Bovine Polyomavirus-1 (Epsilonpolyomavirus bovis): An Emerging Fetal Pathogen of Cattle That Causes Renal Lesions Resembling Polyomavirus-Associated Nephropathy of Humans
Abstract
:1. Introduction
2. Materials and Methods
2.1. Histopathology and Immunohistochemistry
2.2. Transmission Electron Microscopy
2.3. Molecular Virology and Ancillary Testing for Specific Pathogens
2.4. Whole-Genome Sequencing and Genome Characterization
2.5. Phylogenetic Analyses
2.6. Sequence Identity
2.7. Virus Isolation
3. Results
3.1. Case History
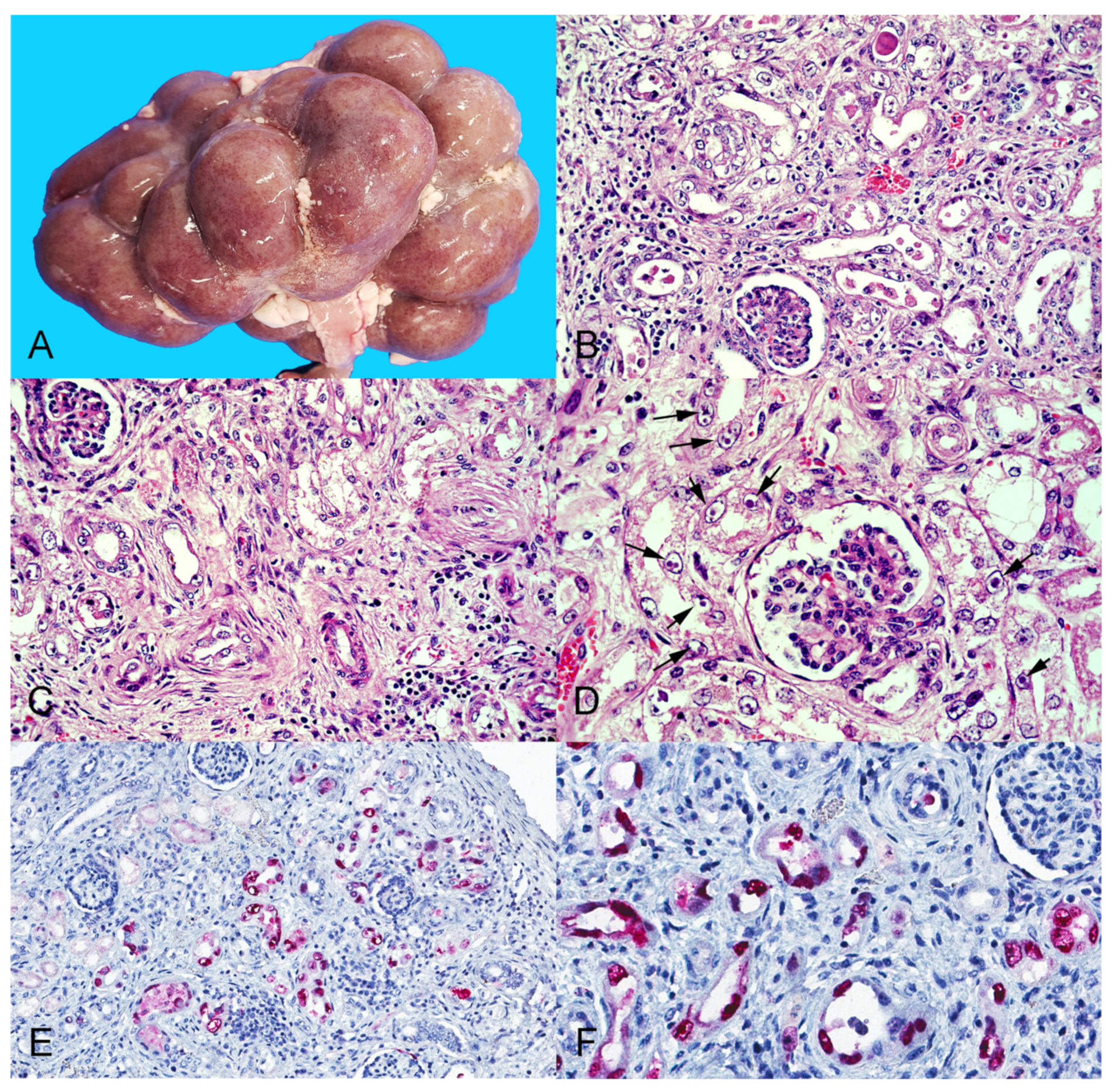
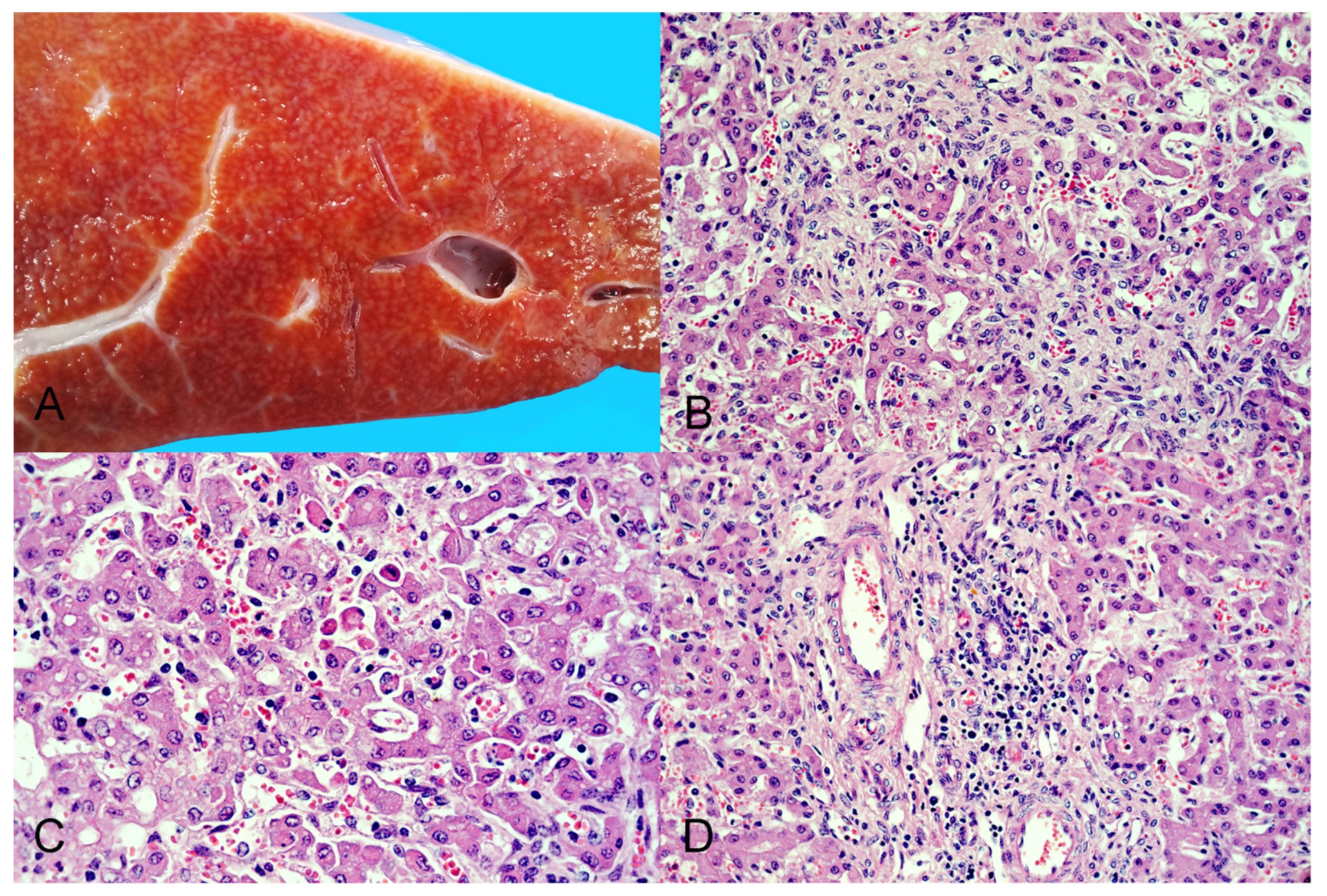
