Dancing with the Devil: A Review of the Importance of Host RNA-Binding Proteins to Alphaviral RNAs during Infection
Abstract
1. Introduction
2. Alphavirus Background
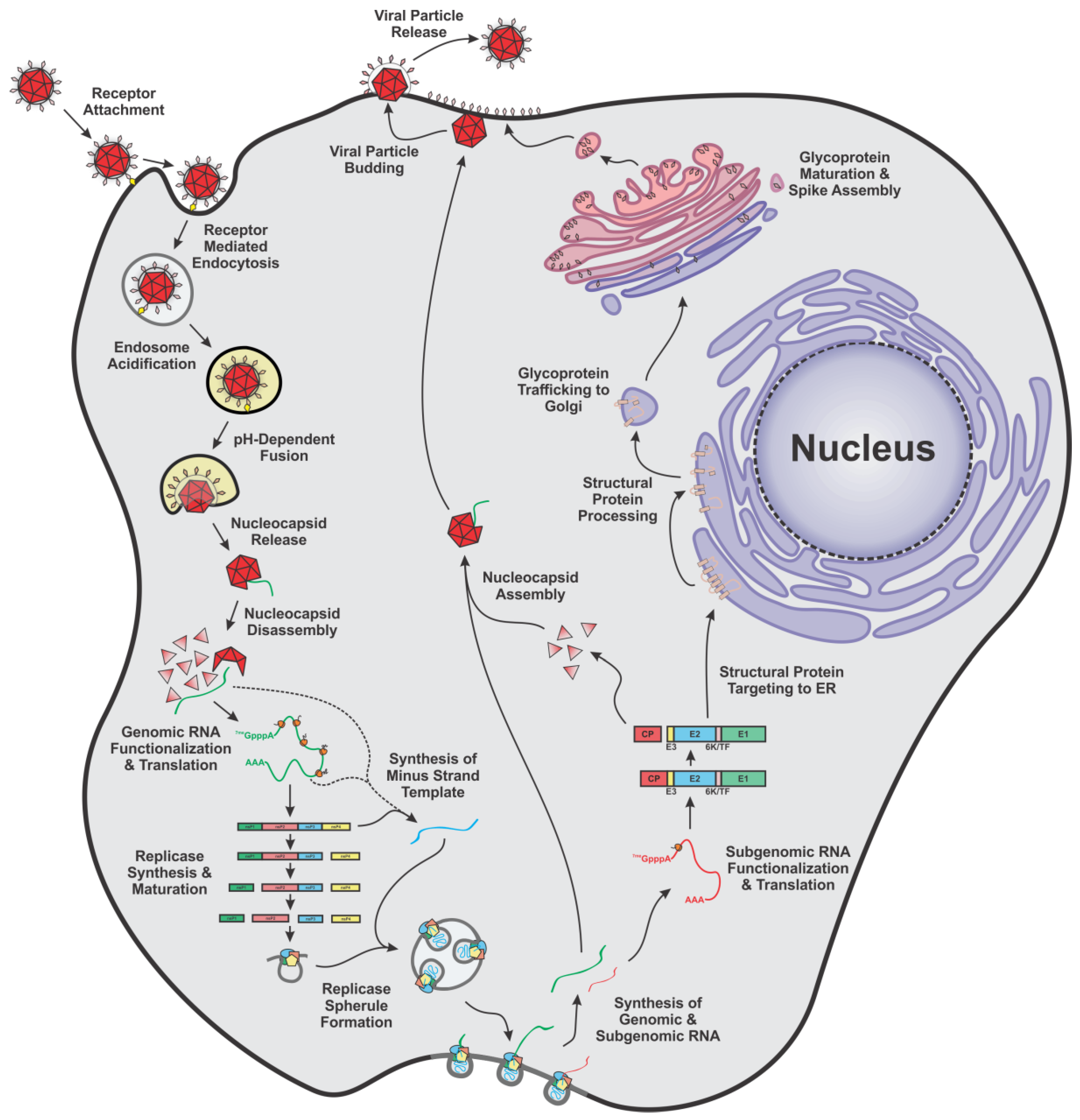
2.1. An Overview of the Molecular Lifecycle
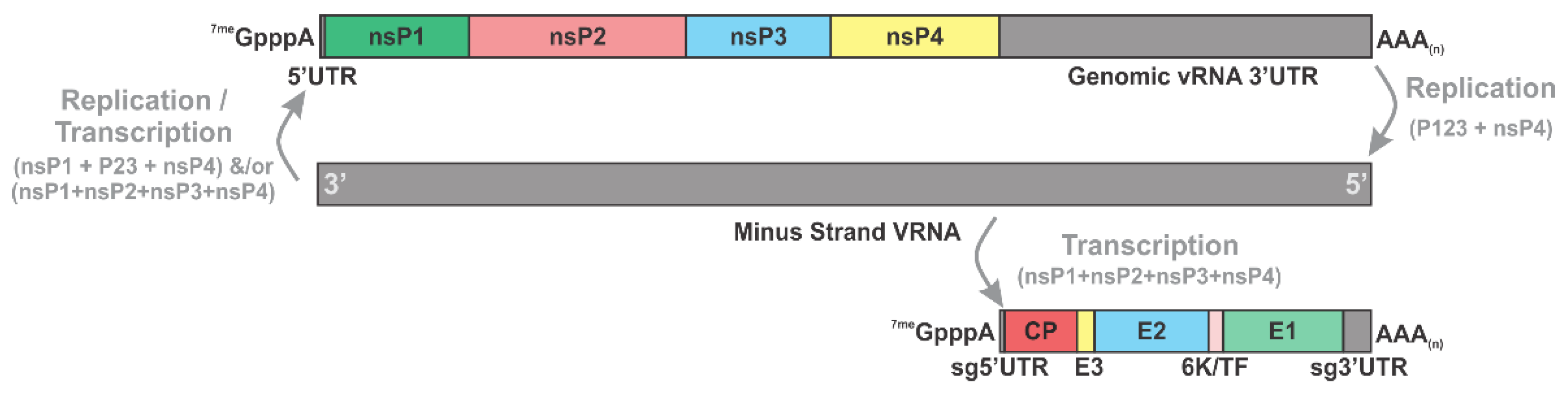
2.2. Genetic Organization of Alphaviruses
3. Discovery Efforts
3.1. Defining the Protein:Protein Interaction Interface of the Alphaviruses with their Host
3.2. Defining the RNA:Protein Interaction Interface of the Alphaviruses with their Host
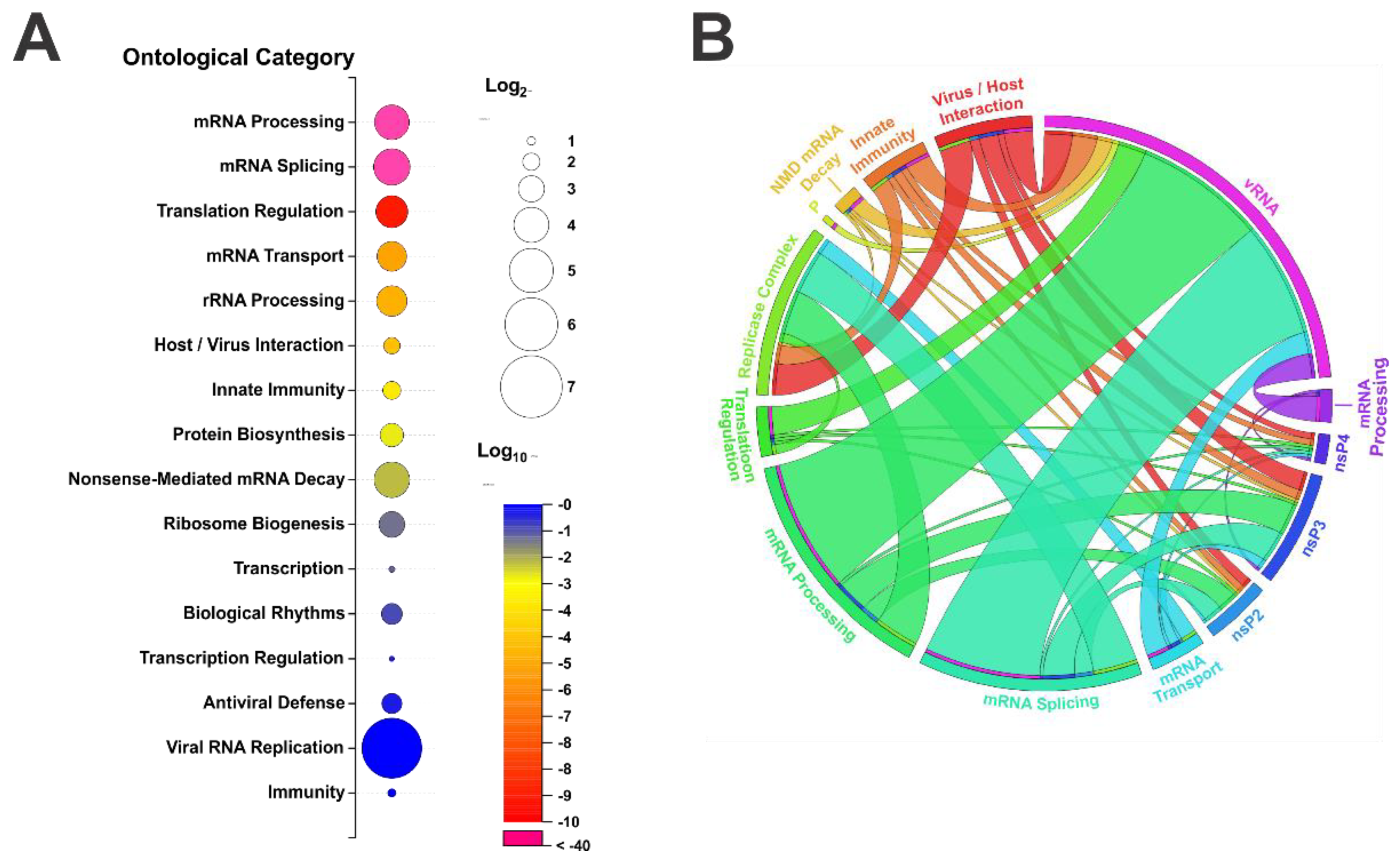
4. Ontological Analysis of Alphavirus—Host Protein Interactants
5. Specific Roles of RBPs during Alphavirus Infection
5.1. RNA Binding Proteins
5.1.1. La: A Possible Replication Regulator of 3′ Minus Strand
5.1.2. HuR/ELAV1: Stabilizing the Alphaviral RNAs from 3′ RNA Decay
5.1.3. G3BP1/2: Regulation of Alphaviral Replicase and Minus Strand Synthesis
5.1.4. TIA1/R: Promotes Infection by Forming Stress Granule Decoy
5.1.5. GEMIN5: Regulation of Protein Expression through 5′ UTR Interaction
5.1.6. FXR1: Interaction with the nsP3 Hypervariable Domain Promotes Infection
5.1.7. DEAD-Box Helicases: Promotion of Spreading Infection to Neighboring Cells
5.2. Heterogenous Nuclear RibonucleoProteins (hnRNPs)
5.2.1. hnRNP A1: Promotion of RNA Replication and Synthesis
5.2.2. hnRNP C: Negative Regulator of Alphaviral Infection
5.2.3. hnRNP K: Phosphorylated Form Regulates RNA Synthesis and Gene Translation
5.2.4. hnRNP M: Regulation of RNA Synthesis and Translation
5.2.5. PCBP1/hnRNP E1: Unknown Promotion of Alphaviral Infection
5.2.6. PTBP1/hnRNP I: Interaction with vRNA Regulations Translation
6. Perspectives and Limits of Understanding
6.1. Picking a Dance Partner: Who or What Decides Viral RNA Binding Specificity?
6.2. Dancing Backwards and in High Heels: Differential Host Functions and Requirements
6.3. Moving to the Beat: Subcellular (Re) Locations of RBPs during Infection
6.4. Taking Center Stage: Perspective Roles of RBPs during Infection
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Weaver, S.C.; Winegar, R.; Manger, I.D.; Forrester, N.L. Alphaviruses: Population genetics and determinants of emergence. Antivir. Res. 2012, 94, 242–257. [Google Scholar] [CrossRef] [PubMed]
- Knipe, D.; Howley, P.; Griffin, D.; Lamb, R.; Martin, M.; Roizman, B.; Straus, S. Fields Virology, Volumes 1 and 2; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013. [Google Scholar]
- Kendrick, K.; Stanek, D.; Blackmore, C. Notes from the field: Transmission of chikungunya virus in the continental United States—Florida, 2014. MMWR. Morb. Mortal. Wkly. Rep. 2014, 63, 1137. [Google Scholar] [PubMed]
- Weaver, S.C. Arrival of chikungunya virus in the new world: Prospects for spread and impact on public health. PLoS Negl. Trop. Dis. 2014, 8, e2921. [Google Scholar] [CrossRef]
- Johansson, M.A.; Powers, A.M.; Pesik, N.; Cohen, N.J.; Staples, J.E. Nowcasting the spread of chikungunya virus in the Americas. PLoS ONE 2014, 9, e104915. [Google Scholar] [CrossRef] [PubMed]
- Campbell, L.P.; Luther, C.; Moo-Llanes, D.; Ramsey, J.M.; Danis-Lozano, R.; Peterson, A.T. Climate change influences on global distributions of dengue and chikungunya virus vectors. Philos Trans. R Soc. Lond B Biol. Sci. 2015, 370, 20140135. [Google Scholar] [CrossRef]
- Sissoko, D.; Malvy, D.; Ezzedine, K.; Renault, P.; Moscetti, F.; Ledrans, M.; Pierre, V. Post-epidemic Chikungunya disease on Reunion Island: Course of rheumatic manifestations and associated factors over a 15-month period. PLoS Negl. Trop. Dis. 2009, 3, e389. [Google Scholar] [CrossRef] [PubMed]
- Levi, L.I.; Vignuzzi, M. Arthritogenic Alphaviruses: A Worldwide Emerging Threat? Microorganisms 2019, 7, 133. [Google Scholar] [CrossRef] [PubMed]
- Marimoutou, C.; Vivier, E.; Oliver, M.; Boutin, J.P.; Simon, F. Morbidity and impaired quality of life 30 months after chikungunya infection: Comparative cohort of infected and uninfected French military policemen in Reunion Island. Medicine 2012, 91, 212–219. [Google Scholar] [CrossRef]
- Kurkela, S.; Manni, T.; Myllynen, J.; Vaheri, A.; Vapalahti, O. Clinical and laboratory manifestations of Sindbis virus infection: Prospective study, Finland, 2002–2003. J. Infect. Dis. 2005, 191, 1820–1829. [Google Scholar] [CrossRef]
- Adouchief, S.; Smura, T.; Sane, J.; Vapalahti, O.; Kurkela, S. Sindbis virus as a human pathogen-epidemiology, clinical picture and pathogenesis. Rev. Med. Virol. 2016, 26, 221–241. [Google Scholar] [CrossRef]
