2022, 322, 198931. [Google Scholar] [CrossRef]
- Li, D.; Yang, W.; Li, L.; Li, P.; Ma, Z.; Zhang, J.; Qi, X.; Ren, J.; Ru, Y.; Niu, Q.; et al. African Swine Fever Virus MGF-505–7R Negatively Regulates cGAS-STING-Mediated Signaling Pathway. J. Immunol. 2021, 206, 1844–1857. [Google Scholar] [CrossRef]
- Konno, H.; Konno, K.; Barber, G.N. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell 2013, 155, 688–698. [Google Scholar] [CrossRef]
- Chiu, Y.H.; Macmillan, J.B.; Chen, Z.J. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 2009, 138, 576–591. [Google Scholar] [CrossRef]
- Ablasser, A.; Bauernfeind, F.; Hartmann, G.; Latz, E.; Fitzgerald, K.A.; Hornung, V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat. Immunol. 2009, 10, 1065–1072. [Google Scholar] [CrossRef]
- Hornung, V.; Ellegast, J.; Kim, S.; Brzozka, K.; Jung, A.; Kato, H.; Poeck, H.; Akira, S.; Conzelmann, K.K.; Schlee, M.; et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science 2006, 314, 994–997. [Google Scholar] [CrossRef]
- Okamoto, M.; Kouwaki, T.; Fukushima, Y.; Oshiumi, H. Regulation of RIG-I Activation by K63-Linked Polyubiquitination. Front Immunol 2017, 8, 1942. [Google Scholar] [CrossRef]
- Ban, J.; Lee, N.R.; Lee, N.J.; Lee, J.K.; Quan, F.S.; Inn, K.S. Human Respiratory Syncytial Virus NS 1 Targets TRIM25 to Suppress RIG-I Ubiquitination and Subsequent RIG-I-Mediated Antiviral Signaling. Viruses 2018, 10, 716. [Google Scholar] [CrossRef]
- Sanchez-Aparicio, M.T.; Feinman, L.J.; Garcia-Sastre, A.; Shaw, M.L. Paramyxovirus V Proteins Interact with the RIG-I/TRIM25 Regulatory Complex and Inhibit RIG-I Signaling. J. Virol. 2018, 92, e01950-17. [Google Scholar] [CrossRef]
- Hu, H.; Sun, S.C. Ubiquitin signaling in immune responses. Cell Res. 2016, 26, 457–483. [Google Scholar] [CrossRef]
- Maelfait, J.; Beyaert, R. Emerging role of ubiquitination in antiviral RIG-I signaling. Microbiol. Mol. Biol. Rev. 2012, 76, 33–45. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, R.; Kim, S.H.; Shah, H.; Zhang, S.; Liang, J.H.; Fang, Y.; Gentili, M.; Leary, C.N.O.; Elledge, S.J.; et al. SARS-CoV-2 hijacks folate and one-carbon metabolism for viral replication. Nat. Commun. 2021, 12, 1676. [Google Scholar] [CrossRef]
- Sanchez-Garcia, F.J.; Perez-Hernandez, C.A.; Rodriguez-Murillo, M.; Moreno-Altamirano, M.M.B. The Role of Tricarboxylic Acid Cycle Metabolites in Viral Infections. Front. Cell Infect. Microbiol. 2021, 11, 725043. [Google Scholar] [CrossRef]
- Yang, S.; Jin, S.; Xian, H.; Zhao, Z.; Wang, L.; Wu, Y.; Zhou, L.; Li, M.; Cui, J. Metabolic enzyme UAP1 mediates IRF3 pyrophosphorylation to facilitate innate immune response. Mol. Cell 2023, 83, 298–313.e8. [Google Scholar] [CrossRef]
- Gonzalez Plaza, J.J.; Hulak, N.; Kausova, G.; Zhumadilov, Z.; Akilzhanova, A. Role of metabolism during viral infections, and crosstalk with the innate immune system. Intractable Rare Dis. Res. 2016, 5, 90–96. [Google Scholar] [CrossRef]
