SARS-CoV-2 Genome Variations in Viral Shedding of an Immunocompromised Patient with Non-Hodgkin’s Lymphoma
Abstract
:1. Introduction
2. Methods
3. Case Presentation
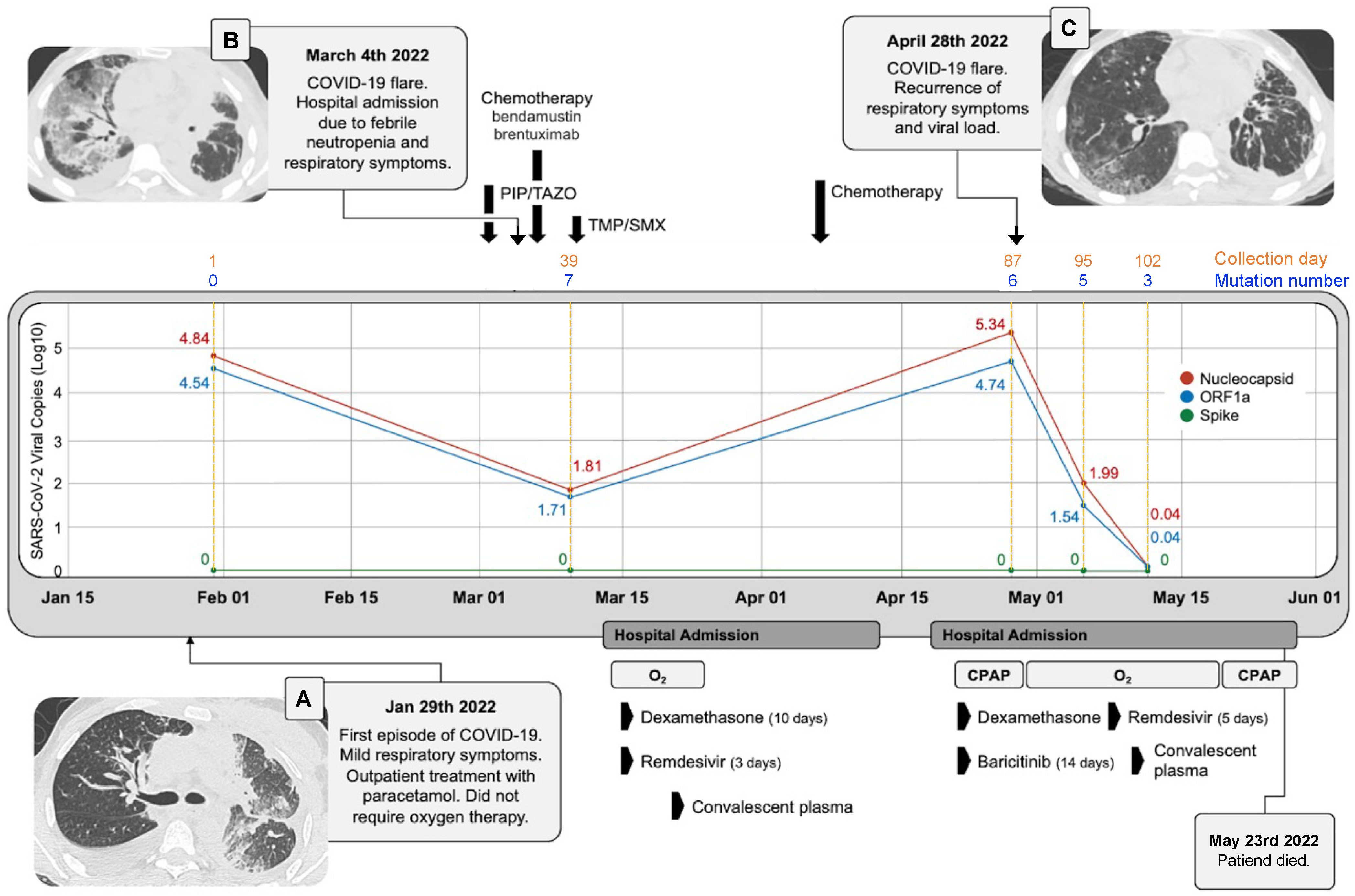
3.1. Clinical Case Description
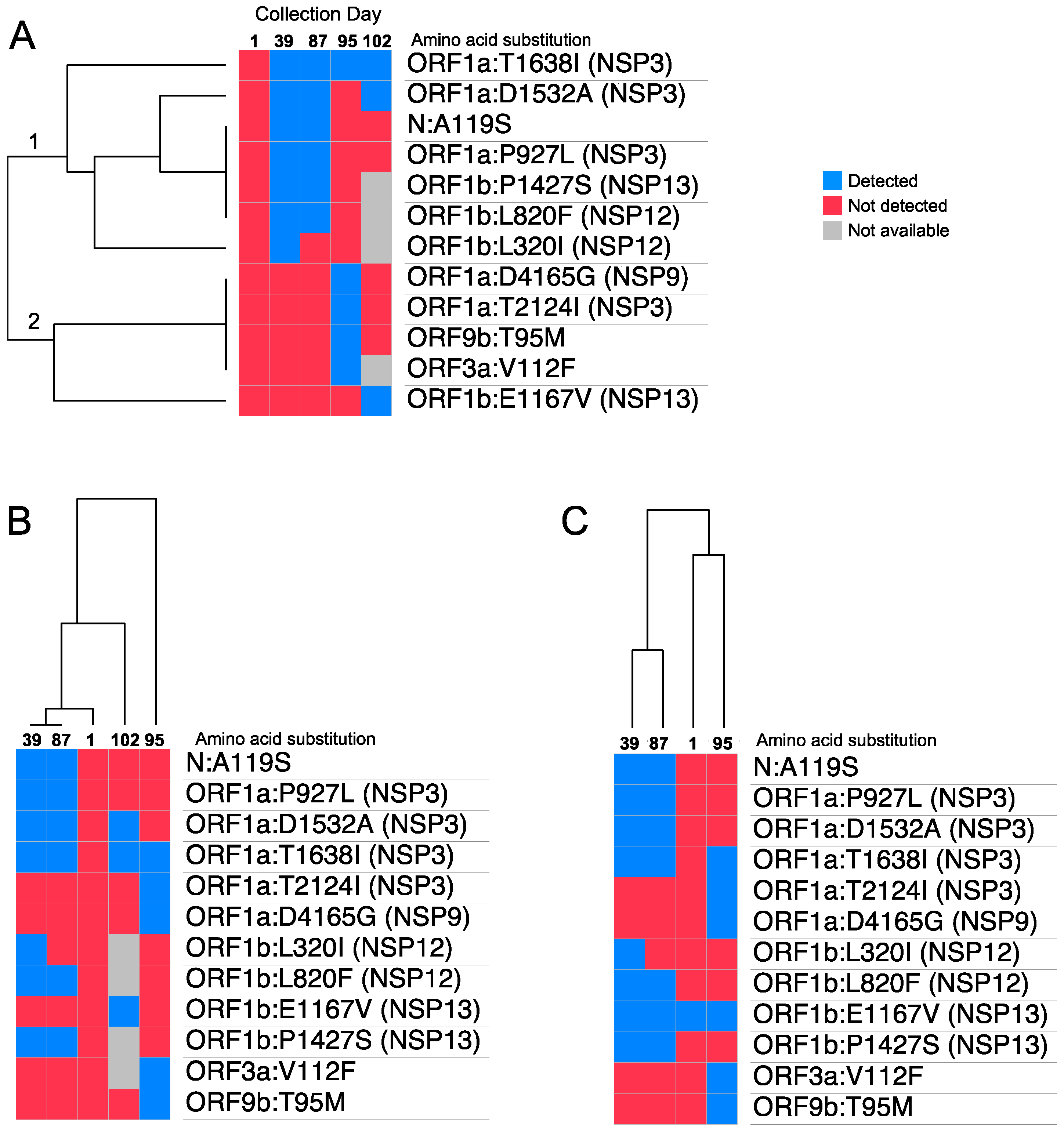
3.2. SARS-CoV-2 Mutational Landscape
3.3. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Seyed Hosseini, E.; Riahi Kashani, N.; Nikzad, H.; Azadbakht, J.; Hassani Bafrani, H.; Haddad Kashani, H. The novel coronavirus Disease-2019 (COVID-19): Mechanism of action, detection and recent therapeutic strategies. Virology 2020, 551, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.L. Author Correction: Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2022, 20, 315. [Google Scholar] [CrossRef] [PubMed]
- Almehdi, A.M.; Khoder, G.; Alchakee, A.S.; Alsayyid, A.T.; Sarg, N.H.; Soliman, S.S.M. SARS-CoV-2 spike protein: Pathogenesis, vaccines, and potential therapies. Infection 2021, 49, 855–876. [Google Scholar] [CrossRef] [PubMed]
- Choi, B.; Choudhary, M.C.; Regan, J.; Sparks, J.A.; Padera, R.F.; Qiu, X.; Solomon, I.H.; Kuo, H.H.; Boucau, J.; Bowman, K.; et al. Persistence and Evolution of SARS-CoV-2 in an Immunocompromised Host. N. Engl. J. Med. 2020, 383, 2291–2293. [Google Scholar] [CrossRef]
- Avanzato, V.A.; Matson, M.J.; Seifert, S.N.; Pryce, R.; Williamson, B.N.; Anzick, S.L.; Barbian, K.; Judson, S.D.; Fischer, E.R.; Martens, C.; et al. Case Study: Prolonged Infectious SARS-CoV-2 Shedding from an Asymptomatic Immunocompromised Individual with Cancer. Cell 2020, 183, 1901–1912.e9. [Google Scholar] [CrossRef]
- Alshukairi, A.N.; El-Kafrawy, S.A.; Dada, A.; Yasir, M.; Yamani, A.H.; Saeedi, M.F.; Aljohaney, A.; AlJohani, N.I.; Bahaudden, H.A.; Alam, I.; et al. Re-infection with a different SARS-CoV-2 clade and prolonged viral shedding in a hematopoietic stem cell transplantation patient. Int. J. Infect. Dis. 2021, 110, 267–271. [Google Scholar] [CrossRef]
- Baang, J.H.; Smith, C.; Mirabelli, C.; Valesano, A.L.; Manthei, D.M.; Bachman, M.A.; Wobus, C.E.; Adams, M.; Washer, L.; Martin, E.T.; et al. Prolonged Severe Acute Respiratory Syndrome Coronavirus 2 Replication in an Immunocompromised Patient. J. Infect. Dis. 2021, 223, 23–27. [Google Scholar] [CrossRef]
