Pathogenesis of Rift Valley Fever Virus in a BALB/c Mouse Model Is Affected by Virus Culture Conditions and Sex of the Animals
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Animals
2.3. Cells
2.4. Virus
2.5. Sequencing
2.6. Challenge Study Design
2.7. Challenge, Monitoring and Sampling
2.8. Viral RNA Measurement
2.9. Histopathological Analysis
2.10. Luminex Analysis
2.11. Statistical Analysis
3. Results
3.1. Sequence Comparison of RVFV Grown in Insect and Mammalian Cell Lines
3.2. RVF Virus Cultivated in Insect Cells Results in a More Rapid Disease Progression, but the Same Dose Is Required for All Animals to Meet Humane Clinical Endpoints, Compared to Mammalian Cell-Grown Virus
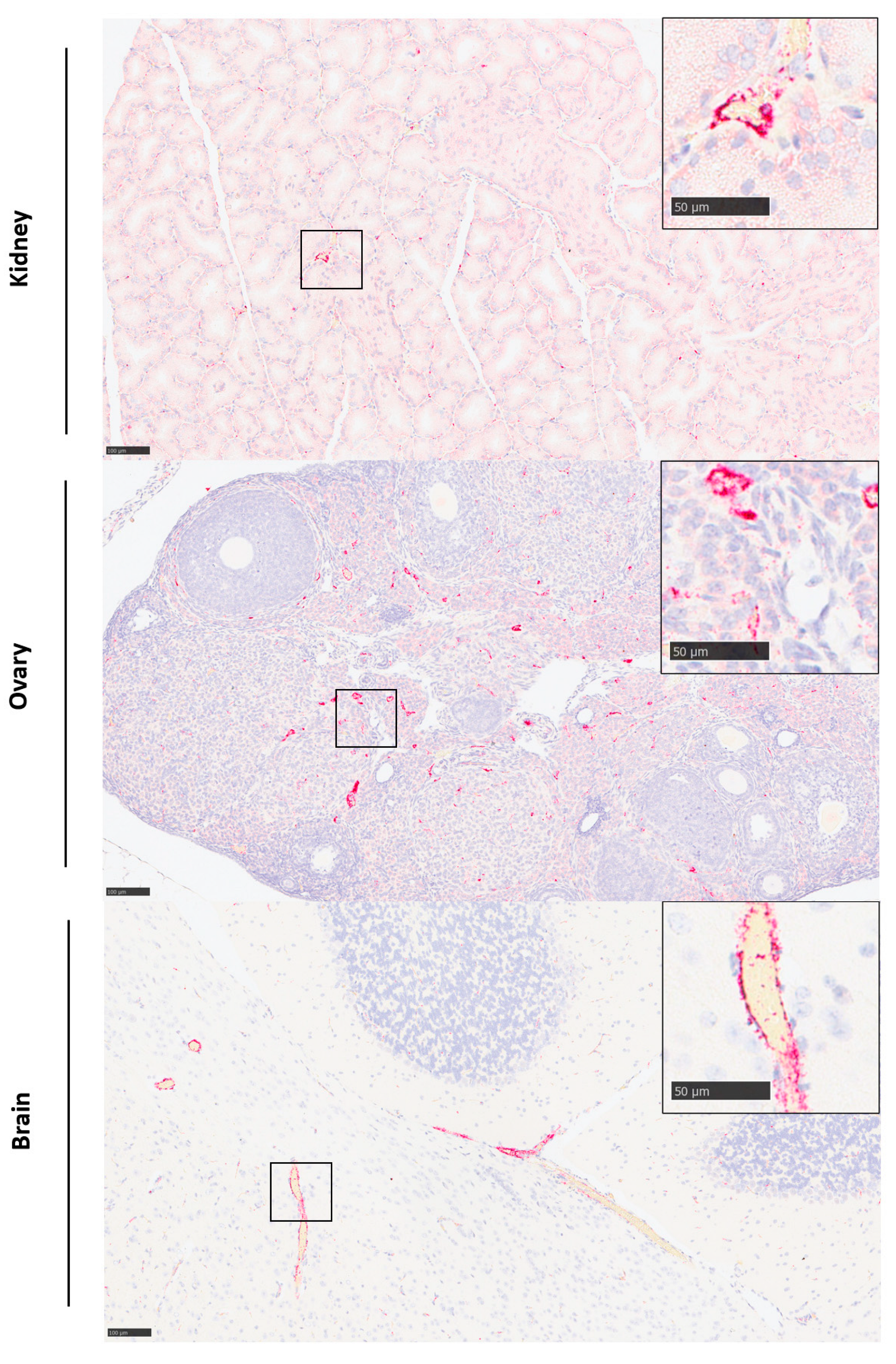
3.3. The Initial Virus Tropism Is to the Liver, before Becoming More Widely Disseminated throughout Other Tissues
3.4. Histopathological Lesions Were Observed in the Liver and Spleen
3.5. Viral RNA Was Detected in a Range of Tissues
3.6. Cytokine, Chemokine and Growth Factor Levels Were Elevated after Challenge with RVF Virus
3.7. Challenge with 10 Pfu Mammalian Cell-Grown RVF Virus Is Reproducible and Results in a More Rapid Disease Progression in Male Mice
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abudurexiti, A.; Adkins, S.; Alioto, D.; Alkhovsky, S.V.; Avsic-Zupanc, T.; Ballinger, M.J.; Bente, D.A.; Beer, M.; Bergeron, E.; Blair, C.D.; et al. Taxonomy of the order Bunyavirales: Update 2019. Arch. Virol. 2019, 164, 1949–1965. [Google Scholar] [CrossRef] [PubMed]
- Daubney, R.; Garnham, P. Enzootic hepatitis of Rift Valley fever: An undescribed virus disease of sheep, cattle and man from East Africa. J. Pathol. Bacteriol. 1931, 34, 8922–8926. [Google Scholar] [CrossRef]
- McMillen, C.M.; Hartman, A.L. Rift Valley fever in animals and humans: Current perspectives. Antivir. Res. 2018, 156, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Outbreak of Rift Valley fever–Yemen, August–October 2000. MMWR Morb. Mortal. Wkly. Rep. 2000, 49, 1065–1066. [Google Scholar]
- Madani, T.A.; Al-Mazrou, Y.Y.; Al-Jeffri, M.H.; Mishkhas, A.A.; Al-Rabeah, A.M.; Turkistani, A.M.; Al-Sayed, M.O.; Abodahish, A.A.; Khan, A.S.; Ksiazek, T.G.; et al. Rift Valley Fever Epidemic in Saudi Arabia: Epidemiological, Clinical, and Laboratory Characteristics. Clin. Infect. Dis. 2003, 37, 1084–1092. [Google Scholar] [CrossRef]
- Coetzer, J.A. The pathology of Rift Valley fever. II. Lesions occurring in field cases in adult cattle, calves and aborted foetuses. Onderstepoort J. Vet. Res. 1982, 49, 11–17. [Google Scholar] [PubMed]