- Ye, L.; Jiang, Y.; Zhang, M. Crosstalk between glucose metabolism, lactate production and immune response modulation. Cytokine Growth Factor Rev. 2022, 68, 81–92. [Google Scholar] [CrossRef]
- Xue, Q.; Liu, H.; Zhu, Z.; Yang, F.; Song, Y.; Li, Z.; Xue, Z.; Cao, W.; Liu, X.; Zheng, H. African Swine Fever Virus Regulates Host Energy and Amino Acid Metabolism To Promote Viral Replication. J. Virol. 2022, 96, e0191921. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, G.; Xu, Z.G.; Tu, H.; Hu, F.; Dai, J.; Chang, Y.; Chen, Y.; Lu, Y.; Zeng, H.; et al. Lactate Is a Natural Suppressor of RLR Signaling by Targeting MAVS. Cell 2019, 178, 176–189.e15. [Google Scholar] [CrossRef]
- Schoggins, J.W.; Rice, C.M. Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 2011, 1, 519–525. [Google Scholar] [CrossRef]
- Li, L.; Fu, J.; Li, J.; Guo, S.; Chen, Q.; Zhang, Y.; Liu, Z.; Tan, C.; Chen, H.; Wang, X. African Swine Fever Virus pI215L Inhibits Type I Interferon Signaling by Targeting Interferon Regulatory Factor 9 for Autophagic Degradation. J. Virol. 2022, 96, e0094422. [Google Scholar] [CrossRef]
- Riera, E.; Garcia-Belmonte, R.; Madrid, R.; Perez-Nunez, D.; Revilla, Y. African swine fever virus ubiquitin-conjugating enzyme pI215L inhibits IFN-I signaling pathway through STAT2 degradation. Front. Microbiol. 2022, 13, 1081035. [Google Scholar] [CrossRef]
- Zhang, K.; Yang, B.; Shen, C.; Zhang, T.; Hao, Y.; Zhang, D.; Liu, H.; Shi, X.; Li, G.; Yang, J.; et al. MGF360–9L Is a Major Virulence Factor Associated with the African Swine Fever Virus by Antagonizing the JAK/STAT Signaling Pathway. mBio 2022, 13, e0233021. [Google Scholar] [CrossRef]
- Li, D.; Zhang, J.; Yang, W.; Li, P.; Ru, Y.; Kang, W.; Li, L.; Ran, Y.; Zheng, H. African swine fever virus protein MGF-505–7R promotes virulence and pathogenesis by inhibiting JAK1- and JAK2-mediated signaling. J. Biol. Chem. 2021, 297, 101190. [Google Scholar] [CrossRef]
- Zhao, C.; Zhao, W. NLRP3 Inflammasome-A Key Player in Antiviral Responses. Front. Immunol. 2020, 11, 211. [Google Scholar] [CrossRef]
- Jo, E.K.; Kim, J.K.; Shin, D.M.; Sasakawa, C. Molecular mechanisms regulating NLRP3 inflammasome activation. Cell. Mol. Immunol. 2016, 13, 148–159. [Google Scholar] [CrossRef]
- Zandi, E.; Rothwarf, D.M.; Delhase, M.; Hayakawa, M.; Karin, M. The IkappaB kinase complex (IKK) contains two kinase subunits, IKKalpha and IKKbeta, necessary for IkappaB phosphorylation and NF-kappaB activation. Cell 1997, 91, 243–252. [Google Scholar] [CrossRef]
- Israel, A. The IKK complex, a central regulator of NF-kappaB activation. Cold Spring Harb. Perspect Biol. 2010, 2, a000158. [Google Scholar] [CrossRef]