- Shoji, K.; Suzuki, A.; Okamoto, M.; Tsinda, E.K.; Sugawara, N.; Sasaki, M.; Nogami, Y.; Kobayashi, M.; Oshitani, H.; Yanai, M.; et al. Prolonged shedding of infectious viruses with haplotype switches of SARS-CoV-2 in an immunocompromised patient. J. infect. Chemother. 2022, 28, 1001–1004. [Google Scholar] [CrossRef]
- Galloway, A.; Park, Y.; Tanukonda, V.; Ho, Y.L.; Nguyen, X.T.; Maripuri, M.; Dey, A.T.; Gerlovin, H.; Posner, D.; Lynch, K.E.; et al. Impact of COVID-19 Severity on Long-term Events in US Veterans using the Veterans Affairs Severity Index for COVID-19 (VASIC). J. Infect. Dis. 2022, 226, 2113–2117. [Google Scholar] [CrossRef]
- Marconi, V.C.; Ramanan, A.V.; de Bono, S.; Kartman, C.E.; Krishnan, V.; Liao, R.; Piruzeli, M.L.B.; Goldman, J.D.; Alatorre-Alexander, J.; Cardoso, A.; et al. Efficacy and safety of baricitinib for the treatment of hospitalised adults with COVID-19 (COV-BARRIER): A randomised, double-blind, parallel-group, placebo-controlled phase 3 trial. Lancet Respir. Med. 2021, 9, 1407–1418. [Google Scholar] [CrossRef]
- Thompson, M.A.; Henderson, J.P.; Shah, P.K.; Rubinstein, S.M.; Joyner, M.J.; Choueiri, T.K.; Flora, D.B.; Griffiths, E.A.; Gulati, A.P.; Hwang, C.; et al. Association of Convalescent Plasma Therapy With Survival in Patients With Hematologic Cancers and COVID-19. JAMA Oncol. 2021, 7, 1167–1175. [Google Scholar] [CrossRef] [PubMed]
- Weinbergerova, B.; Mayer, J.; Kabut, T.; Hrabovsky, S.; Prochazkova, J.; Kral, Z.; Herout, V.; Pacasova, R.; Zdrazilova-Dubska, L.; Husa, P.; et al. Successful early treatment combining remdesivir with high-titer convalescent plasma among COVID-19-infected hematological patients. Hematol. Oncol. 2021, 39, 715–720. [Google Scholar] [CrossRef] [PubMed]
- Magyari, F.; Pinczés, L.I.; Páyer, E.; Farkas, K.; Ujfalusi, S.; Diószegi, Á.; Sik, M.; Simon, Z.; Nagy, G.; Hevessy, Z.; et al. Early administration of remdesivir plus convalescent plasma therapy is effective to treat COVID-19 pneumonia in B-cell depleted patients with hematological malignancies. Ann. Hematol. 2022, 101, 2337–2345. [Google Scholar] [CrossRef] [PubMed]
