Genomic Analysis of Influenza A and B Viruses Carrying Baloxavir Resistance-Associated Substitutions Serially Passaged in Human Epithelial Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells
2.2. Viruses
2.3. Viral Sequence Analysis
2.4. Serial Passaging
2.5. Mapping of AA Substitutions
2.6. Statistical Analysis
3. Results
3.1. Serial Passaging of the WT and MUT Viruses and Alignment of NGS Reads to the Influenza Virus Genome
3.2. Ratio of Nonsynonymous to Synonymous Nucleotide Mutations and Comparison of Mutational Profiles between WT and MUT Genes after Each Passage
3.3. Mapping of AA Substitutions Occurred during Serial Passaging
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Krammer, F.; Smith, G.J.D.; Fouchier, R.A.M.; Peiris, M.; Kedzierska, K.; Doherty, P.C.; Palese, P.; Shaw, M.L.; Treanor, J.; Webster, R.G.; et al. Influenza. Nat. Rev. Dis. Primers 2018, 4, 3. [Google Scholar] [CrossRef] [PubMed]
- Molinari, N.A.; Ortega-Sanchez, I.R.; Messonnier, M.L.; Thompson, W.W.; Wortley, P.M.; Weintraub, E.; Bridges, C.B. The annual impact of seasonal influenza in the US: Measuring disease burden and costs. Vaccine 2007, 25, 5086–5096. [Google Scholar] [CrossRef] [PubMed]
- Dawood, F.S.; Iuliano, A.D.; Reed, C.; Meltzer, M.I.; Shay, D.K.; Cheng, P.Y.; Bandaranayake, D.; Breiman, R.F.; Brooks, W.A.; Buchy, P.; et al. Estimated global mortality associated with the first 12 months of 2009 pandemic influenza A H1N1 virus circulation: A modelling study. Lancet Infect Dis. 2012, 12, 687–695. [Google Scholar] [CrossRef] [PubMed]
- Girard, M.P.; Tam, J.S.; Assossou, O.M.; Kieny, M.P. The 2009 A (H1N1) influenza virus pandemic: A review. Vaccine 2010, 28, 4895–4902. [Google Scholar] [CrossRef] [PubMed]
- Swierczynska, M.; Mirowska-Guzel, D.M.; Pindelska, E. Antiviral Drugs in Influenza. Int. J. Environ. Res. Public Health 2022, 19, 3018. [Google Scholar] [CrossRef] [PubMed]
- Hussain, M.; Galvin, H.D.; Haw, T.Y.; Nutsford, A.N.; Husain, M. Drug resistance in influenza A virus: The epidemiology and management. Infect. Drug Resist. 2017, 10, 121–134. [Google Scholar] [CrossRef] [PubMed]
- Petrova, V.N.; Russell, C.A. The evolution of seasonal influenza viruses. Nat. Rev. Microbiol. 2018, 16, 60. [Google Scholar] [CrossRef]
- Steinhauer, D.A.; Domingo, E.; Holland, J.J. Lack of evidence for proofreading mechanisms associated with an RNA virus polymerase. Gene 1992, 122, 281–288. [Google Scholar] [CrossRef]