- Kurkela, S.; Helve, T.; Vaheri, A.; Vapalahti, O. Arthritis and arthralgia three years after Sindbis virus infection: Clinical follow-up of a cohort of 49 patients. Scand. J. Infect. Dis. 2008, 40, 167–173. [Google Scholar] [CrossRef]
- Zacks, M.A.; Paessler, S. Encephalitic alphaviruses. Vet. Microbiol. 2010, 140, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Carrasco, L.; Sanz, M.A.; González-Almela, E. The Regulation of Translation in Alphavirus-Infected Cells. Viruses 2018, 10, 70. [Google Scholar] [CrossRef]
- Mendes, A.; Kuhn, R.J. Alphavirus Nucleocapsid Packaging and Assembly. Viruses 2018, 10, 138. [Google Scholar] [CrossRef] [PubMed]
- Button, J.M.; Qazi, S.A.; Wang, J.C.; Mukhopadhyay, S. Revisiting an old friend: New findings in alphavirus structure and assembly. Curr. Opin. Virol. 2020, 45, 25–33. [Google Scholar] [CrossRef]
- Schnierle, B.S. Cellular attachment and entry factors for chikungunya virus. Viruses 2019, 11, 1078. [Google Scholar] [CrossRef]
- DeTulleo, L.; Kirchhausen, T. The clathrin endocytic pathway in viral infection. EMBO J. 1998, 17, 4585–4593. [Google Scholar] [CrossRef] [PubMed]
- Dubuisson, J.; Rice, C.M. Sindbis virus attachment: Isolation and characterization of mutants with impaired binding to vertebrate cells. J. Virol. 1993, 67, 3363–3374. [Google Scholar] [CrossRef]
- Helenius, A.; Kartenbeck, J.; Simons, K.; Fries, E. On the entry of semliki forest virus into BHK-21 cells. J. Cell Biol. 1980, 84, 404–420. [Google Scholar] [CrossRef]
- Schmid, S.; Fuchs, R.; Kielian, M.; Helenius, A.; Mellman, I. Acidification of endosome subpopulations in wild-type Chinese hamster ovary cells and temperature-sensitive acidification-defective mutants. J. Cell Biol. 1989, 108, 1291–1300. [Google Scholar] [CrossRef]
- Fan, D.P.; Sefton, B.M. The entry into host cells of Sindbis virus, vesicular stomatitis virus and Sendai virus. Cell 1978, 15, 985–992. [Google Scholar] [CrossRef] [PubMed]
- Wengler, G.; Gros, C.; Wengler, G. Analyses of the role of structural changes in the regulation of uncoating and assembly of alphavirus cores. Virology 1996, 222, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Helenius, A. Semliki Forest virus penetration from endosomes: A morphological study. Biol. Cell 1984, 51, 181–185. [Google Scholar] [CrossRef]
- Wengler, G.; Koschinski, A.; Wengler, G.; Dreyer, F. Entry of alphaviruses at the plasma membrane converts the viral surface proteins into an ion-permeable pore that can be detected by electrophysiological analyses of whole-cell membrane currents. J. Gen. Virol. 2003, 84, 173–181. [Google Scholar] [CrossRef]
- Tuittila, M.T.; Santagati, M.G.; Röyttä, M.; Määttä, J.A.; Hinkkanen, A.E. Replicase complex genes of Semliki Forest virus confer lethal neurovirulence. J. Virol. 2000, 74, 4579–4589. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Rice, C.M. Mutagenesis of the in-frame opal termination codon preceding nsP4 of Sindbis virus: Studies of translational readthrough and its effect on virus replication. J. Virol. 1989, 63, 1326–1337. [Google Scholar] [CrossRef]
- Shirako, Y.; Strauss, J.H. Cleavage between nsP1 and nsP2 initiates the processing pathway of Sindbis virus nonstructural polyprotein P123. Virology 1990, 177, 54–64. [Google Scholar] [CrossRef]
- Lemm, J.A.; Rice, C.M. Assembly of functional Sindbis virus RNA replication complexes: Requirement for coexpression of P123 and P34. J. Virol. 1993, 67, 1905–1915. [Google Scholar] [CrossRef]
- Lemm, J.A.; Rümenapf, T.; Strauss, E.G.; Strauss, J.H.; Rice, C.M. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: A model for the temporal regulation of minus- and plus-strand RNA synthesis. Embo J. 1994, 13, 2925–2934. [Google Scholar] [CrossRef]
- Li, G.; Rice, C.M. The signal for translational readthrough of a UGA codon in Sindbis virus RNA involves a single cytidine residue immediately downstream of the termination codon. J. Virol. 1993, 67, 5062–5067. [Google Scholar] [CrossRef]
- Strauss, J.H.; Strauss, E.G. The alphaviruses: Gene expression, replication, and evolution. Microbiol. Rev. 1994, 58, 491–562. [Google Scholar] [CrossRef]
- Gorchakov, R.; Frolova, E.; Sawicki, S.; Atasheva, S.; Sawicki, D.; Frolov, I. A new role for ns polyprotein cleavage in Sindbis virus replication. J. Virol. 2008, 82, 6218–6231. [Google Scholar] [CrossRef]
- Hardy, W.R.; Hahn, Y.S.; de Groot, R.J.; Strauss, E.G.; Strauss, J.H. Synthesis and processing of the nonstructural polyproteins of several temperature-sensitive mutants of Sindbis virus. Virology 1990, 177, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Froshauer, S.; Kartenbeck, J.; Helenius, A. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J. Cell Biol. 1988, 107, 2075–2086. [Google Scholar] [CrossRef]