- Clark, K.; Nanda, S.; Cohen, P. Molecular control of the NEMO family of ubiquitin-binding proteins. Nat. Rev. Mol. Cell Biol. 2013, 14, 673–685. [Google Scholar] [CrossRef]
- Hadian, K.; Griesbach, R.A.; Dornauer, S.; Wanger, T.M.; Nagel, D.; Metlitzky, M.; Beisker, W.; Schmidt-Supprian, M.; Krappmann, D. NF-kappaB essential modulator (NEMO) interaction with linear and lys-63 ubiquitin chains contributes to NF-kappaB activation. J. Biol. Chem. 2011, 286, 26107–26117. [Google Scholar] [CrossRef]
- Du, M.; Ea, C.K.; Fang, Y.; Chen, Z.J. Liquid phase separation of NEMO induced by polyubiquitin chains activates NF-kappaB. Mol. Cell 2022, 82, 2415–2426.e5. [Google Scholar] [CrossRef]
- Zhou, P.; Dai, J.; Zhang, K.; Wang, T.; Li, L.F.; Luo, Y.; Sun, Y.; Qiu, H.J.; Li, S. The H240R Protein of African Swine Fever Virus Inhibits Interleukin 1beta Production by Inhibiting NEMO Expression and NLRP3 Oligomerization. J. Virol. 2022, 96, e0095422. [Google Scholar] [CrossRef]
- Kwak, Y.T.; Guo, J.; Shen, J.; Gaynor, R.B. Analysis of domains in the IKKalpha and IKKbeta proteins that regulate their kinase activity. J. Biol. Chem. 2000, 275, 14752–14759. [Google Scholar] [CrossRef]
- Yang, J.; Li, S.; Feng, T.; Zhang, X.; Yang, F.; Cao, W.; Chen, H.; Liu, H.; Zhang, K.; Zhu, Z.; et al. African Swine Fever Virus F317L Protein Inhibits NF-kappaB Activation To Evade Host Immune Response and Promote Viral Replication. mSphere 2021, 6, e0065821. [Google Scholar] [CrossRef]
- Chen, H.; Wang, Z.; Gao, X.; Lv, J.; Hu, Y.; Jung, Y.S.; Zhu, S.; Wu, X.; Qian, Y.; Dai, J. ASFV pD345L protein negatively regulates NF-kappaB signalling by inhibiting IKK kinase activity. Vet. Res. 2022, 53, 32. [Google Scholar] [CrossRef]
- Li, J.; Song, J.; Kang, L.; Huang, L.; Zhou, S.; Hu, L.; Zheng, J.; Li, C.; Zhang, X.; He, X.; et al. pMGF505–7R determines pathogenicity of African swine fever virus infection by inhibiting IL-1beta and type I IFN production. PloS Pathog. 2021, 17, e1009733. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-kappaB signaling in inflammation. Signal Transduct Target Ther. 2017, 2, 17023. [Google Scholar] [CrossRef]
- Zhuo, Y.; Guo, Z.; Ba, T.; Zhang, C.; He, L.; Zeng, C.; Dai, H. African Swine Fever Virus MGF360–12L Inhibits Type I Interferon Production by Blocking the Interaction of Importin alpha and NF-kappaB Signaling Pathway. Virol. Sin. 2021, 36, 176–186. [Google Scholar] [CrossRef]
- Barrado-Gil, L.; Del Puerto, A.; Galindo, I.; Cuesta-Geijo, M.A.; Garcia-Dorival, I.; de Motes, C.M.; Alonso, C. African Swine Fever Virus Ubiquitin-Conjugating Enzyme Is an Immunomodulator Targeting NF-kappaB Activation. Viruses 2021, 13, 1160. [Google Scholar] [CrossRef]
- Liu, X.; Ao, D.; Jiang, S.; Xia, N.; Xu, Y.; Shao, Q.; Luo, J.; Wang, H.; Zheng, W.; Chen, N.; et al. African Swine Fever Virus A528R Inhibits TLR8 Mediated NF-kappaB Activity by Targeting p65 Activation and Nuclear Translocation. Viruses 2021, 13, 2046. [Google Scholar] [CrossRef]