- Vahey, M.D.; Fletcher, D.A. Low-Fidelity Assembly of Influenza A Virus Promotes Escape from Host Cells. Cell 2019, 176, 678. [Google Scholar] [CrossRef]
- Smyk, J.M.; Szydlowska, N.; Szulc, W.; Majewska, A. Evolution of Influenza Viruses-Drug Resistance, Treatment Options, and Prospects. Int. J. Mol. Sci. 2022, 23, 12244. [Google Scholar] [CrossRef]
- Hurt, A.C.; Holien, J.K.; Parker, M.; Kelso, A.; Barr, I.G. Zanamivir-resistant influenza viruses with a novel neuraminidase mutation. J. Virol. 2009, 83, 10366–10373. [Google Scholar] [CrossRef] [PubMed]
- Ince, W.L.; Smith, F.B.; O’Rear, J.J.; Thomson, M. Treatment-Emergent Influenza Virus Polymerase Acidic Substitutions Independent of Those at I38 Associated with Reduced Baloxavir Susceptibility and Virus Rebound in Trials of Baloxavir Marboxil. J. Infect. Dis. 2020, 222, 957–961. [Google Scholar] [CrossRef] [PubMed]
- Hickerson, B.T.; Petrovskaya, S.N.; Dickensheets, H.; Donnelly, R.P.; Ince, W.L.; Ilyushina, N.A. Impact of Baloxavir Resistance-Associated Substitutions on Influenza Virus Growth and Drug Susceptibility. J. Virol. 2023, 97, e0015423. [Google Scholar] [CrossRef] [PubMed]
- Hickerson, B.T.; Adams, S.E.; Bovin, N.V.; Donnelly, R.P.; Ilyushina, N.A. Generation and characterization of interferon-beta-resistant H1N1 influenza A virus. Acta Virol. 2022, 66, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Omoto, S.; Speranzini, V.; Hashimoto, T.; Noshi, T.; Yamaguchi, H.; Kawai, M.; Kawaguchi, K.; Uehara, T.; Shishido, T.; Naito, A.; et al. Characterization of influenza virus variants induced by treatment with the endonuclease inhibitor baloxavir marboxil. Sci. Rep. 2018, 8, 9633. [Google Scholar] [CrossRef] [PubMed]
- Noshi, T.; Kitano, M.; Taniguchi, K.; Yamamoto, A.; Omoto, S.; Baba, K.; Hashimoto, T.; Ishida, K.; Kushima, Y.; Hattori, K.; et al. In vitro characterization of baloxavir acid, a first-in-class cap-dependent endonuclease inhibitor of the influenza virus polymerase PA subunit. Antivir. Res. 2018, 160, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Abed, Y.; Baz, M.; Boivin, G. Impact of neuraminidase mutations conferring influenza resistance to neuraminidase inhibitors in the N1 and N2 genetic backgrounds. Antivir. Ther. 2006, 11, 971–976. [Google Scholar] [CrossRef]
- Suzuki, H.; Saito, R.; Masuda, H.; Oshitani, H.; Sato, M.; Sato, I. Emergence of amantadine-resistant influenza A viruses: Epidemiological study. J. Infect. Chemother. 2003, 9, 195–200. [Google Scholar] [CrossRef]