- Kujala, P.; Ikäheimonen, A.; Ehsani, N.; Vihinen, H.; Auvinen, P.; Kääriäinen, L. Biogenesis of the Semliki Forest virus RNA replication complex. J. Virol. 2001, 75, 3873–3884. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.S.; Wan, J.J.; Kielian, M. The Alphavirus Exit Pathway: What We Know and What We Wish We Knew. Viruses 2018, 10, 89. [Google Scholar] [CrossRef]
- Garoff, H.; Huylebroeck, D.; Robinson, A.; Tillman, U.; Liljeström, P. The signal sequence of the p62 protein of Semliki Forest virus is involved in initiation but not in completing chain translocation. J. Cell Biol. 1990, 111, 867–876. [Google Scholar] [CrossRef]
- Liljeström, P.; Garoff, H. Internally located cleavable signal sequences direct the formation of Semliki Forest virus membrane proteins from a polyprotein precursor. J. Virol. 1991, 65, 147–154. [Google Scholar] [CrossRef]
- Zhang, X.; Fugère, M.; Day, R.; Kielian, M. Furin processing and proteolytic activation of Semliki Forest virus. J. Virol. 2003, 77, 2981–2989. [Google Scholar] [CrossRef]
- Klimstra, W.B.; Heidner, H.W.; Johnston, R.E. The furin protease cleavage recognition sequence of Sindbis virus PE2 can mediate virion attachment to cell surface heparan sulfate. J. Virol. 1999, 73, 6299–6306. [Google Scholar] [CrossRef] [PubMed]
- Snyder, J.E.; Kulcsar, K.A.; Schultz, K.L.; Riley, C.P.; Neary, J.T.; Marr, S.; Jose, J.; Griffin, D.E.; Kuhn, R.J. Functional characterization of the alphavirus TF protein. J. Virol. 2013, 87, 8511–8523. [Google Scholar] [CrossRef]
- Firth, A.E.; Chung, B.Y.; Fleeton, M.N.; Atkins, J.F. Discovery of frameshifting in Alphavirus 6K resolves a 20-year enigma. Virol. J. 2008, 5, 108. [Google Scholar] [CrossRef]
- Schmidt, M.; Schmidt, M.F.; Rott, R. Chemical identification of cysteine as palmitoylation site in a transmembrane protein (Semliki Forest virus E1). J. Biol. Chem. 1988, 263, 18635–18639. [Google Scholar] [CrossRef]
- Ivanova, L.; Schlesinger, M.J. Site-directed mutations in the Sindbis virus E2 glycoprotein identify palmitoylation sites and affect virus budding. J. Virol. 1993, 67, 2546–2551. [Google Scholar] [CrossRef]
- Ryan, C.; Ivanova, L.; Schlesinger, M.J. Effects of site-directed mutations of transmembrane cysteines in sindbis virus E1 and E2 glycoproteins on palmitylation and virus replication. Virology 1998, 249, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, J.; Renzi, E.C.; Arnold, R.J.; Trinidad, J.C.; Mukhopadhyay, S. Palmitoylation of Sindbis Virus TF Protein Regulates Its Plasma Membrane Localization and Subsequent Incorporation into Virions. J. Virol. 2017, 91, e02000-16. [Google Scholar] [CrossRef]
- Wilkinson, T.A.; Tellinghuisen, T.L.; Kuhn, R.J.; Post, C.B. Association of sindbis virus capsid protein with phospholipid membranes and the E2 glycoprotein: Implications for alphavirus assembly. Biochemistry 2005, 44, 2800–2810. [Google Scholar] [CrossRef] [PubMed]
- Skoging, U.; Vihinen, M.; Nilsson, L.; Liljeström, P. Aromatic interactions define the binding of the alphavirus spike to its nucleocapsid. Structure 1996, 4, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Owen, K.E.; Choi, H.K.; Lee, H.; Lu, G.; Wengler, G.; Brown, D.T.; Rossmann, M.G.; Kuhn, R.J. Identification of a protein binding site on the surface of the alphavirus nucleocapsid and its implication in virus assembly. Structure 1996, 4, 531–541. [Google Scholar] [CrossRef]
- Hyde, J.L.; Chen, R.; Trobaugh, D.W.; Diamond, M.S.; Weaver, S.C.; Klimstra, W.B.; Wilusz, J. The 5’ and 3’ ends of alphavirus RNAs--Non-coding is not non-functional. Virus Res. 2015, 206, 99–107. [Google Scholar] [CrossRef]
- Hardy, R.W. The role of the 3’ terminus of the Sindbis virus genome in minus-strand initiation site selection. Virology 2006, 345, 520–531. [Google Scholar] [CrossRef]
- Ou, J.H.; Trent, D.W.; Strauss, J.H. The 3’-non-coding regions of alphavirus RNAs contain repeating sequences. J. Mol. Biol. 1982, 156, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Niesters, H.G.; Strauss, J.H. Defined mutations in the 5’ nontranslated sequence of Sindbis virus RNA. J. Virol. 1990, 64, 4162–4168. [Google Scholar] [CrossRef]
- Berben-Bloemheuvel, G.; Kasperaitis, M.A.; van Heugten, H.; Thomas, A.A.; van Steeg, H.; Voorma, H.O. Interaction of initiation factors with the cap structure of chimaeric mRNA containing the 5’-untranslated regions of Semliki Forest virus RNA is related to translational efficiency. Eur. J. Biochem. 1992, 208, 581–587. [Google Scholar] [CrossRef]