- Tarhini, H.; Recoing, A.; Bridier-Nahmias, A.; Rahi, M.; Lambert, C.; Martres, P.; Lucet, J.-C.; Rioux, C.; Bouzid, D.; Lebourgeois, S.; et al. Long-Term Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infectiousness Among Three Immunocompromised Patients: From Prolonged Viral Shedding to SARS-CoV-2 Superinfection. J. Infect. Dis. 2021, 223, 1522–1527. [Google Scholar] [CrossRef]
- Ueda, Y.; Asakura, S.; Wada, S.; Saito, T.; Yano, T. Prolonged COVID-19 in an Immunocompromised Patient Treated with Obinutuzumab and Bendamustine for Follicular Lymphoma. Intern. Med. 2022, 61, 2523–2526. [Google Scholar] [CrossRef]
- Yasuda, H.; Mori, Y.; Chiba, A.; Bai, J.; Murayama, G.; Matsushita, Y.; Miyake, S.; Komatsu, N. Resolution of One-Year Persisting COVID-19 Pneumonia and Development of Immune Thrombocytopenia in a Follicular Lymphoma Patient With Preceding Rituximab Maintenance Therapy: A follow-up Report and Literature Review of Cases With Prolonged Infections. Clin. Lymphoma Myeloma Leuk. 2021, 21, e810–e816. [Google Scholar] [CrossRef]
- Laracy, J.C.; Kamboj, M.; Vardhana, S.A. Long and persistent COVID-19 in patients with hematologic malignancies: From bench to bedside. Curr. Opin. Infect. Dis. 2022, 35, 271–279. [Google Scholar] [CrossRef]
- Ko, K.K.K.; Yingtaweesittikul, H.; Tan, T.T.; Wijaya, L.; Cao, D.Y.; Goh, S.S.; Abdul Rahman, N.B.; Chan, K.X.L.; Tay, H.M.; Sim, J.H.C.; et al. Emergence of SARS-CoV-2 Spike Mutations during Prolonged Infection in Immunocompromised Hosts. Microbiol. Spectr. 2022, 10, e00791-22. [Google Scholar] [CrossRef]
- Sonnleitner, S.T.; Prelog, M.; Sonnleitner, S.; Hinterbichler, E.; Halbfurter, H.; Kopecky, D.B.C.; Almanzar, G.; Koblmüller, S.; Sturmbauer, C.; Feist, L.; et al. Cumulative SARS-CoV-2 Mutations and Corresponding Changes in Immunity in an Immunocompromised Patient Indicate Viral Evolution within the Host. Nat. Commun. 2022, 13, 2560. [Google Scholar] [CrossRef]
- Dudouet, P.; Colson, P.; Aherfi, S.; Levasseur, A.; Beye, M.; Delerce, J.; Burel, E.; Lavrard, P.; Bader, W.; Lagier, J.-C.; et al. SARS-CoV-2 quasi-species analysis from patients with persistent nasopharyngeal shedding. Sci. Rep. 2022, 12, 18721. [Google Scholar] [CrossRef]
