A New Strategy for Efficient Screening and Identification of Monoclonal Antibodies against Oncogenic Avian Herpesvirus Utilizing CRISPR/Cas9-Based Gene-Editing Technology
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Viruses and Cells
2.3. sgRNA Plasmid Construct
2.4. Generation of a pp38-Edited GX0101 Virus
2.5. Immunofluorescence Assay (IFA)
2.6. Reverse-Transcription Quantitative PCR (RT-qPCR)
2.7. Construction of a Pool of mAbs against MDV Proteins
2.8. Cross-Screening of pp38-Specific mAbs
2.9. Protein Expression and Examination of mAb Specificity
2.10. Confocal Microscopy Analysis
2.11. Isotype Characterization of pp38 mAbs
2.12. Reaction Spectrum of pp38 mAbs to MDVs
2.13. Immunoprecipitation (IP)
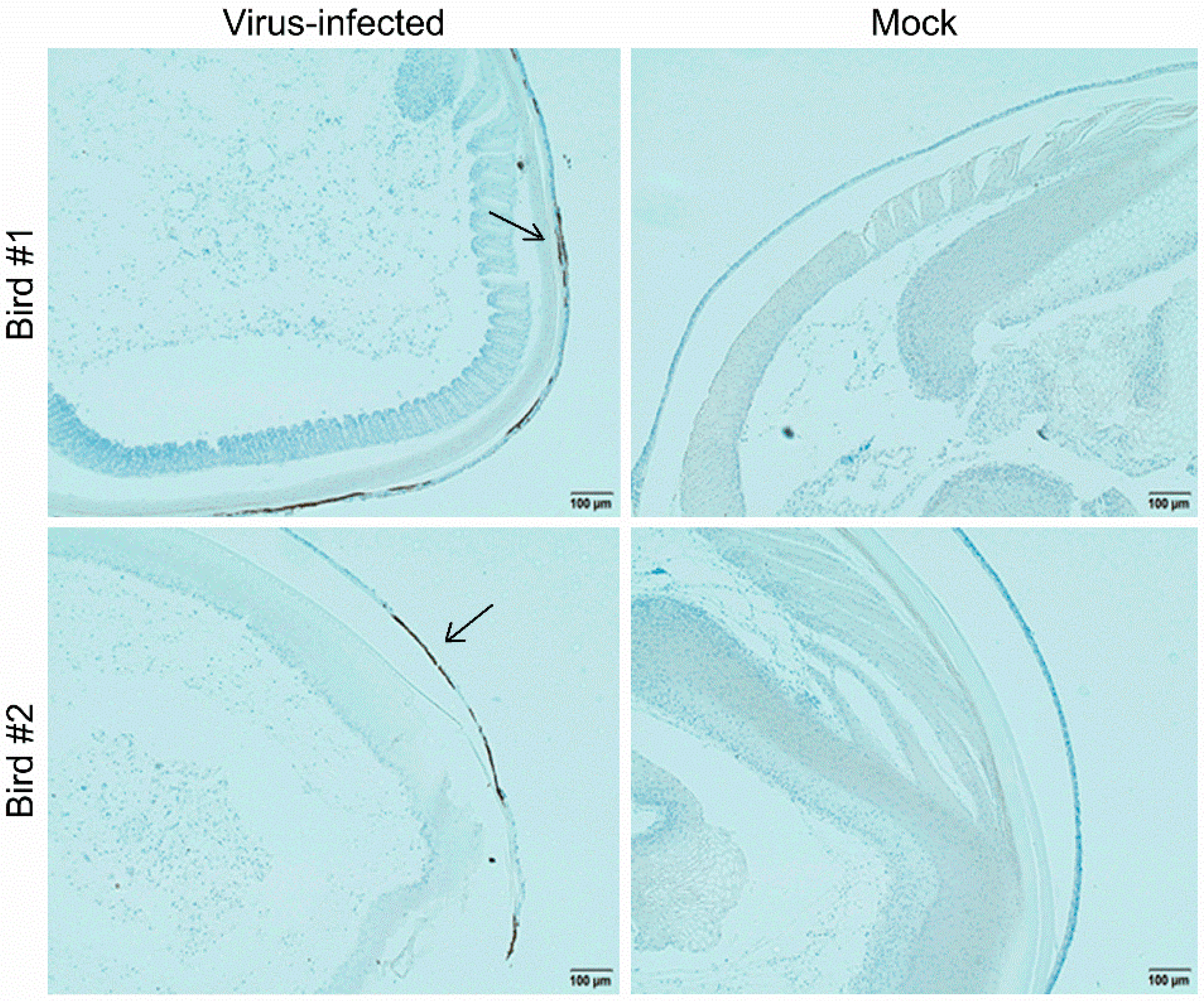
2.14. Immunohistochemistry (IHC)
3. Results
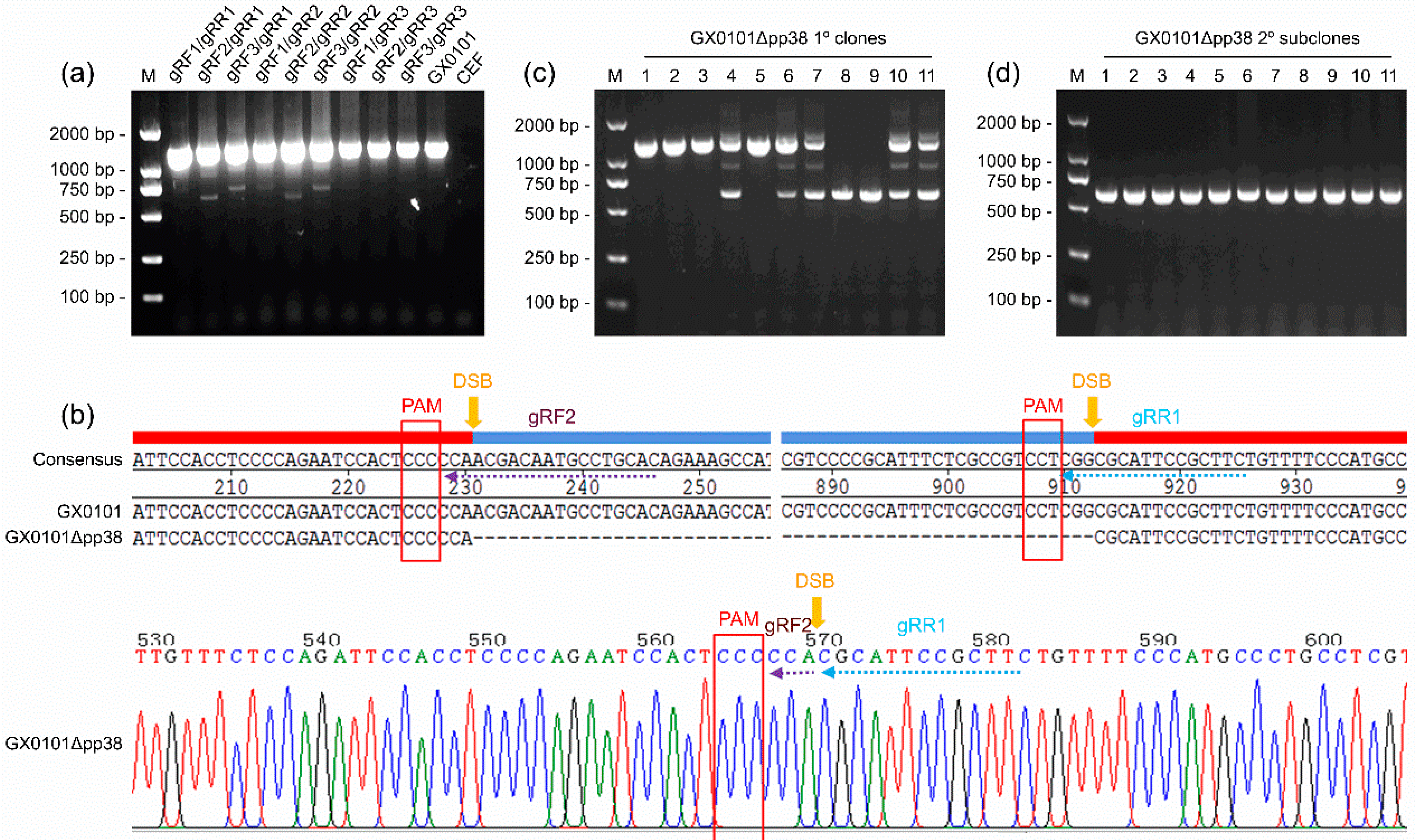
3.1. Generation of a pp38-Deleted MDV Mutant Using the CRISPR/Cas9 System
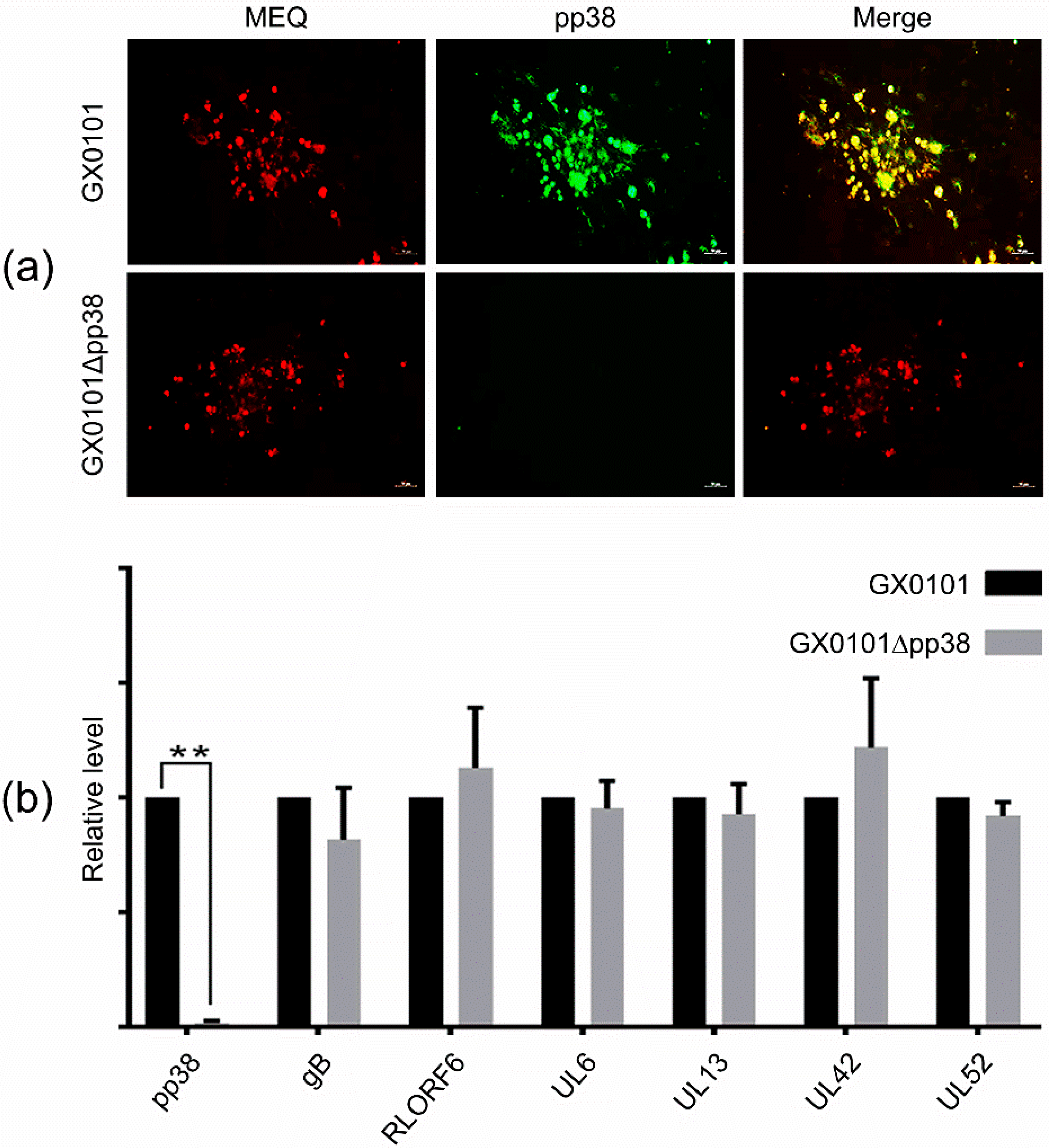
3.2. Identification of the pp38-Deleted MDV Mutant GX0101∆pp38
3.3. Construction of a Pool of mAbs against MDV Proteins
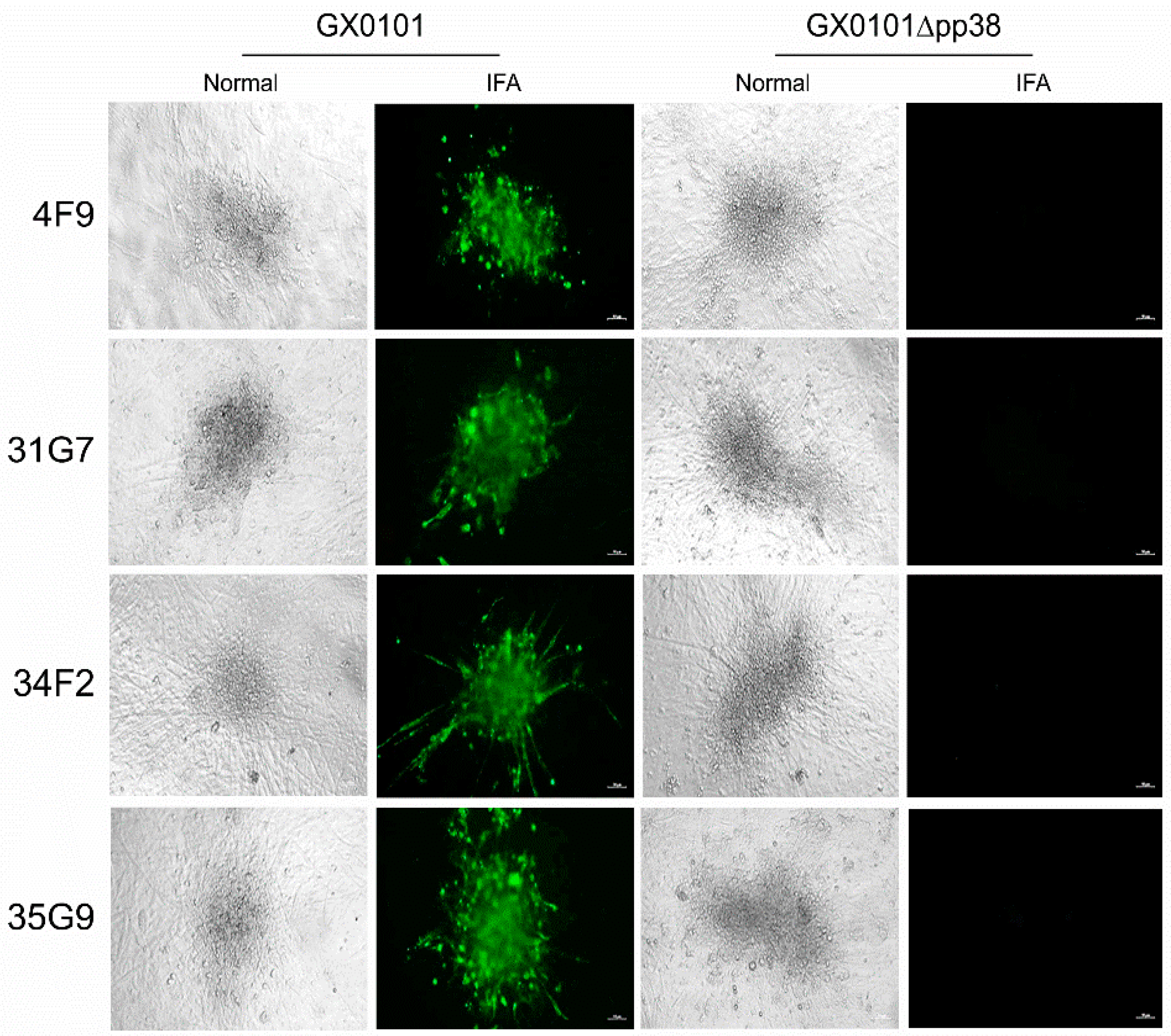
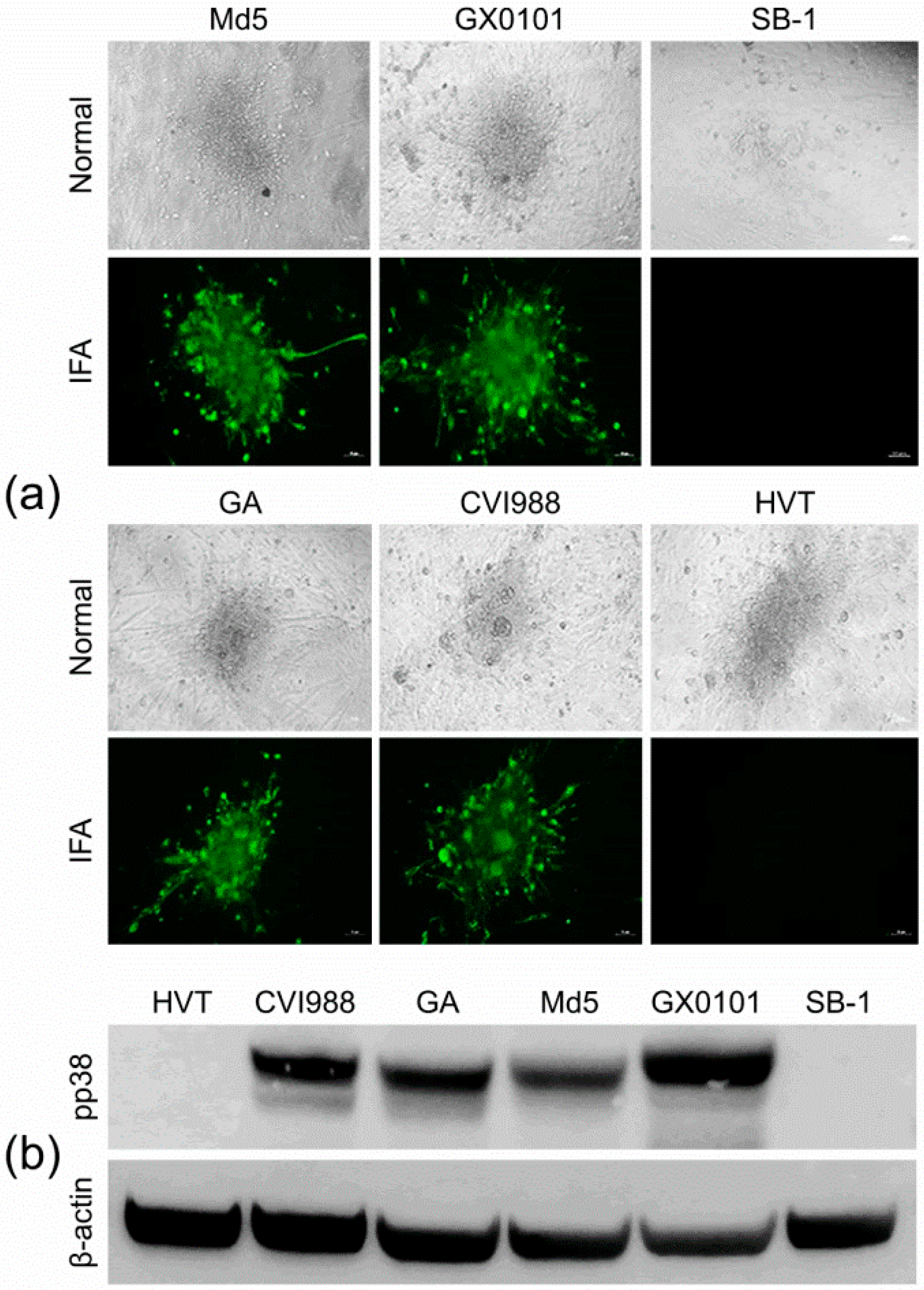
3.4. Cross-Screening and Identification of pp38-Specific mAbs
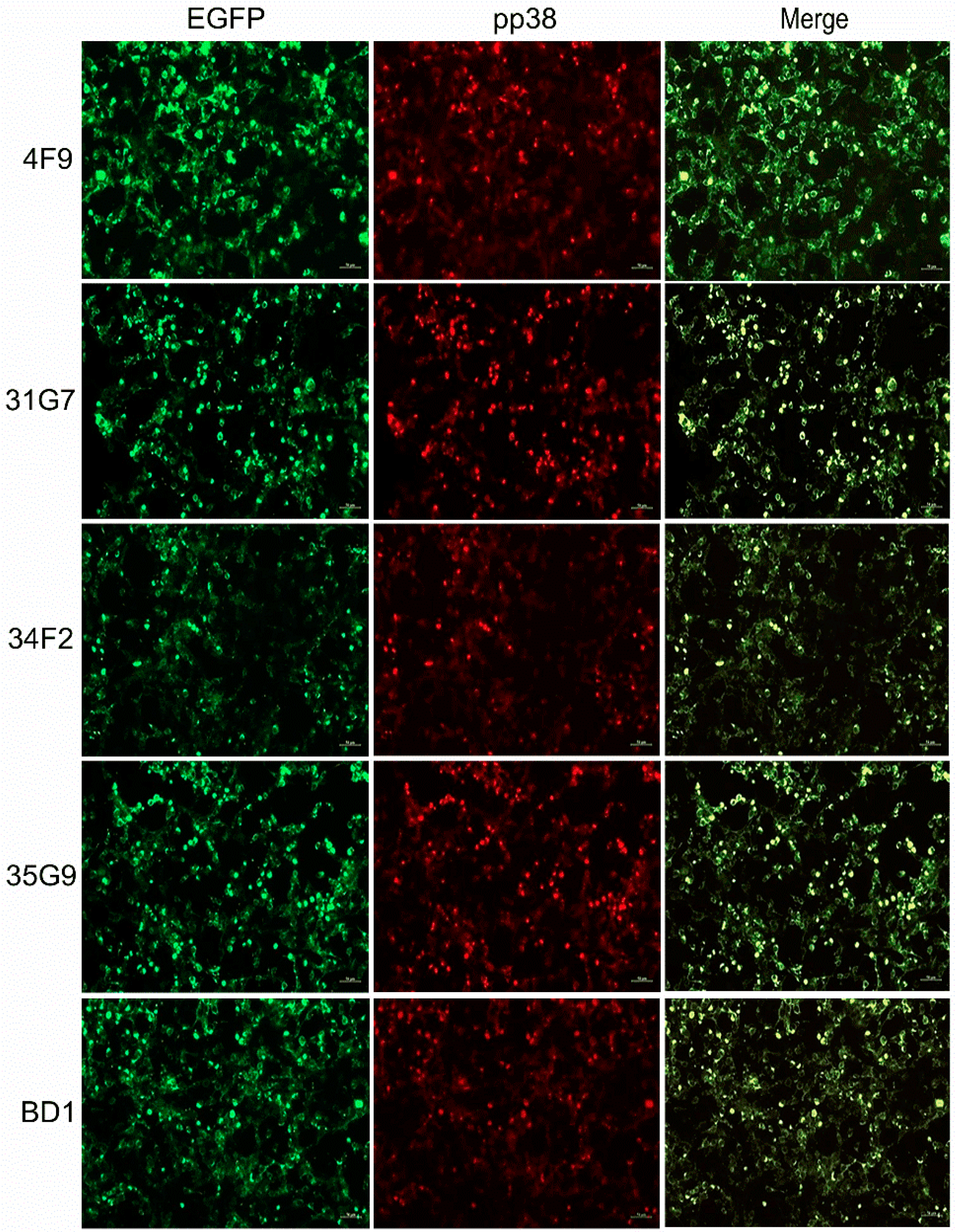
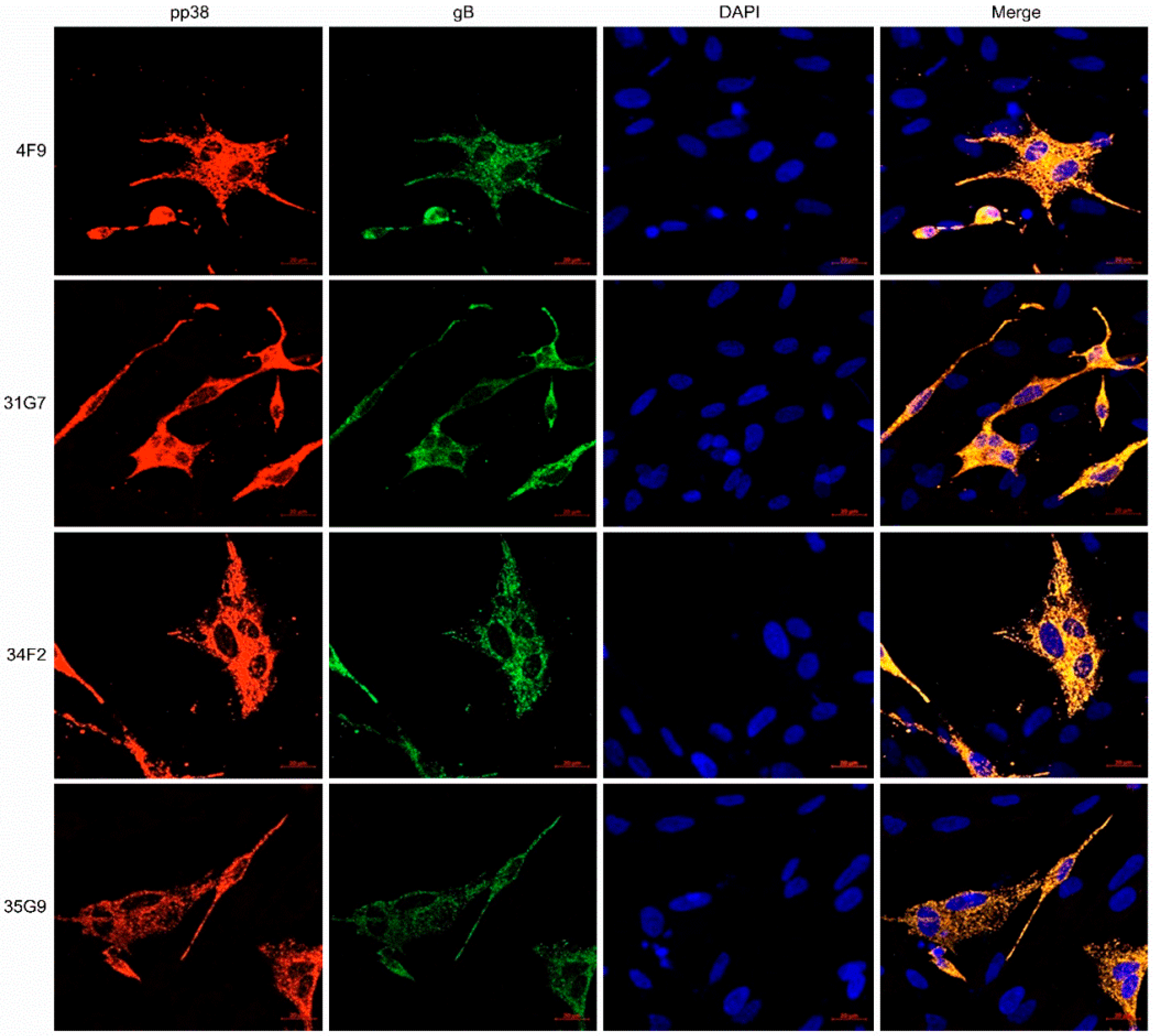
3.5. Characterization of the MDV-1 pp38-Specific mAb 31G7
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Biggs, P.M.; Nair, V. The long view: 40 years of Marek’s disease research and Avian Pathology. Avian Pathol. 2012, 41, 3–9. [Google Scholar] [CrossRef]
- Kennedy, D.A.; Cairns, C.; Jones, M.J.; Bell, A.S.; Salathé, R.M.; Baigent, S.J.; Nair, V.K.; Dunn, P.A.; Read, A.F. Industry-Wide Surveillance of Marek’s Disease Virus on Commercial Poultry Farms. Avian Dis. 2017, 61, 153–164. [Google Scholar] [CrossRef]