- Patel, R.K.; Burnham, A.J.; Gebhart, N.N.; Sokoloski, K.J.; Hardy, R.W. Role for subgenomic mRNA in host translation inhibition during Sindbis virus infection of mammalian cells. Virology 2013, 441, 171–181. [Google Scholar] [CrossRef]
- Frolov, I.; Hardy, R.; Rice, C.M. Cis-acting RNA elements at the 5’ end of Sindbis virus genome RNA regulate minus- and plus-strand RNA synthesis. Rna 2001, 7, 1638–1651. [Google Scholar] [CrossRef] [PubMed]
- Gorchakov, R.; Hardy, R.; Rice, C.M.; Frolov, I. Selection of functional 5’ cis-acting elements promoting efficient sindbis virus genome replication. J. Virol. 2004, 78, 61–75. [Google Scholar] [CrossRef]
- Kulasegaran-Shylini, R.; Atasheva, S.; Gorenstein, D.G.; Frolov, I. Structural and functional elements of the promoter encoded by the 5’ untranslated region of the Venezuelan equine encephalitis virus genome. J. Virol. 2009, 83, 8327–8339. [Google Scholar] [CrossRef] [PubMed]
- Fayzulin, R.; Frolov, I. Changes of the secondary structure of the 5’ end of the Sindbis virus genome inhibit virus growth in mosquito cells and lead to accumulation of adaptive mutations. J. Virol. 2004, 78, 4953–4964. [Google Scholar] [CrossRef]
- Pfeffer, M.; Kinney, R.M.; Kaaden, O.R. The alphavirus 3’-nontranslated region: Size heterogeneity and arrangement of repeated sequence elements. Virology 1998, 240, 100–108. [Google Scholar] [CrossRef]
- Pardigon, N.; Strauss, J.H. Cellular proteins bind to the 3’ end of Sindbis virus minus-strand RNA. J. Virol. 1992, 66, 1007–1015. [Google Scholar] [CrossRef]
- Pardigon, N.; Lenches, E.; Strauss, J.H. Multiple binding sites for cellular proteins in the 3’ end of Sindbis alphavirus minus-sense RNA. J. Virol. 1993, 67, 5003–5011. [Google Scholar] [CrossRef] [PubMed]
- Frolova, E.; Gorchakov, R.; Garmashova, N.; Atasheva, S.; Vergara, L.A.; Frolov, I. Formation of nsP3-specific protein complexes during Sindbis virus replication. J. Virol. 2006, 80, 4122–4134. [Google Scholar] [CrossRef] [PubMed]
- Cristea, I.M.; Carroll, J.W.; Rout, M.P.; Rice, C.M.; Chait, B.T.; MacDonald, M.R. Tracking and elucidating alphavirus-host protein interactions. J. Biol. Chem. 2006, 281, 30269–30278. [Google Scholar] [CrossRef] [PubMed]
- Atasheva, S.; Gorchakov, R.; English, R.; Frolov, I.; Frolova, E. Development of Sindbis viruses encoding nsP2/GFP chimeric proteins and their application for studying nsP2 functioning. J. Virol. 2007, 81, 5046–5057. [Google Scholar] [CrossRef]
- Foy, N.J.; Akhrymuk, M.; Akhrymuk, I.; Atasheva, S.; Bopda-Waffo, A.; Frolov, I.; Frolova, E.I. Hypervariable do-mains of nsP3 proteins of New World and Old World alphaviruses mediate formation of distinct, virus-specific protein complexes. J. Virol. 2013, 87, 1997–2010. [Google Scholar] [CrossRef]
- Varjak, M.; Saul, S.; Arike, L.; Lulla, A.; Peil, L.; Merits, A. Magnetic fractionation and proteomic dissection of cellular organelles occupied by the late replication complexes of Semliki Forest virus. J. Virol. 2013, 87, 10295–10312. [Google Scholar] [CrossRef]
- Bouraï, M.; Lucas-Hourani, M.; Gad, H.H.; Drosten, C.; Jacob, Y.; Tafforeau, L.; Cassonnet, P.; Jones, L.M.; Judith, D.; Couderc, T.; et al. Mapping of Chikungunya virus interactions with host proteins identified nsP2 as a highly connected viral component. J. Virol. 2012, 86, 3121–3134. [Google Scholar] [CrossRef]
- Cristea, I.M.; Rozjabek, H.; Molloy, K.R.; Karki, S.; White, L.L.; Rice, C.M.; Rout, M.P.; Chait, B.T.; MacDonald, M.R. Host factors associated with the Sindbis virus RNA-dependent RNA polymerase: Role for G3BP1 and G3BP2 in virus replication. J. Virol. 2010, 84, 6720–6732. [Google Scholar] [CrossRef]
- Panas, M.D.; Ahola, T.; McInerney, G.M. The C-terminal repeat domains of nsP3 from the Old World alphaviruses bind directly to G3BP. J. Virol. 2014, 88, 5888–5893. [Google Scholar] [CrossRef]
- Scholte, F.E.; Tas, A.; Albulescu, I.C.; Žusinaite, E.; Merits, A.; Snijder, E.J.; van Hemert, M.J. Stress granule components G3BP1 and G3BP2 play a proviral role early in Chikungunya virus replication. J. Virol. 2015, 89, 4457–4469. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.Y.; Reynaud, J.M.; Rasalouskaya, A.; Akhrymuk, I.; Mobley, J.A.; Frolov, I.; Frolova, E.I. New World and Old World Alphaviruses Have Evolved to Exploit Different Components of Stress Granules, FXR and G3BP Proteins, for Assembly of Viral Replication Complexes. PLoS Pathog. 2016, 12, e1005810. [Google Scholar] [CrossRef]
- Götte, B.; Utt, A.; Fragkoudis, R.; Merits, A.; McInerney, G.M. Sensitivity of Alphaviruses to G3BP Deletion Correlates with Efficiency of Replicase Polyprotein Processing. J. Virol. 2020, 94, e01681-19. [Google Scholar] [CrossRef]