3.2. Gross Pathologic Examination
3.3. Histopathology and Immunohistochemistry
3.4. Transmission Electron Microscopy
3.5. Molecular Virology and Ancillary Testing for Specific Pathogens
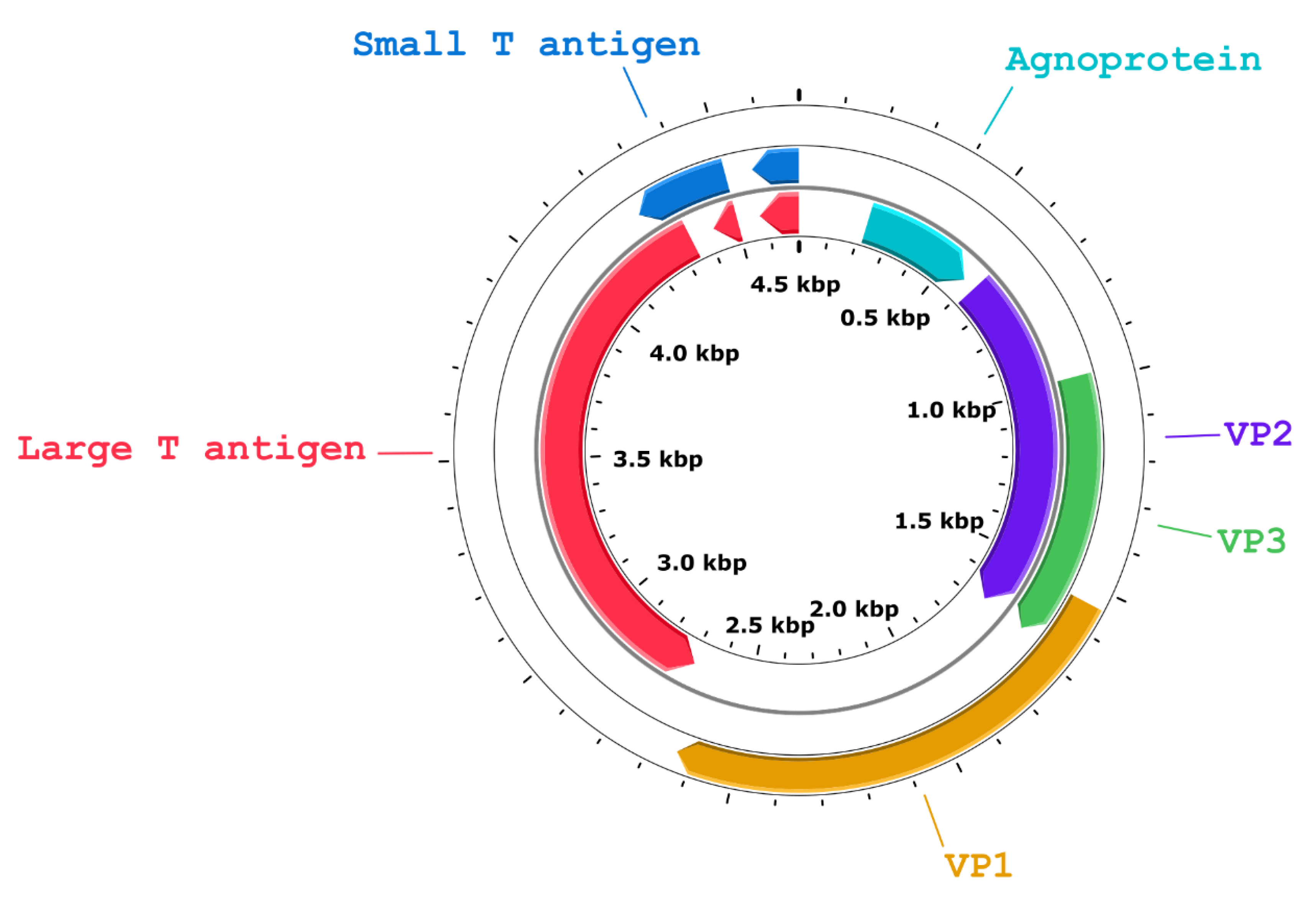
3.6. Whole Genome Sequencing and Genome Characterization
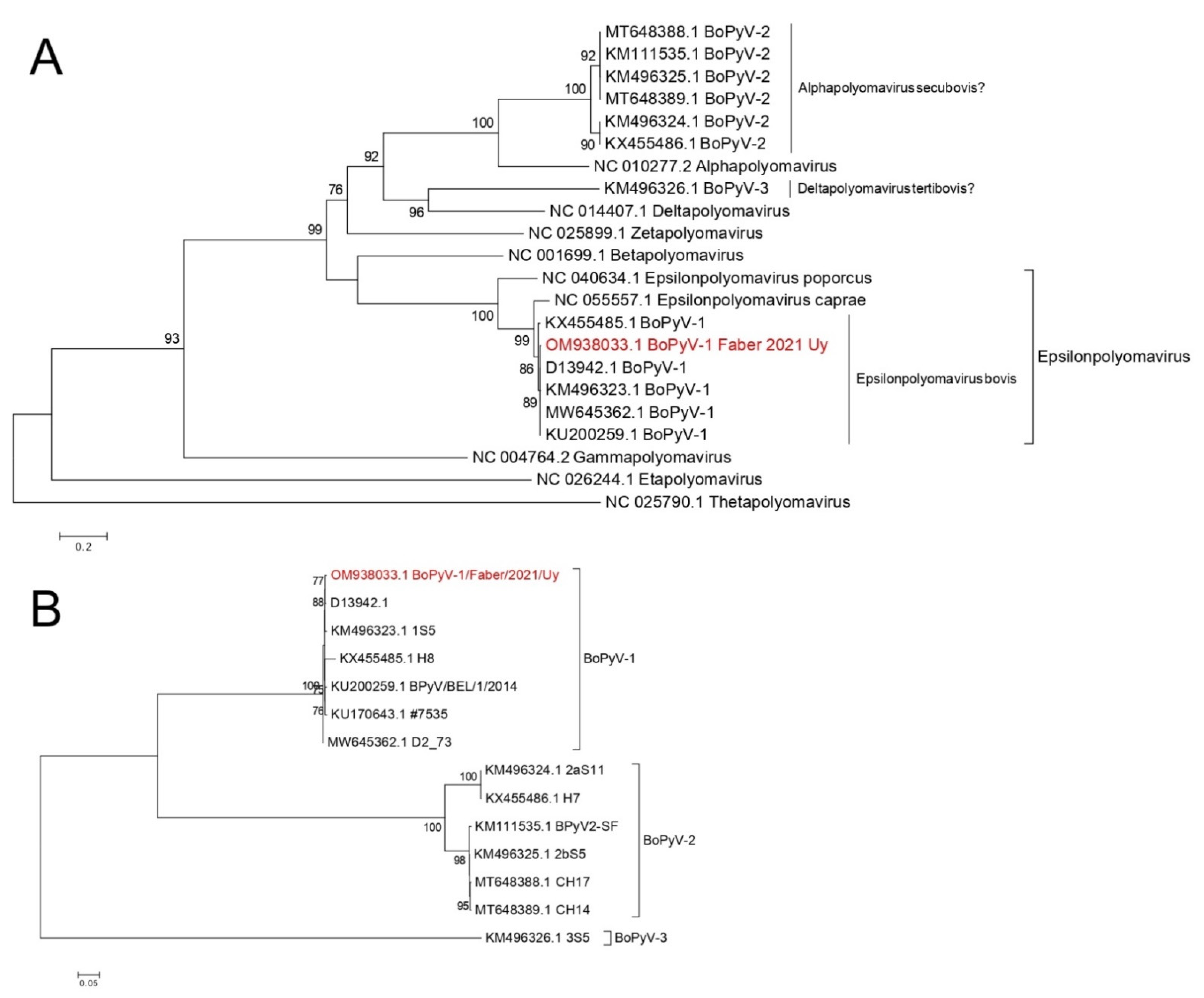
3.7. Phylogenetic Analyses
3.8. Sequence Identity
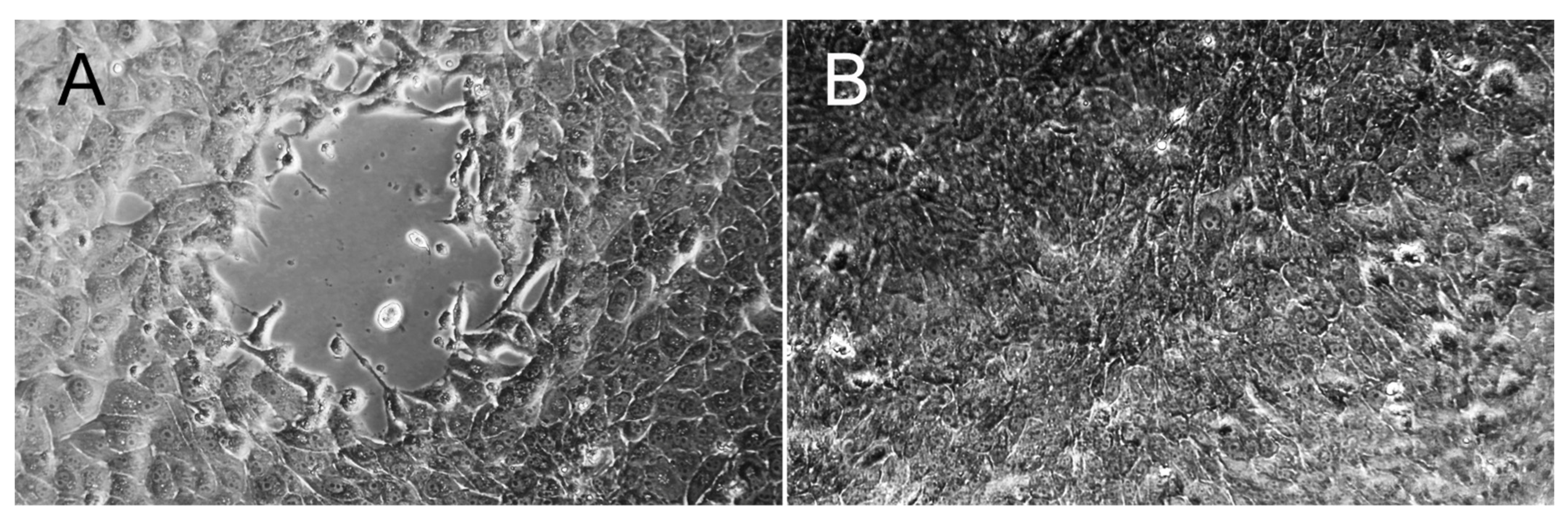
3.9. Virus Isolation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Disclaimer
References
- Moens, U.; Krumbholz, A.; Ehlers, B.; Zell, R.; Johne, R.; Calvignac-Spencer, S.; Lauber, C. Biology, evolution, and