- Gatherer, D.; Depledge, D.P.; Hartley, C.A.; Szpara, M.L.; Vaz, P.K.; Benkő, M.; Brandt, C.R.; Bryant, N.A.; Dastjerdi, A.; Doszpoly, A.; et al. ICTV Virus Taxonomy Profile: Herpesviridae 2021. J. Gen. Virol. 2021, 102, 001673. [Google Scholar] [CrossRef]
- Osterrieder, N.; Kamil, J.P.; Schumacher, D.; Tischer, B.K.; Trapp, S. Marek’s disease virus: From miasma to model. Nat. Rev. Microbiol. 2006, 4, 283–294. [Google Scholar] [CrossRef]
- Witter, R.L.; Calnek, B.W.; Buscaglia, C.; Gimeno, I.M.; Schat, K.A. Classification of Marek’s disease viruses according to pathotype: Philosophy and methodology. Avian Pathol. 2005, 34, 75–90. [Google Scholar] [CrossRef]
- Song, B.; Zeb, J.; Hussain, S.; Aziz, M.U.; Circella, E.; Casalino, G.; Camarda, A.; Yang, G.; Buchon, N.; Sparagano, O. A Review on the Marek’s Disease Outbreak and Its Virulence-Related meq Genovariation in Asia between 2011 and 2021. Animals 2022, 12, 540. [Google Scholar] [CrossRef]
- Spatz, S.J.; Petherbridge, L.; Zhao, Y.; Nair, V. Comparative full-length sequence analysis of oncogenic and vaccine (Rispens) strains of Marek’s disease virus. J. Gen. Virol. 2007, 88, 1080–1096. [Google Scholar] [CrossRef]
- Nair, V. Latency and tumorigenesis in Marek’s disease. Avian Dis. 2013, 57, 360–365. [Google Scholar] [CrossRef]
- Ikuta, K.; Honma, H.; Maotani, K.; Ueda, S.; Kato, S.; Hirai, K. Monoclonal antibodies specific to and cross-reactive with Marek’s disease virus and herpesvirus of turkeys. Biken J. 1982, 25, 171–175. [Google Scholar]
- Silva, R.F.; Lee, L.F. Monoclonal antibody-mediated immunoprecipitation of proteins from cells infected with Marek’s disease virus or turkey herpesvirus. Virology 1984, 136, 307–320. [Google Scholar] [CrossRef]