- Iselin, L.; Palmalux, N.; Kamel, W.; Simmonds, P.; Mohammed, S.; Castello, A. Uncovering viral RNA-host cell interactions on a proteome-wide scale. Trends Biochem. Sci. 2022, 47, 23–38. [Google Scholar] [CrossRef]
- LaPointe, A.T.; Gebhart, N.N.; Meller, M.E.; Hardy, R.W.; Sokoloski, K.J. Identification and Characterization of Sindbis Virus RNA-Host Protein Interactions. J. Virol. 2018, 92, e02171-17. [Google Scholar] [CrossRef]
- Gebhart, N.N.; Hardy, R.W.; Sokoloski, K.J. Comparative analyses of alphaviral RNA:Protein complexes reveals conserved host-pathogen interactions. PLoS ONE 2020, 15, e0238254. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Moreno, M.; Noerenberg, M.; Ni, S.; Järvelin, A.I.; González-Almela, E.; Lenz, C.E.; Bach-Pages, M.; Cox, V.; Avolio, R.; Davis, T.; et al. System-wide Profiling of RNA-Binding Proteins Uncovers Key Regulators of Virus Infection. Mol. Cell 2019, 74, 196–211.e11. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.Y.; Shih, S.R.; Pan, M.; Li, C.; Lue, C.F.; Stollar, V.; Li, M.L. hnRNP A1 interacts with the 5’ untranslated regions of enterovirus 71 and Sindbis virus RNA and is required for viral replication. J. Virol. 2009, 83, 6106–6114. [Google Scholar] [CrossRef]
- Pardigon, N.; Strauss, J.H. Mosquito homolog of the La autoantigen binds to Sindbis virus RNA. J. Virol. 1996, 70, 1173–1181. [Google Scholar] [CrossRef] [PubMed]
- Fleurdépine, S.; Deragon, J.M.; Devic, M.; Guilleminot, J.; Bousquet-Antonelli, C. A bona fide La protein is required for embryogenesis in Arabidopsis thaliana. Nucleic Acids Res. 2007, 35, 3306–3321. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tran, H.; Maurer, F.; Nagamine, Y. Stabilization of urokinase and urokinase receptor mRNAs by HuR is linked to its cytoplasmic accumulation induced by activated mitogen-activated protein kinase-activated protein kinase 2. Mol. Cell Biol. 2003, 23, 7177–7188. [Google Scholar] [CrossRef]
- Doller, A.; Akool el, S.; Huwiler, A.; Müller, R.; Radeke, H.H.; Pfeilschifter, J.; Eberhardt, W. Posttranslational modification of the AU-rich element binding protein HuR by protein kinase Cdelta elicits angiotensin II-induced stabilization and nuclear export of cyclooxygenase 2 mRNA. Mol. Cell Biol. 2008, 28, 2608–2625. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.J.; Cheng, S.; Campbell, C.; Wright, A.; Furneaux, H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem. 1996, 271, 8144–8151. [Google Scholar] [CrossRef] [PubMed]
- Meisner, N.C.; Hintersteiner, M.; Mueller, K.; Bauer, R.; Seifert, J.M.; Naegeli, H.U.; Ottl, J.; Oberer, L.; Guenat, C.; Moss, S.; et al. Identification and mechanistic characterization of low-molecular-weight inhibitors for HuR. Nat. Chem. Biol. 2007, 3, 508–515. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zeng, F.; Liu, Q.; Liu, H.; Liu, Z.; Niu, L.; Teng, M.; Li, X. The structure of the ARE-binding domains of Hu antigen R (HuR) undergoes conformational changes during RNA binding. Acta Cryst. D Biol. Cryst. 2013, 69, 373–380. [Google Scholar] [CrossRef]
- Garneau, N.L.; Sokoloski, K.J.; Opyrchal, M.; Neff, C.P.; Wilusz, C.J.; Wilusz, J. The 3’ untranslated region of sindbis virus represses deadenylation of viral transcripts in mosquito and Mammalian cells. J. Virol. 2008, 82, 880–892. [Google Scholar] [CrossRef]
- Sokoloski, K.J.; Dickson, A.M.; Chaskey, E.L.; Garneau, N.L.; Wilusz, C.J.; Wilusz, J. Sindbis virus usurps the cellular HuR protein to stabilize its transcripts and promote productive infections in mammalian and mosquito cells. Cell Host Microb. 2010, 8, 196–207. [Google Scholar] [CrossRef]
- Barnhart, M.D.; Moon, S.L.; Emch, A.W.; Wilusz, C.J.; Wilusz, J. Changes in cellular mRNA stability, splicing, and polyadenylation through HuR protein sequestration by a cytoplasmic RNA virus. Cell Rep. 2013, 5, 909–917. [Google Scholar] [CrossRef]
- Dickson, A.M.; Anderson, J.R.; Barnhart, M.D.; Sokoloski, K.J.; Oko, L.; Opyrchal, M.; Galanis, E.; Wilusz, C.J.; Morrison, T.E.; Wilusz, J. Dephosphorylation of HuR protein during alphavirus infection is associated with HuR relocalization to the cytoplasm. J. Biol. Chem. 2012, 287, 36229–36238. [Google Scholar] [CrossRef]
- Tourrière, H.; Chebli, K.; Zekri, L.; Courselaud, B.; Blanchard, J.M.; Bertrand, E.; Tazi, J. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell. Biol. 2003, 160, 823–831. [Google Scholar] [CrossRef]
- Hinton, S.D.; Myers, M.P.; Roggero, V.R.; Allison, L.A.; Tonks, N.K. The pseudophosphatase MK-STYX interacts with G3BP and decreases stress granule formation. Biochem. J. 2010, 427, 349–357. [Google Scholar] [CrossRef]