- Kelley, N.; Jeltema, D.; Duan, Y.; He, Y. The NLRP3 Inflammasome: An Overview of Mechanisms of Activation and Regulation. Int. J. Mol. Sci. 2019, 20, 3328. [Google Scholar] [CrossRef]
- Huang, Y.; Xu, W.; Zhou, R. NLRP3 inflammasome activation and cell death. Cell Mol. Immunol. 2021, 18, 2114–2127. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Z.; Ruan, J.; Pan, Y.; Magupalli, V.G.; Wu, H.; Lieberman, J. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 2016, 535, 153–158. [Google Scholar] [CrossRef]
- Kuriakose, T.; Kanneganti, T.D. Pyroptosis in Antiviral Immunity. Curr. Top Microbiol. Immunol. 2019. [Google Scholar] [CrossRef]
- Wen, W.; Li, X.; Wang, H.; Zhao, Q.; Yin, M.; Liu, W.; Chen, H.; Qian, P. Seneca Valley Virus 3C Protease Induces Pyroptosis by Directly Cleaving Porcine Gasdermin D. J. Immunol. 2021, 207, 189–199. [Google Scholar] [CrossRef]
- Yamaoka, Y.; Matsunaga, S.; Jeremiah, S.S.; Nishi, M.; Miyakawa, K.; Morita, T.; Khatun, H.; Shimizu, H.; Okabe, N.; Kimura, H.; et al. Zika virus protease induces caspase-independent pyroptotic cell death by directly cleaving gasdermin D. Biochem. Biophys. Res. Commun. 2021, 534, 666–671. [Google Scholar] [CrossRef]
- Zhao, G.; Li, T.; Liu, X.; Zhang, T.; Zhang, Z.; Kang, L.; Song, J.; Zhou, S.; Chen, X.; Wang, X.; et al. African swine fever virus cysteine protease pS273R inhibits pyroptosis by noncanonically cleaving gasdermin D. J. Biol. Chem. 2022, 298, 101480. [Google Scholar] [CrossRef]
- Huang, L.; Liu, H.; Ye, G.; Liu, X.; Chen, W.; Wang, Z.; Zhao, D.; Zhang, Z.; Feng, C.; Hu, L.; et al. Deletion of African Swine Fever Virus (ASFV) H240R Gene Attenuates the Virulence of ASFV by Enhancing NLRP3-Mediated Inflammatory Responses. J. Virol. 2023, e01227-22. [Google Scholar] [CrossRef]
- Christopher, J.; Burrell, C.R.H.; Murpy, F.A. Pathogenesis of Virus Infections. In Fenner and White’s Medical Virology, 5th ed.; Academic Press: Salt Lake City, UT, USA, 2016; p. 87. [Google Scholar]
- Ding, M.; Dang, W.; Liu, H.; Xu, F.; Huang, H.; Sunkang, Y.; Li, T.; Pei, J.; Liu, X.; Zhang, Y.; et al. Combinational Deletions of MGF360–9L and MGF505–7R Attenuated Highly Virulent African Swine Fever Virus and Conferred Protection against Homologous Challenge. J. Virol. 2022, 96, e0032922. [Google Scholar] [CrossRef]
- Ramirez-Medina, E.; Vuono, E.; Rai, A.; Pruitt, S.; Espinoza, N.; Velazquez-Salinas, L.; Pina-Pedrero, S.; Zhu, J.; Rodriguez, F.; Borca, M.V.; et al. Deletion of E184L, a Putative DIVA Target from the Pandemic Strain of African Swine Fever Virus, Produces a Reduction in Virulence and Protection against Virulent Challenge. J. Virol. 2022, 96, e0141921. [Google Scholar] [CrossRef]