- Bird, B.H.; Githinji, J.W.K.; Macharia, J.M.; Kasiiti, J.L.; Muriithi, R.M.; Gacheru, S.G.; Musaa, J.O.; Towner, J.S.; Reeder, S.A.; Oliver, J.B.; et al. Multiple Virus Lineages Sharing Recent Common Ancestry Were Associated with a Large Rift Valley Fever Outbreak among Livestock in Kenya during 2006–2007. J. Virol. 2008, 82, 11152–11166. [Google Scholar] [CrossRef]
- Ikegami, T.; Makino, S. The Pathogenesis of Rift Valley Fever. Viruses 2011, 3, 493–519. [Google Scholar] [CrossRef]
- Mehand, M.S.; Al-Shorbaji, F.; Millett, P.; Murgue, B. The WHO R&D Blueprint: 2018 review of emerging infectious diseases requiring urgent research and development efforts. Antivir. Res. 2018, 159, 63–67. [Google Scholar] [CrossRef]
- Kitandwe, P.K.; McKay, P.F.; Kaleebu, P.; Shattock, R.J. An Overview of Rift Valley Fever Vaccine Development Strategies. Vaccines 2022, 10, 1794. [Google Scholar] [CrossRef]
- Finch, C.L.; Dowling, W.E.; King, T.H.; Martinez, C.; Nguyen, B.V.; Roozendaal, R.; Rustomjee, R.; Skiadopoulos, M.H.; Vert-Wong, E.; Yellowlees, A.; et al. Bridging Animal and Human Data in Pursuit of Vaccine Licensure. Vaccines 2022, 10, 1384. [Google Scholar] [CrossRef]
- Mims, C.A. Rift Valley Fever virus in mice. I. General features of the infection. Br. J. Exp. Pathol. 1956, 37, 99–109. [Google Scholar] [PubMed]
- Shansky, R.M.; Murphy, A.Z. Considering sex as a biological variable will require a global shift in science culture. Nat. Neurosci. 2021, 24, 457–464. [Google Scholar] [CrossRef] [PubMed]
- Tharmarajah, K.; Everest-Dass, A.; Vider, J.; Liu, X.; Freitas, J.R.; Mostafavi, H.; Bettadapura, J.; von Itzstein, M.; West, N.P.; Taylor, A.; et al. N-Linked Glycans Shape Skin Immune Responses during Arthritis and Myositis after Intradermal Infection with Ross River Virus. J. Virol. 2022, 96, e0099922. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, C.; Pristatsky, P.; Hoang, V.M.; Casimiro, D.R.; Schwartz, R.M.; Rustandi, R.; Ha, S. Characterization of N-glycosylation profiles from mammalian and insect cell derived chikungunya VLP. J. Chromatogr. B 2016, 1032, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Meegan, J.M. The Rift Valley fever epizootic in Egypt 1977-78. 1. Description of the epizzotic and virological studies. Trans. R. Soc. Trop. Med. Hyg. 1979, 73, 618–623. [Google Scholar] [CrossRef] [PubMed]
- Lumley, S.; Horton, D.L.; Hernandez-Triana, L.L.M.; Johnson, N.; Fooks, A.R.; Hewson, R. Rift Valley fever virus: Strategies for maintenance, survival and vertical transmission in mosquitoes. J. Gen. Virol. 2017, 98, 875–887. [Google Scholar] [CrossRef]
- Kafetzopoulou, L.E.; Efthymiadis, K.; Lewandowski, K.; Crook, A.; Carter, D.; Osborne, J.; Aarons, E.; Hewson, R.; Hiscox, J.A.; Carroll, M.W.; et al. Assessment of metagenomic Nanopore and Illumina sequencing for recovering whole genome sequences of chikungunya and dengue viruses directly from clinical samples. Eurosurveillance 2018, 23, pii=1800228. [Google Scholar] [CrossRef]