- Matsuki, H.; Takahashi, M.; Higuchi, M.; Makokha, G.N.; Oie, M.; Fujii, M. Both G3BP1 and G3BP2 contribute to stress granule formation. Genes Cells 2013, 18, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Vognsen, T.; Møller, I.R.; Kristensen, O. Crystal structures of the human G3BP1 NTF2-like domain visualize FxFG Nup repeat specificity. PLoS ONE 2013, 8, e80947. [Google Scholar] [CrossRef]
- Fros, J.J.; Geertsema, C.; Zouache, K.; Baggen, J.; Domeradzka, N.; van Leeuwen, D.M.; Flipse, J.; Vlak, J.M.; Failloux, A.B.; Pijlman, G.P. Mosquito Rasputin interacts with chikungunya virus nsP3 and determines the infection rate in Aedes albopictus. Parasit Vectors 2015, 8, 464. [Google Scholar] [CrossRef]
- Dember, L.M.; Kim, N.D.; Liu, K.Q.; Anderson, P. Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J. Biol. Chem. 1996, 271, 2783–2788. [Google Scholar] [CrossRef] [PubMed]
- Förch, P.; Puig, O.; Kedersha, N.; Martínez, C.; Granneman, S.; Séraphin, B.; Anderson, P.; Valcárcel, J. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol. Cell 2000, 6, 1089–1098. [Google Scholar] [CrossRef] [PubMed]
- Kedersha, N.L.; Gupta, M.; Li, W.; Miller, I.; Anderson, P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 1999, 147, 1431–1442. [Google Scholar] [CrossRef]
- McInerney, G.M.; Kedersha, N.L.; Kaufman, R.J.; Anderson, P.; Liljeström, P. Importance of eIF2alpha phosphorylation and stress granule assembly in alphavirus translation regulation. Mol. Biol. Cell 2005, 16, 3753–3763. [Google Scholar] [CrossRef] [PubMed]
- Battle, D.J.; Lau, C.K.; Wan, L.; Deng, H.; Lotti, F.; Dreyfuss, G. The Gemin5 protein of the SMN complex identifies snRNAs. Mol. Cell 2006, 23, 273–279. [Google Scholar] [CrossRef]
- Bradrick, S.S.; Gromeier, M. Identification of gemin5 as a novel 7-methylguanosine cap-binding protein. PLoS ONE 2009, 4, e7030. [Google Scholar] [CrossRef]
- Francisco-Velilla, R.; Fernandez-Chamorro, J.; Ramajo, J.; Martinez-Salas, E. The RNA-binding protein Gemin5 binds directly to the ribosome and regulates global translation. Nucleic Acids Res. 2016, 44, 8335–8351. [Google Scholar] [CrossRef]
- Estañ, M.C.; Fernández-Núñez, E.; Zaki, M.S.; Esteban, M.I.; Donkervoort, S.; Hawkins, C.; Caparros-Martin, J.A.; Saade, D.; Hu, Y.; Bolduc, V.; et al. Recessive mutations in muscle-specific isoforms of FXR1 cause congenital multi-minicore myopathy. Nat. Commun. 2019, 10, 797. [Google Scholar] [CrossRef] [PubMed]
- Meshram, C.D.; Shiliaev, N.; Frolova, E.I.; Frolov, I.; López, S. Hypervariable Domain of nsP3 of Eastern Equine Encephalitis Virus Is a Critical Determinant of Viral Virulence. J. Virol. 2020, 94, e00617–e00620. [Google Scholar] [CrossRef] [PubMed]
- Meshram, C.D.; Phillips, A.T.; Lukash, T.; Shiliaev, N.; Frolova, E.I.; Frolov, I.; López, S. Mutations in Hypervariable Domain of Venezuelan Equine Encephalitis Virus nsP3 Protein Differentially Affect Viral Replication. J. Virol. 2020, 94, e01841-19. [Google Scholar] [CrossRef]
- Jarmoskaite, I.; Russell, R. RNA helicase proteins as chaperones and remodelers. Annu Rev. Biochem. 2014, 83, 697–725. [Google Scholar] [CrossRef] [PubMed]
- Schwer, B. A new twist on RNA helicases: DExH/D box proteins as RNPases. Nat. Struct. Biol. 2001, 8, 113–116. [Google Scholar] [CrossRef] [PubMed]
- Rocak, S.; Linder, P. DEAD-box proteins: The driving forces behind RNA metabolism. Nat. Rev. Mol. Cell Biol. 2004, 5, 232–241. [Google Scholar] [CrossRef]
- Amaya, M.; Brooks-Faulconer, T.; Lark, T.; Keck, F.; Bailey, C.; Raman, V.; Narayanan, A. Venezuelan equine encephalitis virus non-structural protein 3 (nsP3) interacts with RNA helicases DDX1 and DDX3 in infected cells. Antivir. Res. 2016, 131, 49–60. [Google Scholar] [CrossRef]
- Lawrence, P.; Conderino, J.S.; Rieder, E. Redistribution of demethylated RNA helicase A during foot-and-mouth disease virus infection: Role of Jumonji C-domain containing protein 6 in RHA demethylation. Virology 2014, 452–453, 1–11. [Google Scholar] [CrossRef]
- Lin, L.; Li, Y.; Pyo, H.M.; Lu, X.; Raman, S.N.; Liu, Q.; Brown, E.G.; Zhou, Y. Identification of RNA helicase A as a cellular factor that interacts with influenza A virus NS1 protein and its role in the virus life cycle. J. Virol. 2012, 86, 1942–1954. [Google Scholar] [CrossRef]
- Li, Y.; Masaki, T.; Shimakami, T.; Lemon, S.M. hnRNP L and NF90 interact with hepatitis C virus 5’-terminal untranslated RNA and promote efficient replication. J. Virol. 2014, 88, 7199–7209. [Google Scholar] [CrossRef]
- Sheng, C.; Yao, Y.; Chen, B.; Wang, Y.; Chen, J.; Xiao, M. RNA helicase is involved in the expression and replication of classical swine fever virus and interacts with untranslated region. Virus Res. 2013, 171, 257–261. [Google Scholar] [CrossRef] [PubMed]