- Zhang, Y.; Ke, J.; Zhang, J.; Yang, J.; Yue, H.; Zhou, X.; Qi, Y.; Zhu, R.; Miao, F.; Li, Q.; et al. African Swine Fever Virus Bearing an I226R Gene Deletion Elicits Robust Immunity in Pigs to African Swine Fever. J. Virol. 2021, 95, e0119921. [Google Scholar] [CrossRef]
- Gladue, D.P.; Ramirez-Medina, E.; Vuono, E.; Silva, E.; Rai, A.; Pruitt, S.; Espinoza, N.; Velazquez-Salinas, L.; Borca, M.V. Deletion of the A137R Gene from the Pandemic Strain of African Swine Fever Virus Attenuates the Strain and Offers Protection against the Virulent Pandemic Virus. J. Virol. 2021, 95, e0113921. [Google Scholar] [CrossRef]
- Wang, T.; Sun, Y.; Luo, Y.; Qiu, H.J. Prevention, control and vaccine development of African swine fever: Challenges and countermeasures. Sheng Wu Gong Cheng Xue Bao 2018, 34, 1931–1942. [Google Scholar] [CrossRef]
- Arias, M.; de la Torre, A.; Dixon, L.; Gallardo, C.; Jori, F.; Laddomada, A.; Martins, C.; Parkhouse, R.M.; Revilla, Y.; Rodriguez, F.A.J.; et al. Approaches and Perspectives for Development of African Swine Fever Virus Vaccines. Vaccines 2017, 5, 35. [Google Scholar] [CrossRef]
- Wang, Z.; Ai, Q.; Huang, S.; Ou, Y.; Gao, Y.; Tong, T.; Fan, H. Immune Escape Mechanism and Vaccine Research Progress of African Swine Fever Virus. Vaccines 2022, 10, 344. [Google Scholar] [CrossRef]
- Latz, E.; Schoenemeyer, A.; Visintin, A.; Fitzgerald, K.A.; Monks, B.G.; Knetter, C.F.; Lien, E.; Nilsen, N.J.; Espevik, T.; Golenbock, D.T. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat. Immunol. 2004, 5, 190–198. [Google Scholar] [CrossRef]
- Fiola, S.; Gosselin, D.; Takada, K.; Gosselin, J. TLR9 contributes to the recognition of EBV by primary monocytes and plasmacytoid dendritic cells. J. Immunol. 2010, 185, 3620–3631. [Google Scholar] [CrossRef]
- Gil, S.; Sepulveda, N.; Albina, E.; Leitao, A.; Martins, C. The low-virulent African swine fever virus (ASFV/NH/P68) induces enhanced expression and production of relevant regulatory cytokines (IFNalpha, TNFalpha and IL12p40) on porcine macrophages in comparison to the highly virulent ASFV/L60. Arch. Virol. 2008, 153, 1845–1854. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, J.; Zhang, Y.; Yang, J.; Wang, L.; Qi, Y.; Han, X.; Zhou, X.; Miao, F.; Chen, T.; et al. Cytokine Storm in Domestic Pigs Induced by Infection of Virulent African Swine Fever Virus. Front. Vet. Sci. 2020, 7, 601641. [Google Scholar] [CrossRef]
- Weber, S.; Hakobyan, A.; Zakaryan, H.; Doerfler, W. Intracellular African swine fever virus DNA remains unmethylated in infected Vero cells. Epigenomics 2018, 10, 289–299. [Google Scholar] [CrossRef]
- Bao, W.; Xia, H.; Liang, Y.; Ye, Y.; Lu, Y.; Xu, X.; Duan, A.; He, J.; Chen, Z.; Wu, Y.; et al. Toll-like Receptor 9 Can be Activated by Endogenous Mitochondrial DNA to Induce Podocyte Apoptosis. Sci. Rep. 2016, 6, 22579. [Google Scholar] [CrossRef]