- Dharan, N.J.; Gubareva, L.V.; Meyer, J.J.; Okomo-Adhiambo, M.; McClinton, R.C.; Marshall, S.A.; St George, K.; Epperson, S.; Brammer, L.; Klimov, A.I.; et al. Infections with oseltamivir-resistant influenza A(H1N1) virus in the United States. JAMA 2009, 301, 1034–1041. [Google Scholar] [CrossRef]
- Okomo-Adhiambo, M.; Sleeman, K.; Ballenger, K.; Nguyen, H.T.; Mishin, V.P.; Sheu, T.G.; Smagala, J.; Li, Y.; Klimov, A.I.; Gubareva, L.V. Neuraminidase inhibitor susceptibility testing in human influenza viruses: A laboratory surveillance perspective. Viruses 2010, 2, 2269–2289. [Google Scholar] [CrossRef]
- Hayden, F.G.; Sugaya, N.; Hirotsu, N.; Lee, N.; de Jong, M.D.; Hurt, A.C.; Ishida, T.; Sekino, H.; Yamada, K.; Portsmouth, S.; et al. Baloxavir Marboxil for Uncomplicated Influenza in Adults and Adolescents. N. Engl. J. Med. 2018, 379, 913–923. [Google Scholar] [CrossRef] [PubMed]
- Checkmahomed, L.; M’Hamdi, Z.; Carbonneau, J.; Venable, M.C.; Baz, M.; Abed, Y.; Boivin, G. Impact of the Baloxavir-Resistant Polymerase Acid I38T Substitution on the Fitness of Contemporary Influenza A(H1N1)pdm09 and A(H3N2) Strains. J. Infect. Dis. 2020, 221, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Chesnokov, A.; Patel, M.C.; Mishin, V.P.; De La Cruz, J.A.; Lollis, L.; Nguyen, H.T.; Dugan, V.; Wentworth, D.E.; Gubareva, L.V. Replicative Fitness of Seasonal Influenza A Viruses With Decreased Susceptibility to Baloxavir. J. Infect. Dis. 2020, 221, 367–371. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.C.; Pascua, P.N.Q.; Fabrizio, T.P.; Marathe, B.M.; Seiler, P.; Barman, S.; Webby, R.J.; Webster, R.G.; Govorkova, E.A. Influenza A and B viruses with reduced baloxavir susceptibility display attenuated in vitro fitness but retain ferret transmissibility. Proc. Natl. Acad. Sci. USA 2020, 117, 8593–8601. [Google Scholar] [CrossRef] [PubMed]
- Koszalka, P.; Tilmanis, D.; Roe, M.; Vijaykrishna, D.; Hurt, A.C. Baloxavir marboxil susceptibility of influenza viruses from the Asia-Pacific, 2012–2018. Antivir. Res. 2019, 164, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Hickerson, B.T.; Adams, S.E.; Barman, S.; Miller, L.; Lugovtsev, V.Y.; Webby, R.J.; Ince, W.L.; Donnelly, R.P.; Ilyushina, N.A. Pleiotropic Effects of Influenza H1, H3, and B Baloxavir-Resistant Substitutions on Replication, Sensitivity to Baloxavir, and Interferon Expression. Antimicrob. Agents Chemother. 2022, 66, e0000922. [Google Scholar] [CrossRef] [PubMed]