- Lewandowski, K.; Xu, Y.; Pullan, S.T.; Lumley, S.F.; Foster, D.; Sanderson, N.; Vaughan, A.; Morgan, M.; Bright, N.; Kavanagh, J.; et al. Metagenomic Nanopore Sequencing of Influenza Virus Direct from Clinical Respiratory Samples. J. Clin. Microbiol. 2019, 58, e00963-19. [Google Scholar] [CrossRef]
- Greninger, A.L.; Naccache, S.N.; Federman, S.; Yu, G.; Mbala, P.; Bres, V.; Stryke, D.; Bouquet, J.; Somasekar, S.; Linnen, J.M.; et al. Rapid metagenomic identification of viral pathogens in clinical samples by real-time nanopore sequencing analysis. Genome Med. 2015, 7, 99. [Google Scholar] [CrossRef]
- Penedos, A.R.; Myers, R.; Hadef, B.; Aladin, F.; Brown, K.E. Assessment of the Utility of Whole Genome Sequencing of Measles Virus in the Characterisation of Outbreaks. PLoS ONE 2015, 10, e0143081. [Google Scholar] [CrossRef] [PubMed]
- Drosten, C.; Göttig, S.; Schilling, S.; Asper, M.; Panning, M.; Schmitz, H.; Günther, S. Rapid Detection and Quantification of RNA of Ebola and Marburg Viruses, Lassa Virus, Crimean-Congo Hemorrhagic Fever Virus, Rift Valley Fever Virus, Dengue Virus, and Yellow Fever Virus by Real-Time Reverse Transcription-PCR. J. Clin. Microbiol. 2002, 40, 2323–2330. [Google Scholar] [CrossRef] [PubMed]
- Findlay-Wilson, S.; Flett, L.; Salguero, F.J.; Ruedas-Torres, I.; Fotheringham, S.; Easterbrook, L.; Graham, V.; Dowall, S. Establishment of a Nipah Virus Disease Model in Hamsters, including a Comparison of Intranasal and Intraperitoneal Routes of Challenge. Pathogens 2023, 12, 976. [Google Scholar] [CrossRef] [PubMed]
- Dowall, S.D.; Graham, V.A.; Fletcher, T.; Hewson, R. Use and reliability of multiplex bead-based assays (Luminex) at Containment Level 4. Methods 2019, 158, 17–21. [Google Scholar] [CrossRef]
- Dowall, S.D.; Graham, V.A.; Tipton, T.R.; Hewson, R. Multiplex cytokine profiling with highly pathogenic material: Use of formalin solution in luminex analysis. J. Immunol. Methods 2009, 348, 30–35. [Google Scholar] [CrossRef]
- Morrill, J.C.; Ikegami, T.; Yoshikawa-Iwata, N.; Lokugamage, N.; Won, S.; Terasaki, K.; Zamoto-Niikura, A.; Peters, C.J.; Makino, S. Rapid accumulation of virulent rift valley Fever virus in mice from an attenuated virus carrying a single nucleotide substitution in the m RNA. PLoS ONE 2010, 5, e9986. [Google Scholar] [CrossRef]
- Bui, T.T.; Moi, M.L.; Nabeshima, T.; Takemura, T.; Nguyen, T.T.; Nguyen, L.N.; Pham, H.T.T.; Nguyen, T.T.T.; Manh, D.H.; Dumre, S.P.; et al. A single amino acid substitution in the NS4B protein of Dengue virus confers enhanced virus growth and fitness in human cells in vitro through IFN-dependent host response. J. Gen. Virol. 2018, 99, 1044–1057. [Google Scholar] [CrossRef]