- Jary, A.; Leducq, V.; Malet, I.; Marot, S.; Klement-Frutos, E.; Teyssou, E.; Abdi, B.; Wirden, M.; Pourcher, V.; Caumes, E.; et al. Evolution of viral quasispecies during SARS-CoV-2 infection. Clin. Microbiol. Infect. 2020, 26, 1560.e1–1560.e4. [Google Scholar] [CrossRef] [PubMed]
- Caccuri, F.; Messali, S.; Bortolotti, D.; Di Silvestre, D.; De Palma, A.; Cattaneo, C.; Bertelli, A.; Zani, A.; Milanesi, M.; Giovanetti, M.; et al. Competition for dominance within replicating quasispecies during prolonged SARS-CoV-2 infection in an immunocompromised host. Virus Evolut. 2022, 8, veac042. [Google Scholar] [CrossRef] [PubMed]
- Freire-Neto, F.P.; Teixeira, D.G.; da Cunha, D.C.S.; Morais, I.C.; Tavares, C.P.M.; Gurgel, G.P.; Medeiros, S.D.N.; dos Santos, D.C.; Sales, A.D.O.; Jeronimo, S.M.B. SARS-CoV-2 reinfections with BA.1 (Omicron) variant among fully vaccinated individuals in northeastern Brazil. PLoS Negl. Trop. Dis. 2022, 16, e0010337. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Zhou, J.; Guo, J.; Hao, C.; Zheng, M.; Zhang, R.; Huang, Q.; Yao, X.; Li, R.; Jin, Y. Reinfection in patients with COVID-19: A systematic review. Glob. Health Res. Policy 2022, 7, 1–20. [Google Scholar] [CrossRef]
- Van der Moeren, N.; Selhorst, P.; Ha, M.; Heireman, L.; Van Gaal, P.J.; Breems, D.; Meysman, P.; Laukens, K.; Verstrepen, W.; Van Gasse, N.; et al. Viral Evolution and Immunology of SARS-CoV-2 in a Persistent Infection after Treatment with Rituximab. Viruses 2022, 14, 752. [Google Scholar] [CrossRef]
- Sepulcri, C.; Dentone, C.; Mikulska, M.; Bruzzone, B.; Lai, A.; Fenoglio, D.; Bozzano, F.; Bergna, A.; Parodi, A.; Altosole, T.; et al. The Longest Persistence of Viable SARS-CoV-2 With Recurrence of Viremia and Relapsing Symptomatic COVID-19 in an Immunocompromised Patient-A Case Study. Open Forum Infect. Dis. 2021, 8, ofab217. [Google Scholar] [CrossRef]
- de Silva, T.I.; Liu, G.; Lindsey, B.B.; Dong, D.; Moore, S.C.; Hsu, N.S.; Shah, D.; Wellington, D.; Mentzer, A.J.; Angyal, A.; et al. The impact of viral mutations on recognition by SARS-CoV-2 specific T cells. iScience 2021, 24, 103353. [Google Scholar] [CrossRef]