- Horiuchi, M.; Kodama, H.; Mikami, T. Identification of virus-specific antigens in cultured cells infected with Marek’s disease virus serotype 2 and CVI-988 strain. Nihon Juigaku Zasshi 1990, 52, 985–994. [Google Scholar] [CrossRef]
- Teng, M.; Yao, Y.; Nair, V.; Luo, J. Latest Advances of Virology Research Using CRISPR/Cas9-Based Gene-Editing Technology and Its Application to Vaccine Development. Viruses 2021, 13, 779. [Google Scholar] [CrossRef]
- Yao, Y.; Bassett, A.; Nair, V. Targeted editing of avian herpesvirus vaccine vector using CRISPR/Cas9 nucleases. J. Vaccine Technol. 2016, 1, 1–7. [Google Scholar]
- Zhang, Y.; Tang, N.; Sadigh, Y.; Baigent, S.; Shen, Z.; Nair, V.; Yao, Y. Application of CRISPR/Cas9 Gene Editing System on MDV-1 Genome for the Study of Gene Function. Viruses 2018, 10, 279. [Google Scholar] [CrossRef]
- Zhang, Y.; Luo, J.; Tang, N.; Teng, M.; Reddy, V.; Moffat, K.; Shen, Z.; Nair, V.; Yao, Y. Targeted Editing of the pp38 Gene in Marek’s Disease Virus-Transformed Cell Lines Using CRISPR/Cas9 System. Viruses 2019, 11, 391. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tang, N.; Luo, J.; Teng, M.; Moffat, K.; Shen, Z.; Watson, M.; Nair, V.; Yao, Y. Marek’s Disease Virus-Encoded MicroRNA 155 Ortholog Critical for the Induction of Lymphomas Is Not Essential for the Proliferation of Transformed Cell Lines. J. Virol. 2019, 93, e00713-19. [Google Scholar] [CrossRef]
- Luo, J.; Teng, M.; Zai, X.; Tang, N.; Zhang, Y.; Mandviwala, A.; Reddy, V.; Baigent, S.; Yao, Y.; Nair, V. Efficient Mutagenesis of Marek’s Disease Virus-Encoded microRNAs Using a CRISPR/Cas9-Based Gene Editing System. Viruses 2020, 12, 466. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, W.; Tang, N.; Moffat, K.; Nair, V.; Yao, Y. Targeted Deletion of Glycoprotein B Gene by CRISPR/Cas9 Nuclease Inhibits Gallid herpesvirus Type 3 in Dually Infected Marek’s Disease Virus-Transformed Lymphoblastoid Cell Line MSB-1. J. Virol. 2022, 96, e0202721. [Google Scholar] [CrossRef] [PubMed]