- Modak, A.; Mishra, S.R.; Awasthi, M.; Sreedevi, S.; Sobha, A.; Aravind, A.; Kuppusamy, K.; Sreekumar, E. Higher-temperature-adapted dengue virus serotype 2 strain exhibits enhanced virulence in AG129 mouse model. FASEB J. 2023, 37, e23062. [Google Scholar] [CrossRef]
- Shabman, R.S.; Morrison, T.E.; Moore, C.; White, L.; Suthar, M.S.; Hueston, L.; Rulli, N.; Lidbury, B.; Ting, J.P.; Mahalingam, S.; et al. Differential induction of type I interferon responses in myeloid dendritic cells by mosquito and mammalian-cell-derived alphaviruses. J. Virol. 2007, 81, 237–247. [Google Scholar] [CrossRef]
- Näslund, J.; Lagerqvist, N.; Lundkvist, Å.; Evander, M.; Ahlm, C.; Bucht, G. Kinetics of Rift Valley Fever Virus in experimentally infected mice using quantitative real-time RT-PCR. J. Virol. Methods 2008, 151, 277–282. [Google Scholar] [CrossRef]
- Reed, C.; Lin, K.; Wilhelmsen, C.; Friedrich, B.; Nalca, A.; Keeney, A.; Donnelly, G.; Shamblin, J.; Hensley, L.E.; Olinger, G.; et al. Aerosol Exposure to Rift Valley Fever Virus Causes Earlier and More Severe Neuropathology in the Murine Model, which Has Important Implications for Therapeutic Development. PLoS Neglected Trop. Dis. 2013, 7, e2156. [Google Scholar] [CrossRef]
- Smith, D.R.; Steele, K.E.; Shamblin, J.; Honko, A.; Johnson, J.; Reed, C.; Kennedy, M.; Chapman, J.L.; Hensley, L.E. The pathogenesis of Rift Valley fever virus in the mouse model. Virology 2010, 407, 256–267. [Google Scholar] [CrossRef] [PubMed]
- Gray, K.K.; Worthy, M.N.; Juelich, T.L.; Agar, S.L.; Poussard, A.; Ragland, D.; Freiberg, A.N.; Holbrook, M.R. Chemotactic and Inflammatory Responses in the Liver and Brain Are Associated with Pathogenesis of Rift Valley Fever Virus Infection in the Mouse. PLoS Negl. Trop. Dis. 2012, 6, e1529. [Google Scholar] [CrossRef] [PubMed]
- Nair, N.; Osterhaus, A.D.M.E.; Rimmelzwaan, G.F.; Prajeeth, C.K. Rift Valley Fever Virus—Infection, Pathogenesis and Host Immune Responses. Pathogens 2023, 12, 1174. [Google Scholar] [CrossRef] [PubMed]
- Msimang, V.; Thompson, P.N.; van Vuren, P.J.; Tempia, S.; Cordel, C.; Kgaladi, J.; Khosa, J.; Burt, F.J.; Liang, J.; Rostal, M.K.; et al. Rift Valley Fever Virus Exposure amongst Farmers, Farm Workers, and Veterinary Professionals in Central South Africa. Viruses 2019, 11, 140. [Google Scholar] [CrossRef]
- Le Coupanec, A.; Babin, D.; Fiette, L.; Jouvion, G.; Ave, P.; Misse, D.; Bouloy, M.; Choumet, V. Aedes Mosquito Saliva Modulates Rift Valley Fever Virus Pathogenicity. PLoS Neglected Trop. Dis. 2013, 7, e2237. [Google Scholar] [CrossRef] [PubMed]