- Jing, R.; Hu, Z.K.; Lin, F.; He, S.; Zhang, S.S.; Ge, W.Y.; Dai, H.J.; Du, X.K.; Lin, J.Y.; Pan, L.H. Mitophagy-Mediated mtDNA Release Aggravates Stretching-Induced Inflammation and Lung Epithelial Cell Injury via the TLR9/MyD88/NF-kappaB Pathway. Front. Cell Dev. Biol. 2020, 8, 819. [Google Scholar] [CrossRef]
- Taffoni, C.; Steer, A.; Marines, J.; Chamma, H.; Vila, I.K.; Laguette, N. Nucleic Acid Immunity and DNA Damage Response: New Friends and Old Foes. Front. Immunol. 2021, 12, 660560. [Google Scholar] [CrossRef]
- Simoes, M.; Martins, C.; Ferreira, F. Host DNA damage response facilitates African swine fever virus infection. Vet. Microbiol. 2013, 165, 140–147. [Google Scholar] [CrossRef]
- Karki, R.; Kanneganti, T.D. The ‘cytokine storm’: Molecular mechanisms and therapeutic prospects. Trends Immunol. 2021, 42, 681–705. [Google Scholar] [CrossRef]
- Kalinina, O.; Golovkin, A.; Zaikova, E.; Aquino, A.; Bezrukikh, V.; Melnik, O.; Vasilieva, E.; Karonova, T.; Kudryavtsev, I.; Shlyakhto, E. Cytokine Storm Signature in Patients with Moderate and Severe COVID-19. Int. J. Mol. Sci. 2022, 23, 8879. [Google Scholar] [CrossRef]
- Sun, W.; Liu, S.; Huang, X.; Yuan, R.; Yu, J. Cytokine storms and pyroptosis are primarily responsible for the rapid death of mice infected with pseudorabies virus. R. Soc. Open Sci. 2021, 8, 210296. [Google Scholar] [CrossRef]
- Karki, R.; Sharma, B.R.; Tuladhar, S.; Williams, E.P.; Zalduondo, L.; Samir, P.; Zheng, M.; Sundaram, B.; Banoth, B.; Malireddi, R.K.S.; et al. Synergism of TNF-alpha and IFN-gamma Triggers Inflammatory Cell Death, Tissue Damage, and Mortality in SARS-CoV-2 Infection and Cytokine Shock Syndromes. Cell 2021, 184, 149–168.e17. [Google Scholar] [CrossRef]
- Pandian, N.; Kanneganti, T.D. PANoptosis: A Unique Innate Immune Inflammatory Cell Death Modality. J. Immunol. 2022, 209, 1625–1633. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, L.; Zhu, Z.; Deng, J.; Tian, K.; Li, X. Antagonisms of ASFV towards Host Defense Mechanisms: Knowledge Gaps in Viral Immune Evasion and Pathogenesis. Viruses 2023, 15, 574. https://doi.org/10.3390/v15020574
Yu L, Zhu Z, Deng J, Tian K, Li X. Antagonisms of ASFV towards Host Defense Mechanisms: Knowledge Gaps in Viral Immune Evasion and Pathogenesis. Viruses. 2023; 15(2):574. https://doi.org/10.3390/v15020574
Chicago/Turabian StyleYu, Liangzheng, Zhenbang Zhu, Junhua Deng, Kegong Tian, and Xiangdong Li. 2023. "Antagonisms of ASFV towards Host Defense Mechanisms: Knowledge Gaps in Viral Immune Evasion and Pathogenesis" Viruses 15, no. 2: 574. https://doi.org/10.3390/v15020574
APA StyleYu, L., Zhu, Z., Deng, J., Tian, K., & Li, X. (2023). Antagonisms of ASFV towards Host Defense Mechanisms: Knowledge Gaps in Viral Immune Evasion and Pathogenesis. Viruses, 15(2), 574. https://doi.org/10.3390/v15020574