- Tang, N.; Zhang, Y.; Pedrera, M.; Chang, P.; Baigent, S.; Moffat, K.; Shen, Z.; Nair, V.; Yao, Y. A simple and rapid approach to develop recombinant avian herpesvirus vectored vaccines using CRISPR/Cas9 system. Vaccine 2018, 36, 716–722. [Google Scholar] [CrossRef] [PubMed]
- Tang, N.; Zhang, Y.; Sadigh, Y.; Moffat, K.; Shen, Z.; Nair, V.; Yao, Y. Generation of A Triple Insert Live Avian Herpesvirus Vectored Vaccine Using CRISPR/Cas9-Based Gene Editing. Vaccines 2020, 8, 97. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Liao, Y.; Yang, S.; Jin, J.; Lu, W.; Teng, M.; Luo, J.; Zhang, G.; Sun, A.; Zhuang, G. Deletion of miR-M8 and miR-M13 eliminates the bursa atrophy induced by Marek’s disease virus infection. Vet. Microbiol. 2022, 268, 109409. [Google Scholar] [CrossRef] [PubMed]
- Niikura, M.; Matsuura, Y.; Endoh, D.; Onuma, M.; Mikami, T. Expression of the Marek’s disease virus (MDV) homolog of glycoprotein B of herpes simplex virus by a recombinant baculovirus and its identification as the B antigen (gp100, gp60, gp49) of MDV. J. Virol. 1992, 66, 2631–2638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ono, M.; Kawaguchi, Y.; Maeda, K.; Kamiya, N.; Tohya, Y.; Kai, C.; Niikura, M.; Mikami, T. Nucleotide sequence analysis of Marek’s disease virus (MDV) serotype 2 homolog of MDV serotype 1 pp38, an antigen associated with transformed cells. Virology 1994, 201, 142–146. [Google Scholar] [CrossRef] [PubMed]
- Ono, M.; Jang, H.K.; Maeda, K.; Kawaguchi, Y.; Tohya, Y.; Niikura, M.; Mikami, T. Preparation of monoclonal antibodies against Marek’s disease virus serotype 1 glycoprotein D expressed by a recombinant baculovirus. Virus Res. 1995, 38, 219–230. [Google Scholar] [CrossRef]