- Cartwright, H.N.; Barbeau, D.J.; Doyle, J.D.; Klein, E.; Heise, M.T.; Ferris, M.T.; McElroy, A.K. Genetic diversity of collaborative cross mice enables identification of novel rift valley fever virus encephalitis model. PLoS Pathog. 2022, 18, e1010649. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-R.; Liu, M.-T.; Lei, H.-Y.; Liu, C.-C.; Wu, J.-M.; Tung, Y.-C.; Lin, Y.-S.; Yeh, T.-M.; Chen, S.-H.; Liu, H.-S. MCP-1, a highly expressed chemokine in dengue haemorrhagic fever/dengue shock syndrome patients, may cause permeability change, possibly through reduced tight junctions of vascular endothelium cells. J. Gen. Virol. 2006, 87, 3623–3630. [Google Scholar] [CrossRef]
- Nfon, C.K.; Marszal, P.; Zhang, S.; Weingartl, H.M. Innate Immune Response to Rift Valley Fever Virus in Goats. PLoS Neglected Trop. Dis. 2012, 6, e1623. [Google Scholar] [CrossRef]
- Ganaie, S.S.; Schwarz, M.M.; McMillen, C.M.; Price, D.A.; Feng, A.X.; Albe, J.R.; Wang, W.; Miersch, S.; Orvedahl, A.; Cole, A.R.; et al. Lrp1 is a host entry factor for Rift Valley fever virus. Cell 2021, 184, 5163–5178.e24. [Google Scholar] [CrossRef]
- Abbo, S.R.; Nguyen, W.; Abma-Henkens, M.H.C.; van de Kamer, D.; Savelkoul, N.H.A.; Geertsema, C.; Le, T.T.T.; Tang, B.; Yan, K.; Dumenil, T.; et al. Comparative Efficacy of Mayaro Virus-Like Particle Vaccines Produced in Insect or Mammalian Cells. J. Virol. 2023, 97, e0160122. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.M.; Pajerowski, J.D.; Daniels, C.L.; McHugh, P.M.; Flynn, J.A.; Balliet, J.W.; Casimiro, D.R.; Subramanian, S. Enhanced production of Chikungunya virus-like particles using a high-pH adapted spodoptera frugiperda insect cell line. PLoS ONE 2014, 9, e94401. [Google Scholar] [CrossRef]
- Schön, K.; Lindenwald, D.L.; Monteiro, J.T.; Glanz, J.; Jung, K.; Becker, S.C.; Lepenies, B. Vector and Host C-Type Lectin Receptor (CLR)–Fc Fusion Proteins as a Cross-Species Comparative Approach to Screen for CLR–Rift Valley Fever Virus Interactions. Int. J. Mol. Sci. 2022, 23, 3243. [Google Scholar] [CrossRef] [PubMed]
- Gregor, K.M.; Michaely, L.M.; Gutjahr, B.; Rissmann, M.; Keller, M.; Dornbusch, S.; Naccache, F.; Schön, K.; Jansen, S.; Heitmann, A.; et al. Rift Valley fever virus detection in susceptible hosts with special emphasis in insects. Sci. Rep. 2021, 11, 9822. [Google Scholar] [CrossRef] [PubMed]
- Brennan, B.; Welch, S.R.; Elliott, R.M. The Consequences of Reconfiguring the Ambisense S Genome Segment of Rift Valley Fever Virus on Viral Replication in Mammalian and Mosquito Cells and for Genome Packaging. PLoS Pathog. 2014, 10, e1003922. [Google Scholar] [CrossRef]
- Vaughn, V.M.; Streeter, C.C.; Miller, D.J.; Gerrard, S.R. Restriction of Rift Valley Fever Virus Virulence in Mosquito Cells. Viruses 2010, 2, 655–675. [Google Scholar] [CrossRef]