- Takizawa, N.; Momose, F. A novel E198K substitution in the PA gene of influenza A virus with reduced susceptibility to baloxavir acid. Arch. Virol. 2022, 167, 1565–1570. [Google Scholar] [CrossRef]
- Jones, J.C.; Zagribelnyy, B.; Pascua, P.N.Q.; Bezrukov, D.S.; Barman, S.; Okda, F.; Webby, R.J.; Ivanenkov, Y.A.; Govorkova, E.A. Influenza A virus polymerase acidic protein E23G/K substitutions weaken key baloxavir drug-binding contacts with minimal impact on replication and transmission. PLoS Pathog. 2022, 18, e1010698. [Google Scholar] [CrossRef]
- Jester, B.; Schwerzmann, J.; Mustaquim, D.; Aden, T.; Brammer, L.; Humes, R.; Shult, P.; Shahangian, S.; Gubareva, L.; Xu, X.; et al. Mapping of the US Domestic Influenza Virologic Surveillance Landscape. Emerg. Infect. Dis. 2018, 24, 1300–1306. [Google Scholar] [CrossRef]
- Van Poelvoorde, L.A.E.; Saelens, X.; Thomas, I.; Roosens, N.H. Next-Generation Sequencing: An Eye-Opener for the Surveillance of Antiviral Resistance in Influenza. Trends Biotechnol. 2020, 38, 360–367. [Google Scholar] [CrossRef]
- Roosenhoff, R.; Schutten, M.; Reed, V.; Clinch, B.; van der Linden, A.; Fouchier, R.A.M.; Fraaij, P.L.A. Secondary substitutions in the hemagglutinin and neuraminidase genes associated with neuraminidase inhibitor resistance are rare in the Influenza Resistance Information Study (IRIS). Antivir. Res. 2021, 189, 105060. [Google Scholar] [CrossRef] [PubMed]
- Ilyushina, N.A.; Lugovtsev, V.Y.; Samsonova, A.P.; Sheikh, F.G.; Bovin, N.V.; Donnelly, R.P. Generation and characterization of interferon-lambda 1-resistant H1N1 influenza A viruses. PLoS ONE 2017, 12, e0181999. [Google Scholar] [CrossRef] [PubMed]
- Matrosovich, M.N.; Matrosovich, T.Y.; Gray, T.; Roberts, N.A.; Klenk, H.D. Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc. Natl. Acad. Sci. USA 2004, 101, 4620–4624. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, E.; Neumann, G.; Kawaoka, Y.; Hobom, G.; Webster, R.G. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. USA 2000, 97, 6108–6113. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, E.; Stech, J.; Guan, Y.; Webster, R.G.; Perez, D.R. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 2001, 146, 2275–2289. [Google Scholar] [CrossRef] [PubMed]
- Simonyan, V.; Chumakov, K.; Dingerdissen, H.; Faison, W.; Goldweber, S.; Golikov, A.; Gulzar, N.; Karagiannis, K.; Vinh Nguyen Lam, P.; Maudru, T.; et al. High-performance integrated virtual environment (HIVE): A robust infrastructure for next-generation sequence data analysis. Database 2016, 2016, baw022. [Google Scholar] [CrossRef]