- Wu, P.; Lee, L.F.; Reed, W.M. Serological characteristics of a membrane glycoprotein gp82 of Marek’s disease virus. Avian Dis. 1997, 41, 824–831. [Google Scholar] [CrossRef]
- Xing, Z.; Xie, Q.; Morgan, R.W.; Schat, K.A. A monoclonal antibody to ICP4 of MDV recognizing ICP4 of serotype 1 and 3 MDV strains. Acta Virol. 1999, 43, 113–120. [Google Scholar]
- Jang, H.K. Characterization and localization of the unique Marek’s disease virus type 2 ORF873 gene product. J. Vet. Sci. 2004, 5, 207–213. [Google Scholar] [CrossRef]
- Liu, J.L.; Lee, L.F.; Ye, Y.; Qian, Z.; Kung, H.J. Nucleolar and nuclear localization properties of a herpesvirus bZIP oncoprotein, MEQ. J. Virol. 1997, 71, 3188–3196. [Google Scholar] [CrossRef]
- Lee, L.F.; Liu, J.L.; Cui, X.P.; Kung, H.J. Marek’s disease virus latent protein MEQ: Delineation of an epitope in the BR1 domain involved in nuclear localization. Virus Genes 2003, 27, 211–218. [Google Scholar] [CrossRef]
- Kurokawa, A.; Yamamoto, Y. Development of monoclonal antibodies specific to Marek disease virus-EcoRI-Q (Meq) for the immunohistochemical diagnosis of Marek disease using formalin-fixed, paraffin-embedded samples. J. Vet. Diagn. Investig. 2022, 34, 458–464. [Google Scholar] [CrossRef]
- Jarosinski, K.W.; Margulis, N.G.; Kamil, J.P.; Spatz, S.J.; Nair, V.K.; Osterrieder, N. Horizontal transmission of Marek’s disease virus requires US2, the UL13 protein kinase, and gC. J. Virol. 2007, 81, 10575–10587. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Reed, W.M.; Yoshida, S.; Sui, D.; Lee, L.F. Identification and characterization of glycoprotein H of MDV-1 GA strain. Acta Virol. 1999, 43, 152–158. [Google Scholar] [PubMed]