- Bouloy, M.; Janzen, C.; Vialat, P.; Khun, H.; Pavlovic, J.; Huerre, M.; Haller, O. Genetic Evidence for an Interferon-Antagonistic Function of Rift Valley Fever Virus Nonstructural Protein NSs. J. Virol. 2001, 75, 1371–1377. [Google Scholar] [CrossRef]
- Habjan, M.; Pichlmair, A.; Elliott, R.M.; Överby, A.K.; Glatter, T.; Gstaiger, M.; Superti-Furga, G.; Unger, H.; Weber, F. NSs Protein of Rift Valley Fever Virus Induces the Specific Degradation of the Double-Stranded RNA-Dependent Protein Kinase. J. Virol. 2009, 83, 4365–4375. [Google Scholar] [CrossRef]
- Weingartl, H.M.; Zhang, S.; Marszal, P.; McGreevy, A.; Burton, L.; Wilson, W.C. Rift Valley fever virus incorporates the 78 kDa glycoprotein into virions matured in mosquito C6/36 cells. PLoS ONE 2014, 9, e87385. [Google Scholar] [CrossRef]
- Terasaki, K.; Kalveram, B.; Johnson, K.N.; Juelich, T.; Smith, J.K.; Zhang, L.; Freiberg, A.N.; Makino, S. Rift Valley fever virus 78kDa envelope protein attenuates virus replication in macrophage-derived cell lines and viral virulence in mice. PLoS Neglected Trop. Dis. 2021, 15, e0009785. [Google Scholar] [CrossRef]
- Jones, B.G.; Sealy, R.E.; Penkert, R.R.; Surman, S.L.; Maul, R.W.; Neale, G.; Xu, B.; Gearhart, P.J.; Hurwitz, J.L. Complex sex-biased antibody responses: Estrogen receptors bind estrogen response elements centered within immunoglobulin heavy chain gene enhancers. Int. Immunol. 2019, 31, 141–156. [Google Scholar] [CrossRef]
- Surman, S.L.; Jones, B.G.; Penkert, R.R.; Sealy, R.E.; Marion, T.; Thomas, P.G.; Neale, G.; Xu, B.; Hurwitz, J.L. How Estrogen, Testosterone, and Sex Differences Influence Serum Immunoglobulin Isotype Patterns in Mice and Humans. Viruses 2023, 15, 482. [Google Scholar] [CrossRef] [PubMed]
- Cartwright, H.N.; Barbeau, D.J.; McElroy, A.K. Rift Valley Fever Virus Is Lethal in Different Inbred Mouse Strains Independent of Sex. Front. Microbiol. 2020, 11, 1962. [Google Scholar] [CrossRef] [PubMed]
- Snyder-Keller, A.; Kramer, L.D.; Zink, S.; Bolivar, V.J. Mouse Strain and Sex-Dependent Differences in Long-term Behavioral Abnormalities and Neuropathologies after Developmental Zika Infection. J. Neurosci. 2019, 39, 5393–5403. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, J.M.; Sholar, P.W.; Bilbo, S.D. Sex differences in microglial colonization of the developing rat brain. J. Neurochem. 2011, 120, 948–963. [Google Scholar] [CrossRef] [PubMed]
- Huber, S.A. Increased susceptibility of male BALB/c mice to coxsackievirus B3-induced myocarditis: Role for CD1d. Med. Microbiol. Immunol. 2004, 194, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Roberts, B.J.; Moussawi, M.; Huber, S.A. Sex differences in TLR2 and TLR4 expression and their effect on coxsackievirus-induced autoimmune myocarditis. Exp. Mol. Pathol. 2013, 94, 58–64. [Google Scholar] [CrossRef]