- Barbezange, C.; Jones, L.; Blanc, H.; Isakov, O.; Celniker, G.; Enouf, V.; Shomron, N.; Vignuzzi, M.; van der Werf, S. Seasonal Genetic Drift of Human Influenza A Virus Quasispecies Revealed by Deep Sequencing. Front. Microbiol. 2018, 9, 2596. [Google Scholar] [CrossRef]
- Nobusawa, E.; Sato, K. Comparison of the mutation rates of human influenza A and B viruses. J. Virol. 2006, 80, 3675–3678. [Google Scholar] [CrossRef]
- Jeffares, D.C.; Tomiczek, B.; Sojo, V.; dos Reis, M. A beginners guide to estimating the non-synonymous to synonymous rate ratio of all protein-coding genes in a genome. Methods Mol. Biol. 2015, 1201, 65–90. [Google Scholar] [CrossRef]
- Bhatt, S.; Holmes, E.C.; Pybus, O.G. The genomic rate of molecular adaptation of the human influenza A virus. Mol. Biol. Evol. 2011, 28, 2443–2451. [Google Scholar] [CrossRef]
- Ilyushina, N.A.; Lee, N.; Lugovtsev, V.Y.; Kan, A.; Bovin, N.V.; Donnelly, R.P. Adaptation of influenza B virus by serial passage in human airway epithelial cells. Virology 2020, 549, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Liu, Y.; Yang, J.; Huang, X.; Han, K.; Zhao, D.; Bi, K.; Li, Y. Two Genetically Similar H9N2 Influenza A Viruses Show Different Pathogenicity in Mice. Front. Microbiol. 2016, 7, 1737. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.I.; Lee, I.; Park, S.; Bae, J.Y.; Yoo, K.; Cheong, H.J.; Noh, J.Y.; Hong, K.W.; Lemey, P.; Vrancken, B.; et al. Phylogenetic relationships of the HA and NA genes between vaccine and seasonal influenza A(H3N2) strains in Korea. PLoS ONE 2017, 12, e0172059. [Google Scholar] [CrossRef] [PubMed]
- Mohr, P.G.; Deng, Y.M.; McKimm-Breschkin, J.L. The neuraminidases of MDCK grown human influenza A(H3N2) viruses isolated since 1994 can demonstrate receptor binding. Virol. J. 2015, 12, 67. [Google Scholar] [CrossRef] [PubMed]
- Patel, D.; Schultz, L.W.; Umland, T.C. Influenza A polymerase subunit PB2 possesses overlapping binding sites for polymerase subunit PB1 and human MAVS proteins. Virus Res. 2013, 172, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Ives, J.A.; Carr, J.A.; Mendel, D.B.; Tai, C.Y.; Lambkin, R.; Kelly, L.; Oxford, J.S.; Hayden, F.G.; Roberts, N.A. The H274Y mutation in the influenza A/H1N1 neuraminidase active site following oseltamivir phosphate treatment leave virus severely compromised both in vitro and in vivo. Antivir. Res. 2002, 55, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Abed, Y.; Goyette, N.; Boivin, G. A reverse genetics study of resistance to neuraminidase inhibitors in an influenza A/H1N1 virus. Antivir. Ther. 2004, 9, 577–581. [Google Scholar] [CrossRef]