- Dorange, F.; Tischer, B.K.; Vautherot, J.F.; Osterrieder, N. Characterization of Marek’s disease virus serotype 1 (MDV-1) deletion mutants that lack UL46 to UL49 genes: MDV-1 UL49, encoding VP22, is indispensable for virus growth. J. Virol. 2002, 76, 1959–1970. [Google Scholar] [CrossRef] [Green Version]
- Dorange, F.; El Mehdaoui, S.; Pichon, C.; Coursaget, P.; Vautherot, J.F. Marek’s disease virus (MDV) homologues of herpes simplex virus type 1 UL49 (VP22) and UL48 (VP16) genes: High-level expression and characterization of MDV-1 VP22 and VP16. J. Gen. Virol. 2000, 81, 2219–2230. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.F.; Heidari, M.; Sun, A.; Zhang, H.; Lupiani, B.; Reddy, S. Identification and in vitro characterization of a Marek’s disease virus-encoded Ribonucleotide reductase. Avian Dis. 2013, 57, 178–187. [Google Scholar] [CrossRef]
- Cui, Z.Z.; Lee, L.F.; Liu, J.L.; Kung, H.J. Structural analysis and transcriptional mapping of the Marek’s disease virus gene encoding pp38, an antigen associated with transformed cells. J. Virol. 1991, 65, 6509–6515. [Google Scholar] [CrossRef]
- Ikuta, K.; Nakajima, K.; Naito, M.; Ann, S.H.; Ueda, S.; Kato, S.; Hirai, K. Identification of Marek’s disease virus-specific antigens in Marek’s disease lymphoblastoid cell lines using monoclonal antibody against virus-specific phosphorylated polypeptides. Int. J. Cancer 1985, 35, 257–264. [Google Scholar] [CrossRef]
- Li, D.; Green, P.F.; Skinner, M.A.; Jiang, C.; Ross, N. Use of recombinant pp38 antigen of Marek’s disease virus to identify serotype 1-specific antibodies in chicken sera by western blotting. J. Virol. Methods 1994, 50, 185–195. [Google Scholar] [CrossRef]
- Zhizhong, C.; Zhi, Z.; Aijian, Q.; Lucy, L.F. Analyzing the H19- and T65-epitopes in 38 kd phosphorylated protein of Marek’s disease viruses and comparing chicken immunological reactions to viruses point-mutated in the epitopes. Sci. China C Life Sci. 2004, 47, 82–91. [Google Scholar]