- Geurs, T.L.; Hill, E.B.; Lippold, D.M.; French, A.R. Sex differences in murine susceptibility to systemic viral infections. J. Autoimmun. 2012, 38, J245–J253. [Google Scholar] [CrossRef]
- Yee Mon, K.J.; Goldsmith, E.; Watson, N.B.; Wang, J.; Smith, N.L.; Rudd, B.D. Differential Sensitivity to IL-12 Drives Sex-Specific Differences in the CD8+ T Cell Response to Infection. ImmunoHorizons 2019, 3, 121–132. [Google Scholar] [CrossRef]
- Humeniuk, P.; Barrett, A.; Axelsson, H.; Corciulo, C.; Drevinge, C.; Pons, A.D.C.; Angeletti, D.; Scheffler, J.M.; Islander, U. Profiling of innate and adaptive immune cells during influenza virus infection reveals sex bias in invariant natural killer T (iNKT) cells. Immun. Inflamm. Dis. 2023, 11, e837. [Google Scholar] [CrossRef]
- Robinson, D.P.; Lorenzo, M.E.; Jian, W.; Klein, S.L. Elevated 17beta-estradiol protects females from influenza A virus pathogenesis by suppressing inflammatory responses. PLoS Pathog. 2011, 7, e1002149. [Google Scholar] [CrossRef] [PubMed]
- Celestino, I.; Checconi, P.; Amatore, D.; De Angelis, M.; Coluccio, P.; Dattilo, R.; Fegatelli, D.A.; Clemente, A.M.; Matarrese, P.; Torcia, M.G.; et al. Differential Redox State Contributes to Sex Disparities in the Response to Influenza Virus Infection in Male and Female Mice. Front. Immunol. 2018, 9, 1747. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, J.; Otte, A.; Thiele, S.; Lotter, H.; Shu, Y.; Gabriel, G. Sex differences in H7N9 influenza A virus pathogenesis. Vaccine 2015, 33, 6949–6954. [Google Scholar] [CrossRef]
- Channappanavar, R.; Fett, C.; Mack, M.; Ten Eyck, P.P.; Meyerholz, D.K.; Perlman, S. Sex-Based Differences in Susceptibility to Severe Acute Respiratory Syndrome Coronavirus Infection. J. Immunol. 2017, 198, 4046–4053. [Google Scholar] [CrossRef]
- Viveiros, A.; Gheblawi, M.; Aujla, P.K.; Sosnowski, D.K.; Seubert, J.M.; Kassiri, Z.; Oudit, G.Y. Sex- and age-specific regulation of ACE2: Insights into severe COVID-19 susceptibility. J. Mol. Cell. Cardiol. 2021, 164, 13–16. [Google Scholar] [CrossRef] [PubMed]
- Viveiros, A.; Rasmuson, J.; Vu, J.; Mulvagh, S.L.; Yip, C.Y.Y.; Norris, C.M.; Oudit, G.Y. Sex differences in COVID-19: Candidate pathways, genetics of ACE2, and sex hormones. Am. J. Physiol. Circ. Physiol. 2021, 320, H296–H304. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, H.; Klein, S.L. Sex Differences in Immunity to Viral Infections. Front. Immunol. 2021, 12, 720952. [Google Scholar] [CrossRef]
- Davis, M.A.; Voss, K.; Turnbull, J.B.; Gustin, A.T.; Knoll, M.; Muruato, A.; Hsiang, T.-Y.; Dinnonn III, K.H.; Leist, S.R.; Nickel, K.; et al. A C57BL/6 mouse model of SARS-CoV-2 infection recapitulates age- and sex-based differences in human COVID-19 disease and recovery. Vaccines 2023, 11, 47. [Google Scholar] [CrossRef]