- Matkovic, R.; Bernard, E.; Fontanel, S.; Eldin, P.; Chazal, N.; Hassan Hersi, D.; Merits, A.; Péloponèse, J.M., Jr.; Briant, L. The Host DHX9 DExH-Box Helicase Is Recruited to Chikungunya Virus Replication Complexes for Optimal Genomic RNA Translation. J. Virol. 2019, 93, e01764-18. [Google Scholar] [CrossRef] [PubMed]
- Geuens, T.; Bouhy, D.; Timmerman, V. The hnRNP family: Insights into their role in health and disease. Hum. Genet. 2016, 135, 851–867. [Google Scholar] [CrossRef]
- Shi, S.T.; Huang, P.; Li, H.P.; Lai, M.M. Heterogeneous nuclear ribonucleoprotein A1 regulates RNA synthesis of a cytoplasmic virus. Embo J. 2000, 19, 4701–4711. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.S.; Seol, S.K.; Song, O.K.; Park, J.H.; Jang, S.K. An RNA-binding protein, hnRNP A1, and a scaffold protein, septin 6, facilitate hepatitis C virus replication. J. Virol. 2007, 81, 3852–3865. [Google Scholar] [CrossRef]
- Paranjape, S.M.; Harris, E. Y box-binding protein-1 binds to the dengue virus 3’-untranslated region and mediates antiviral effects. J. Biol. Chem. 2007, 282, 30497–30508. [Google Scholar] [CrossRef]
- Zhao, X.; Rush, M.; Schwartz, S. Identification of an hnRNP A1-dependent splicing silencer in the human papillomavirus type 16 L1 coding region that prevents premature expression of the late L1 gene. J. Virol. 2004, 78, 10888–10905. [Google Scholar] [CrossRef]
- Pettit Kneller, E.L.; Connor, J.H.; Lyles, D.S. hnRNPs Relocalize to the cytoplasm following infection with vesicular stomatitis virus. J. Virol. 2009, 83, 770–780. [Google Scholar] [CrossRef] [PubMed]
- Gui, H.; Lu, C.W.; Adams, S.; Stollar, V.; Li, M.L. hnRNP A1 interacts with the genomic and subgenomic RNA promoters of Sindbis virus and is required for the synthesis of G and SG RNA. J. Biomed. Sci. 2010, 17, 59. [Google Scholar] [CrossRef]
- Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 2015, 518, 560–564. [Google Scholar] [CrossRef]
- Herrmann, C.; Dybas, J.M.; Liddle, J.C.; Price, A.M.; Hayer, K.E.; Lauman, R.; Purman, C.E.; Charman, M.; Kim, E.T.; Garcia, B.A.; et al. Adenovirus-mediated ubiquitination alters protein-RNA binding and aids viral RNA processing. Nat. Microbiol. 2020, 5, 1217–1231. [Google Scholar] [CrossRef] [PubMed]
- Dechtawewat, T.; Songprakhon, P.; Limjindaporn, T.; Puttikhunt, C.; Kasinrerk, W.; Saitornuang, S.; Yenchitsomanus, P.T.; Noisakran, S. Role of human heterogeneous nuclear ribonucleoprotein C1/C2 in dengue virus replication. Virol. J. 2015, 12, 14. [Google Scholar] [CrossRef]
- Ertel, K.J.; Brunner, J.E.; Semler, B.L. Mechanistic consequences of hnRNP C binding to both RNA termini of poliovirus negative-strand RNA intermediates. J. Virol. 2010, 84, 4229–4242. [Google Scholar] [CrossRef]
- Moumen, A.; Masterson, P.; O’Connor, M.J.; Jackson, S.P. hnRNP K: An HDM2 target and transcriptional coactivator of p53 in response to DNA damage. Cell 2005, 123, 1065–1078. [Google Scholar] [CrossRef] [PubMed]
- Burnham, A.J.; Gong, L.; Hardy, R.W. Heterogeneous nuclear ribonuclear protein K interacts with Sindbis virus nonstructural proteins and viral subgenomic mRNA. Virology 2007, 367, 212–221. [Google Scholar] [CrossRef]
- Cao, P.; Luo, W.W.; Li, C.; Tong, Z.; Zheng, Z.Q.; Zhou, L.; Xiong, Y.; Li, S. The heterogeneous nuclear ribonucleoprotein hnRNPM inhibits RNA virus-triggered innate immunity by antagonizing RNA sensing of RIG-I-like receptors. PLoS Pathog. 2019, 15, e1007983. [Google Scholar] [CrossRef] [PubMed]
- West, K.O.; Scott, H.M.; Torres-Odio, S.; West, A.P.; Patrick, K.L.; Watson, R.O. The Splicing Factor hnRNP M Is a Critical Regulator of Innate Immune Gene Expression in Macrophages. Cell Rep. 2019, 29, 1594–1609.e1595. [Google Scholar] [CrossRef]
- Walter, B.L.; Parsley, T.B.; Ehrenfeld, E.; Semler, B.L. Distinct poly(rC) binding protein KH domain determinants for poliovirus translation initiation and viral RNA replication. J. Virol. 2002, 76, 12008–12022. [Google Scholar] [CrossRef]
- Gamarnik, A.V.; Andino, R. Two functional complexes formed by KH domain containing proteins with the 5’ noncoding region of poliovirus RNA. Rna 1997, 3, 882–892. [Google Scholar]
- Collier, B.; Goobar-Larsson, L.; Sokolowski, M.; Schwartz, S. Translational inhibition in vitro of human papillomavirus type 16 L2 mRNA mediated through interaction with heterogenous ribonucleoprotein K and poly(rC)-binding proteins 1 and 2. J. Biol. Chem. 1998, 273, 22648–22656. [Google Scholar] [CrossRef]