- Bloom, J.D.; Gong, L.I.; Baltimore, D. Permissive secondary mutations enable the evolution of influenza oseltamivir resistance. Science 2010, 328, 1272–1275. [Google Scholar] [CrossRef]
- Hurt, A.C.; Hardie, K.; Wilson, N.J.; Deng, Y.M.; Osbourn, M.; Leang, S.K.; Lee, R.T.; Iannello, P.; Gehrig, N.; Shaw, R.; et al. Characteristics of a widespread community cluster of H275Y oseltamivir-resistant A(H1N1)pdm09 influenza in Australia. J. Infect. Dis. 2012, 206, 148–157. [Google Scholar] [CrossRef]
- Butler, J.; Hooper, K.A.; Petrie, S.; Lee, R.; Maurer-Stroh, S.; Reh, L.; Guarnaccia, T.; Baas, C.; Xue, L.; Vitesnik, S.; et al. Estimating the fitness advantage conferred by permissive neuraminidase mutations in recent oseltamivir-resistant A(H1N1)pdm09 influenza viruses. PLoS Pathog. 2014, 10, e1004065. [Google Scholar] [CrossRef]
- Pflug, A.; Guilligay, D.; Reich, S.; Cusack, S. Structure of influenza A polymerase bound to the viral RNA promoter. Nature 2014, 516, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Pascua, P.N.Q.; Jones, J.C.; Webby, R.J.; Govorkova, E.A. Effect of E23G/K, F36V, N37T, E119D, and E199G polymerase acidic protein substitutions on the replication and baloxavir susceptibility of influenza B viruses. Antivir. Res. 2022, 208, 105455. [Google Scholar] [CrossRef] [PubMed]


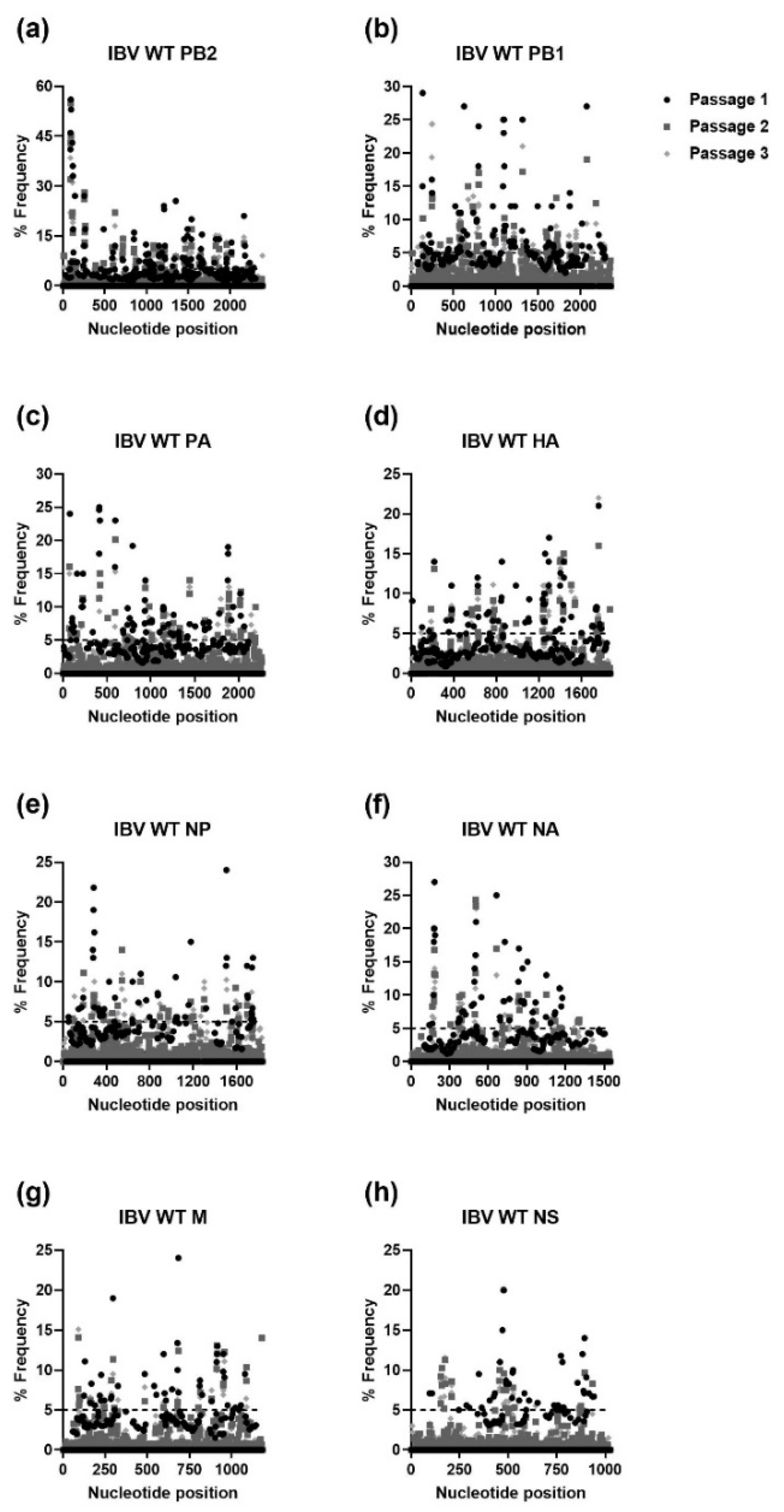
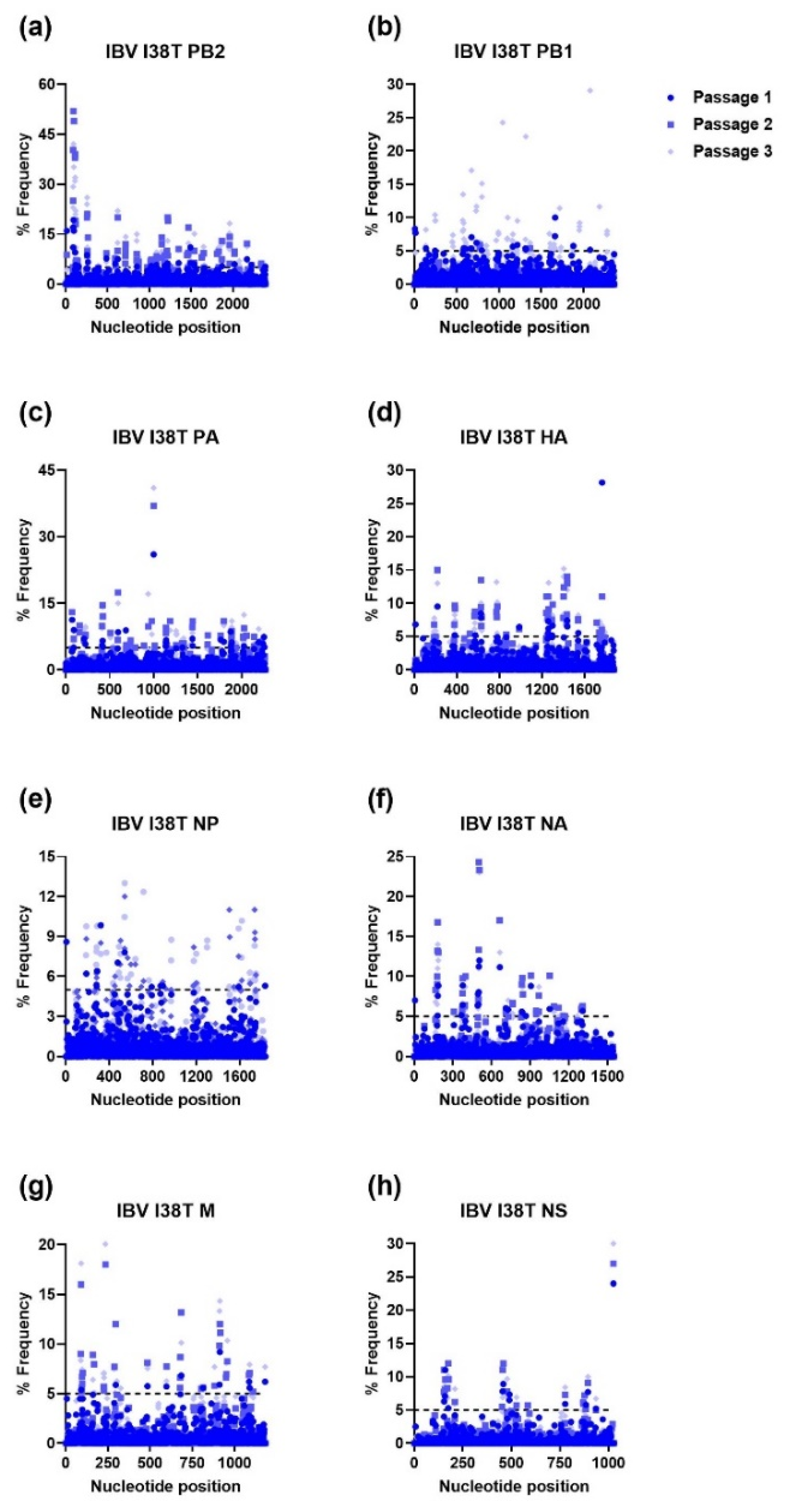
| Gene | Passage | H1N1 | H3N2 | IBV | |||||
|---|---|---|---|---|---|---|---|---|---|
| WT | I38L | I38T | E199D | WT | I38T | WT | I38T | ||
| PB2 | 1 | 2/2 * (– †) | 2/2 (–) | 1/1 (–) | 1/1 (–) | 0/0 (–) | 87/100 (6.7) | 95/104 (10.6) | 28/31 (9.3) |
| 2 | 1/1 (–) | 0/0 (–) | 1/1 (–) | 1/1 (–) | 6/13 (0.9) | 33/43 (3.3) | 71/80 (7.9) | 71/78 (10.1) | |
| 3 | 0/0 (–) | 0/0 (–) | 0/0 (–) | 1/1 (–) | 0/1 (–) | 6/12 (1.0) | 67/74 (9.6) | 65/72 (9.3) | |
| PB1 | 1 | 0/0 (–) | 0/0 (–) | 0/0 (–) | 1/1 (–) | 1/2 (1.0) | 79/102 (3.4) | 79/98 (4.2) | 13/16 (4.3) |
| 2 | 0/0 (–) | 0/0 (–) | 0/0 (–) | 0/0 (–) | 0/1 (–) | 32/41 (3.6) | 49/57 (6.1) | 53/65 (4.4) | |
| 3 | 0/0 (–) | 0/0 (–) | 0/0 (–) | 0/0 (–) | 3/8 (0.6) | 5/6 (5.0) | 56/65 (6.2) | 54/63 (6.0) | |
| PA | 1 | 0/0 (–) | 1/1 (–) | 0/0 (–) | 1/1 (–) | 0/0 (–) | 81/98 (4.8) | 69/82 (5.3) | 25/27 (12.5) |
| 2 | 0/0 (–) | 1/1 (–) | 0/0 (–) | 0/0 (–) | 10/13 (3.3) | 26/34 (3.3) | 55/60 (11) | 49/54 (9.8) | |