- Stelzig, K.E.; Canepa-Escaro, F.; Schiliro, M.; Berdnikovs, S.; Prakash, Y.S.; Chiarella, S.E. Estrogen regulates the expression of SARS-CoV-2 receptor ACE2 in differentiated airway epithelial cells. Am. J. Physiol. Lung Cell Mol. Physiol. 2020, 318, L1280–L1281. [Google Scholar] [CrossRef]
- Flanagan, K.L.; Fink, A.L.; Plebanski, M.; Klein, S.L. Sex and Gender Differences in the Outcomes of Vaccination over the Life Course. Annu. Rev. Cell Dev. Biol. 2017, 33, 577–599. [Google Scholar] [CrossRef]
- Fink, A.L.; Engle, K.; Ursin, R.L.; Tang, W.-Y.; Klein, S.L. Biological sex affects vaccine efficacy and protection against influenza in mice. Proc. Natl. Acad. Sci. USA 2018, 115, 12477–12482. [Google Scholar] [CrossRef] [PubMed]


| Genome Segment | Segment Position | Mammalian Cell Base | Insect Cell Base | Reference Genome Base 1 |
|---|---|---|---|---|
| S | 522 | A (99.9%) | W (78.8% A/21.2% T) | A |
| M | 273 | G (1.3% A/98.7% G) | R (43.95% A/56.05% G) | G |
| M | 843 | Y (45.8% C/54.2% T) | T (99.95%) | T |
| M | 855 | G (2.9% A/97.1% G) | R (46.55% A/53.45% G) | G |
| M | 1315 | Y (76.6% C/23.4% T) | C (99.95%) | C |
| L | 5739 | Y (24.05% C/75.95% T) | T (100%) | T |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Graham, V.A.; Easterbrook, L.; Kennedy, E.; Rayner, E.; Findlay-Wilson, S.; Flett, L.; Wise, E.L.; Treagus, S.; Fotheringham, S.; Kempster, S.; et al. Pathogenesis of Rift Valley Fever Virus in a BALB/c Mouse Model Is Affected by Virus Culture Conditions and Sex of the Animals. Viruses 2023, 15, 2369. https://doi.org/10.3390/v15122369
Graham VA, Easterbrook L, Kennedy E, Rayner E, Findlay-Wilson S, Flett L, Wise EL, Treagus S, Fotheringham S, Kempster S, et al. Pathogenesis of Rift Valley Fever Virus in a BALB/c Mouse Model Is Affected by Virus Culture Conditions and Sex of the Animals. Viruses. 2023; 15(12):2369. https://doi.org/10.3390/v15122369
Chicago/Turabian StyleGraham, Victoria A., Linda Easterbrook, Emma Kennedy, Emma Rayner, Stephen Findlay-Wilson, Lucy Flett, Emma Louise Wise, Samantha Treagus, Susan Fotheringham, Sarah Kempster, and et al. 2023. "Pathogenesis of Rift Valley Fever Virus in a BALB/c Mouse Model Is Affected by Virus Culture Conditions and Sex of the Animals" Viruses 15, no. 12: 2369. https://doi.org/10.3390/v15122369
APA StyleGraham, V. A., Easterbrook, L., Kennedy, E., Rayner, E., Findlay-Wilson, S., Flett, L., Wise, E. L., Treagus, S., Fotheringham, S., Kempster, S., Almond, N., & Dowall, S. (2023). Pathogenesis of Rift Valley Fever Virus in a BALB/c Mouse Model Is Affected by Virus Culture Conditions and Sex of the Animals. Viruses, 15(12), 2369. https://doi.org/10.3390/v15122369