- Cousineau, S.E.; Rheault, M.; Sagan, S.M. Poly(rC)-Binding Protein 1 Limits Hepatitis C Virus Virion Assembly and Secretion. Viruses 2022, 14, 291. [Google Scholar] [CrossRef] [PubMed]
- Dai, S.; Wang, C.; Zhang, C.; Feng, L.; Zhang, W.; Zhou, X.; He, Y.; Xia, X.; Chen, B.; Song, W. PTB: Not just a polypyrimidine tract-binding protein. J. Cell Physiol. 2022, 237, 2357–2373. [Google Scholar] [CrossRef]
- Singh, R.; Valcárcel, J.; Green, M.R. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science 1995, 268, 1173–1176. [Google Scholar] [CrossRef] [PubMed]
- Oberstrass, F.C.; Auweter, S.D.; Erat, M.; Hargous, Y.; Henning, A.; Wenter, P.; Reymond, L.; Amir-Ahmady, B.; Pitsch, S.; Black, D.L.; et al. Structure of PTB Bound to RNA: Specific Binding and Implications for Splicing Regulation. Science 2005, 309, 2054–2057. [Google Scholar] [CrossRef]
- Florez, P.M.; Sessions, O.M.; Wagner, E.J.; Gromeier, M.; Garcia-Blanco, M.A. The polypyrimidine tract binding protein is required for efficient picornavirus gene expression and propagation. J. Virol. 2005, 79, 6172–6179. [Google Scholar] [CrossRef]
- Markovtsov, V.; Nikolic, J.M.; Goldman, J.A.; Turck, C.W.; Chou, M.Y.; Black, D.L. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol. Cell Biol. 2000, 20, 7463–7479. [Google Scholar] [CrossRef] [PubMed]
- Shereen, M.A.; Bashir, N.; Su, R.; Liu, F.; Wu, K.; Luo, Z.; Wu, J. Zika virus dysregulates the expression of astrocytic genes involved in neurodevelopment. PLoS Negl. Trop. Dis. 2021, 15, e0009362. [Google Scholar] [CrossRef]
- Vavougios, G.D. SARS-CoV-2 dysregulation of PTBP1 and YWHAE/Z gene expression: A primer of neurodegeneration. Med. Hypotheses 2020, 144, 110212. [Google Scholar] [CrossRef]
- Sola, I.; Galán, C.; Mateos-Gómez, P.A.; Palacio, L.; Zúñiga, S.; Cruz, J.L.; Almazán, F.; Enjuanes, L. The Polypyrimidine Tract-Binding Protein Affects Coronavirus RNA Accumulation Levels and Relocalizes Viral RNAs to Novel Cytoplasmic Domains Different from Replication-Transcription Sites. J. Virol. 2011, 85, 5136–5149. [Google Scholar] [CrossRef]
- Zang, W.Q.; Li, B.; Huang, P.Y.; Lai, M.M.; Yen, T.S. Role of polypyrimidine tract binding protein in the function of the hepatitis B virus posttranscriptional regulatory element. J. Virol. 2001, 75, 10779–10786. [Google Scholar] [CrossRef]
- Zhang, K.; Shang, G.; Padavannil, A.; Wang, J.; Sakthivel, R.; Chen, X.; Kim, M.; Thompson, M.G.; García-Sastre, A.; Lynch, K.W.; et al. Structural-functional interactions of NS1-BP protein with the splicing and mRNA export machineries for viral and host gene expression. Proc. Natl. Acad. Sci. USA 2018, 115, E12218–E12227. [Google Scholar] [CrossRef] [PubMed]
- Westcott, C.E.; Qazi, S.; Maiocco, A.M.; Mukhopadhyay, S.; Sokoloski, K.J. Binding of hnRNP I–vRNA Regulates Sindbis Virus Structural Protein Expression to Promote Particle Infectivity. Viruses 2022, 14, 1423. [Google Scholar]
- Sokoloski, K.J.; Nease, L.M.; May, N.A.; Gebhart, N.N.; Jones, C.E.; Morrison, T.E.; Hardy, R.W. Identification of Interactions between Sindbis Virus Capsid Protein and Cytoplasmic vRNA as Novel Virulence Determinants. PLoS Pathog. 2017, 13, e1006473. [Google Scholar] [CrossRef]
- Xu, Y.; Wu, W.; Han, Q.; Wang, Y.; Li, C.; Zhang, P.; Xu, H. Post-translational modification control of RNA-binding protein hnRNPK function. Open Biol. 2019, 9, 180239. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Liu, S.; Li, Y.; Yang, G.; Luo, Y.; Li, S.; Du, H.; Zhao, Y.; Wang, D.; Chen, J.; et al. The Nuclear Matrix Protein SAFA Surveils Viral RNA and Facilitates Immunity by Activating Antiviral Enhancers and Super-enhancers. Cell Host Microbe 2019, 26, 369–384.e368. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Westcott, C.E.; Isom, C.M.; Karki, D.; Sokoloski, K.J. Dancing with the Devil: A Review of the Importance of Host RNA-Binding Proteins to Alphaviral RNAs during Infection. Viruses 2023, 15, 164. https://doi.org/10.3390/v15010164
Westcott CE, Isom CM, Karki D, Sokoloski KJ. Dancing with the Devil: A Review of the Importance of Host RNA-Binding Proteins to Alphaviral RNAs during Infection. Viruses. 2023; 15(1):164. https://doi.org/10.3390/v15010164
Chicago/Turabian StyleWestcott, Claire E., Cierra M. Isom, Deepa Karki, and Kevin J. Sokoloski. 2023. "Dancing with the Devil: A Review of the Importance of Host RNA-Binding Proteins to Alphaviral RNAs during Infection" Viruses 15, no. 1: 164. https://doi.org/10.3390/v15010164
APA StyleWestcott, C. E., Isom, C. M., Karki, D., & Sokoloski, K. J. (2023). Dancing with the Devil: A Review of the Importance of Host RNA-Binding Proteins to Alphaviral RNAs during Infection. Viruses, 15(1), 164. https://doi.org/10.3390/v15010164