| Primer Pair | Primer Name | Sequence (5′-3′) |
|---|---|---|
| 1 | pp38-F | CCGAAAGACAAAACCCAAAT |
| pp38-R | ATGTAACCAGCATATAAGAACGC | |
| 2 | gB-F | TCTAGGGCATGGCACACGAC |
| gB-R | GAATACGGAAACACAGAGCGG | |
| 3 | meq-F | AGCCGGAGAGGCTTTATGC |
| meq-R | GGCCCGAATACAAGGAATCC | |
| 4 | LORF6-F | AATGCGGATCATCAGGGTCTC |
| LORF6-R | GAGAGGCTTTATGCTCGTCTTACC | |
| 5 | UL6-F | GAATTCGATGTCGGCAGTAAGC |
| UL6-R | TACATTCCCCACGCTCACCAC | |
| 6 | UL13-F | ACCCTCGGTGACGTTAACAAAG |
| UL13-R | TTCATGGATCTCGGCAAAGC | |
| 7 | UL42-F | TTCGTCAGCCCTCATCGTG |
| UL42-R | AAATGCGTTAGTATCTTCCAGTGC | |
| 8 | UL52-F | AATGGGTTATCTCTGAAGGGTCG |
| UL52-R | AGTCAGACCTCGTTTACCCCTTG |
| gRNA/Target | Oligo/Primer | Sequence (5′-3′) * |
|---|---|---|
| gRF1 | gRF1–5p | CACCGCGTGCATGTCATTTTTCGC |
| gRF1–3p | AAACGCGAAAAATGACATGCACGC | |
| gRF2 | gRF2–5p | CACCGGTGCAGGCATTGTCGTTGG |
| gRF2–3p | AAACCCAACGACAATGCCTGCACC | |
| gRF3 | gRF3–5p | CACCGGGTATGTTAGTCGGTAGAA |
| gRF3–3p | AAACTTCTACCGACTAACATACCC | |
| gRR1 | gRR1–5p | CACCGAACAGAAGCGGAATGCGCCG |
| gRR1–3p | AAACCGGCGCATTCCGCTTCTGTTC | |
| gRR2 | gRR2–5p | CACCGTCGCCGACGAGGCAGGGCAT |
| gRR2–3p | AAACATGCCCTGCCTCGTCGGCGAC | |
| gRR3 | gRR3–5p | CACCGCGAAGGGCTGACGGCGTCTT |
| gRR3–3p | AAACAAGACGCCGTCAGCCCTTCGC | |
| pp38 | pp38F | TGGTGGGGAGATAGTCTCGG |
| pp38R | TGATCGGTGGTGTAACCGTG |
| No. | Hybridomas | Specificity | ||
|---|---|---|---|---|
| MDV-1/GX0101 | MDV-3/HVT | MDV-2/SB-1 | ||
| 1 | 4B1 | + | + | + |
| 2 | 4B5 | + | + | + |
| 3 | 4B8 | + | + | + |
| 4 | 4F11 | + | + | + |
| 5 | 5D5 | + | + | + |
| 6 | 5D10 | + | + | + |
| 7 | 5E8 | + | + | + |
| 8 | 5E10 | + | + | + |
| 9 | 5F6 | + | + | + |
| 10 | 6B1 | + | + | + |
| 11 | 6E4 | + | + | + |
| 12 | 6G9 | + | + | + |
| 13 | 6G11 | + | + | + |