| 3 | 1/1 (–) | 1/1 (–) | 0/0 (–) | 0/0 (–) | 2/3 (2.0) | 1/1 (–) | 49/53 (12.3) | 47/51 (11.8) | |
| HA | 1 | 0/0 (–) | 0/0 (–) | 0/0 (–) | 0/0 (–) | 0/0 (–) | 72/87 (4.8) | 54/60 (9.0) | 11/13 (5.5) |
| 2 | 0/0 (–) | 0/0 (–) | 0/0 (–) | 0/0 (–) | 3/3 (–) | 35/44 (3.9) | 39/41 (19.5) | 40/42 (20.0) | |
| 3 | 0/0 (–) | 0/0 (–) | 0/0 (–) | 0/0 (–) | 0/0 (–) | 3/3 (N/A) | 38/41 (12.7) | 42/45 (14.0) | |
| NP | 1 | 0/0 (–) | 0/0 (–) | 0/0 (–) | 0/0 (–) | 0/0 (–) | 45/60 (3.0) | 44/56 (3.7) | 8/11 (2.7) |
| 2 | 0/0 (–) | 0/0 (–) | 0/0 (–) | 0/0 (–) | 4/6 (2.0) | 21/27 (3.5) | 23/32 (2.6) | 29/37 (3.6) | |
| 3 | 0/0 (–) | 0/0 (–) | 0/0 (–) | 0/0 (–) | 0/0 (–) | 1/1 (–) | 28/35 (4) | 30/39 (3.3) | |
| NA | 1 | 0/0 (–) | 0/0 (–) | 0/0 (–) | 0/0 (–) | 1/1 (–) | 39/47 (4.9) | 45/47 (22.5) | 21/23 (10.5) |
| 2 | 0/0 (–) | 0/0 (–) | 0/0 (–) | 0/0 (–) | 5/6 (5.0) | 26/33 (3.7) | 34/38 (8.5) | 36/45 (4.0) | |
| 3 | 0/0 (–) | 0/0 (–) ‡ | 0/0 (–) | 0/0 (–) | 0/0 (–) | 4/5 (4.0) | 25/28 (8.3) | 30/35 (6.0) | |
| M | 1 | 0/0 (–) | 0/0 (–) | 0/0 (–) | 0/0 (–) | 0/1 (–) | 16/24 (2.0) | 36/44 (4.5) | 10/14 (2.5) |
| 2 | 0/0 (–) | 0/0 (–) | 0/0 (–) | 0/0 (–) | 7/8 (7.0) | 10/13 (3.3) | 23/30 (3.3) | 30/35 (6.0) | |
| 3 | 0/0 (–) | 0/0 (–) | 0/0 (–) | 0/0 (–) | 0/1 (–) | 0/1 (–) | 28/32 (7.0) | 31/36 (6.2) | |
| NS | 1 | 0/0 (–) | 0/0 (–) | 0/1 (–) | 0/0 (–) | 0/0 (–) | 17/18 (17.0) | 55/63 (6.9) | 10/14 (2.5) |
| 2 | 0/0 (–) | 0/0 (–) | 0/0 (–) | 0/0 (–) | 0/0 (–) | 10/14 (2.5) | 20/23 (6.7) | 17/21 (4.3) | |
| 3 | 0/0 (–) | 0/0 (–) | 0/0 (–) | 0/0 (–) | 0/0 (–) | 1/1 (–) | 17/18 (17.0) | 16/20 (4.0) | |
| Gene | Passage | H1N1 | H3N2 | IBV | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT | I38L | I38T | E199D | WT | I38T | WT | I38T | ||||||||||
| 5% | 30% | 5% | 30% | 5% | 30% | 5% | 30% | 5% | 30% | 5% | 30% | 5% | 30% | 5% | 30% | ||
| PB2 | 1 | 2 * | – ‡ | 2 | – | 1 | – | 1 | – | 0 | – | 87 | M735L | 95 | Q24K §, T25K, Y30N, N31K, I32K | 28 | – |
| 2 | 1 | – | 0 | – | 1 | – | 1 | – | 6 | – | 33 | – | 71 | Q24K, T25K | 71 | Q24K, T25K, Y30N | |
| 3 | 0 | – | 0 | – | 0 | – | 1 | – | 0 | – | 6 | – | 67 | Q24K, T25K, Y30N | 65 | Q24K, T25K, Y30N | |
| PB1 | 1 | 0 | – | 0 | – | 0 | – | 1 | – | 1 | – | 79 | R187K | 79 | – | 13 | – |
| 2 | 0 | – | 0 | – | 0 | – | 0 | – | 0 | – | 32 | – | 49 | – | 53 | – | |
| 3 | 0 | – | 0 | – | 0 | – | 0 | – | 3 | – | 5 | – | 56 | – | 54 | – | |
| PA | 1 | 0 | – | 1 | – | 0 | – | 1 | – | 0 | – | 81 | – | 69 | – | 25 | – |