- Voloch, C.M.; da Silva Francisco, R., Jr.; de Almeida, L.G.P.; Cardoso, C.C.; Brustolini, O.J.; Gerber, A.L.; Guimarães, A.P.d.C.; Mariani, D.; da Costa, R.M.; Ferreira, O.C., Jr.; et al. Genomic characterization of a novel SARS-CoV-2 lineage from Rio de Janeiro, Brazil. J. Virol. 2021, 95, e00119-21. [Google Scholar] [CrossRef]
- Zhang, M.; Li, L.; Luo, M.; Liang, B. Genomic characterization and evolution of SARS-CoV-2 of a Canadian population. PLoS ONE 2021, 16, e0247799. [Google Scholar] [CrossRef]
- Corey, L.; Beyrer, C.; Cohen, M.S.; Michael, N.L.; Bedford, T.; Rolland, M. SARS-CoV-2 Variants in Patients with Immunosuppression. N. Engl. J. Med. 2021, 385, 562–566. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Villaseñor-Echavarri, R.; Gomez-Romero, L.; Martin-Onraet, A.; Herrera, L.A.; Escobar-Arrazola, M.A.; Ramirez-Vega, O.A.; Barrientos-Flores, C.; Mendoza-Vargas, A.; Hidalgo-Miranda, A.; Vilar-Compte, D.; et al. SARS-CoV-2 Genome Variations in Viral Shedding of an Immunocompromised Patient with Non-Hodgkin’s Lymphoma. Viruses 2023, 15, 377. https://doi.org/10.3390/v15020377
Villaseñor-Echavarri R, Gomez-Romero L, Martin-Onraet A, Herrera LA, Escobar-Arrazola MA, Ramirez-Vega OA, Barrientos-Flores C, Mendoza-Vargas A, Hidalgo-Miranda A, Vilar-Compte D, et al. SARS-CoV-2 Genome Variations in Viral Shedding of an Immunocompromised Patient with Non-Hodgkin’s Lymphoma. Viruses. 2023; 15(2):377. https://doi.org/10.3390/v15020377
Chicago/Turabian StyleVillaseñor-Echavarri, Rodrigo, Laura Gomez-Romero, Alexandra Martin-Onraet, Luis A. Herrera, Marco A. Escobar-Arrazola, Oscar A. Ramirez-Vega, Corazón Barrientos-Flores, Alfredo Mendoza-Vargas, Alfredo Hidalgo-Miranda, Diana Vilar-Compte, and et al. 2023. "SARS-CoV-2 Genome Variations in Viral Shedding of an Immunocompromised Patient with Non-Hodgkin’s Lymphoma" Viruses 15, no. 2: 377. https://doi.org/10.3390/v15020377
APA StyleVillaseñor-Echavarri, R., Gomez-Romero, L., Martin-Onraet, A., Herrera, L. A., Escobar-Arrazola, M. A., Ramirez-Vega, O. A., Barrientos-Flores, C., Mendoza-Vargas, A., Hidalgo-Miranda, A., Vilar-Compte, D., & Cedro-Tanda, A. (2023). SARS-CoV-2 Genome Variations in Viral Shedding of an Immunocompromised Patient with Non-Hodgkin’s Lymphoma. Viruses, 15(2), 377. https://doi.org/10.3390/v15020377