| 14 | 10B2 | + | + | + |
| 15 | 27H3 | + | + | + |
| 16 | 1F5 | + | + | - |
| 17 | 2E2 | + | + | - |
| 18 | 2F3 | + | + | - |
| 19 | 2G4 | + | + | - |
| 20 | 2G6 | + | + | - |
| 21 | 3B7 | + | + | - |
| 22 | 3G3 | + | + | - |
| 23 | 4B5 | + | + | - |
| 24 | 4E7 | + | + | - |
| 25 | 4E11 | + | + | - |
| 26 | 6C4 | + | + | - |
| 27 | 4F9 | + | - | - |
| 28 | 10B1 | + | - | - |
| 29 | 10G9 | + | - | - |
| 30 | 10F9 | + | - | - |
| 31 | 21G9 | + | - | - |
| 32 | 31G7 | + | - | - |
| 33 | 34F2 | + | - | - |
| 34 | 35G9 | + | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teng, M.; Zhou, Z.-Y.; Yao, Y.; Nair, V.; Zhang, G.-P.; Luo, J. A New Strategy for Efficient Screening and Identification of Monoclonal Antibodies against Oncogenic Avian Herpesvirus Utilizing CRISPR/Cas9-Based Gene-Editing Technology. Viruses 2022, 14, 2045. https://doi.org/10.3390/v14092045
Teng M, Zhou Z-Y, Yao Y, Nair V, Zhang G-P, Luo J. A New Strategy for Efficient Screening and Identification of Monoclonal Antibodies against Oncogenic Avian Herpesvirus Utilizing CRISPR/Cas9-Based Gene-Editing Technology. Viruses. 2022; 14(9):2045. https://doi.org/10.3390/v14092045
Chicago/Turabian StyleTeng, Man, Zi-Yu Zhou, Yongxiu Yao, Venugopal Nair, Gai-Ping Zhang, and Jun Luo. 2022. "A New Strategy for Efficient Screening and Identification of Monoclonal Antibodies against Oncogenic Avian Herpesvirus Utilizing CRISPR/Cas9-Based Gene-Editing Technology" Viruses 14, no. 9: 2045. https://doi.org/10.3390/v14092045
APA StyleTeng, M., Zhou, Z.-Y., Yao, Y., Nair, V., Zhang, G.-P., & Luo, J. (2022). A New Strategy for Efficient Screening and Identification of Monoclonal Antibodies against Oncogenic Avian Herpesvirus Utilizing CRISPR/Cas9-Based Gene-Editing Technology. Viruses, 14(9), 2045. https://doi.org/10.3390/v14092045