| 2 | 0 | – | 1 | – | 0 | – | 0 | – | 10 | G372E | 26 | – | 55 | – | 49 | E329G | |
| 3 | 1 | – | 1 | D394N † | 0 | – | 0 | – | 2 | G372E | 1 | – | 49 | – | 47 | E329G | |
| HA | 1 | 0 | – | 0 | – | 0 | – | 1 | – | 0 | – | 72 | R139K | 54 | – | 11 | – |
| 2 | 0 | – | 0 | – | 0 | – | 0 | – | 3 | – | 35 | – | 39 | – | 40 | – | |
| 3 | 0 | – | 0 | – | 0 | – | 0 | – | 0 | – | 3 | – | 38 | – | 42 | – | |
| NP | 1 | 0 | – | 0 | – | 0 | – | 0 | – | 0 | – | 45 | – | 44 | – | 8 | – |
| 2 | 0 | – | 0 | – | 0 | – | 0 | – | 4 | – | 21 | – | 23 | – | 29 | – | |
| 3 | 0 | – | 0 | – | 0 | – | 0 | – | 0 | – | 1 | – | 28 | – | 30 | – | |
| NA | 1 | 0 | – | 0 | – | 0 | – | 0 | – | 1 | – | 39 | E221K | 45 | – | 21 | – |
| 2 | 0 | – | 0 | – | 0 | – | 0 | – | 5 | – | 26 | – | 34 | – | 36 | – | |
| 3 | 0 | – | 0 | – | 0 | – | 0 | – | 0 | – | 4 | – | 25 | – | 30 | – | |
| M | 1 | 0 | – | 0 | – | 0 | – | 0 | – | 0 | – | 16 | – | 36 | – | 10 | – |
| 2 | 0 | – | 0 | – | 0 | – | 0 | – | 7 | – | 10 | – | 23 | – | 30 | – | |
| 3 | 0 | – | 0 | – | 0 | – | 0 | – | 0 | – | 0 | – | 28 | – | 31 | – | |
| NS | 1 | 0 | – | 0 | – | 0 | – | 0 | – | 0 | – | 17 | Q21K | 55 | – | 10 | – |
| 2 | 0 | – | 0 | – | 0 | – | 0 | – | 0 | – | 10 | – | 20 | – | 17 | – | |
| 3 | 0 | – | 0 | – | 0 | – | 0 | – | 0 | – | 1 | – | 17 | – | 16 | – | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hickerson, B.T.; Huang, B.K.; Petrovskaya, S.N.; Ilyushina, N.A. Genomic Analysis of Influenza A and B Viruses Carrying Baloxavir Resistance-Associated Substitutions Serially Passaged in Human Epithelial Cells. Viruses 2023, 15, 2446. https://doi.org/10.3390/v15122446
Hickerson BT, Huang BK, Petrovskaya SN, Ilyushina NA. Genomic Analysis of Influenza A and B Viruses Carrying Baloxavir Resistance-Associated Substitutions Serially Passaged in Human Epithelial Cells. Viruses. 2023; 15(12):2446. https://doi.org/10.3390/v15122446
Chicago/Turabian StyleHickerson, Brady T., Bruce K. Huang, Svetlana N. Petrovskaya, and Natalia A. Ilyushina. 2023. "Genomic Analysis of Influenza A and B Viruses Carrying Baloxavir Resistance-Associated Substitutions Serially Passaged in Human Epithelial Cells" Viruses 15, no. 12: 2446. https://doi.org/10.3390/v15122446
APA StyleHickerson, B. T., Huang, B. K., Petrovskaya, S. N., & Ilyushina, N. A. (2023). Genomic Analysis of Influenza A and B Viruses Carrying Baloxavir Resistance-Associated Substitutions Serially Passaged in Human Epithelial Cells. Viruses, 15(12), 2446. https://doi.org/10.3390/v15122446