medical importance of polyomaviruses: An update. Infect. Genet. Evol. 2017, 54, 18–38. [Google Scholar] [CrossRef] [PubMed]
- Imperiale, M.J.; Jiang, M. Polyomavirus persistence. Annu. Rev. Virol. 2016, 3, 517–532. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Shuda, M.; Chang, Y.; Moore, P.S. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 2008, 319, 1096–1100. [Google Scholar] [CrossRef] [PubMed]
- Dela Cruz, F.N.; Giannitti, F.; Li, L.; Woods, L.W.; Del Valle, L.; Delwart, E.; Pesavento, P.A. Novel polyomavirus associated with brain tumors in free-ranging raccoons, western United States. Emerg. Infect. Dis. 2013, 19, 77–84. [Google Scholar] [CrossRef]
- Giannitti, F.; Higgins, R.J.; Pesavento, P.A.; Dela Cruz, F.; Clifford, D.L.; Piazza, M.; Parker Struckhoff, A.; Del Valle, L.; Bollen, A.W.; Puschner, B.; et al. Temporal and geographic clustering of polyomavirus-associated olfactory tumors in 10 free-ranging raccoons (Procyon lotor). Vet. Pathol. 2014, 51, 832–845. [Google Scholar] [CrossRef]
- Church, M.E.; Estrada, M.; Leutenegger, C.M.; Dela Cruz, F.N.; Pesavento, P.A.; Woolard, K.D. BRD4 is associated with raccoon polyomavirus genome and mediates viral gene transcription and maintenance of a stem cell state in neuroglial tumour cells. J. Gen. Virol. 2016, 97, 2939–2948. [Google Scholar] [CrossRef]
- Mazziotta, C.; Pellielo, G.; Tognon, M.; Martini, F.; Rotondo, J.C. Significantly low levels of IgG antibodies against oncogenic Merkel cell polyomavirus in sera from females affected by spontaneous abortion. Front. Microbiol. 2021, 12, 789991. [Google Scholar] [CrossRef]
- Reissig, M.; Kelly, T.J.; Daniel, R.W.; Rangan, S.R.; Shah, K.V. Identification of the stumptailed macaque virus as a new papovavirus. Infect. Immun. 1976, 14, 225–231. [Google Scholar] [CrossRef]
- Wognum, A.W.; Sol, C.J.A.; van der Noordaa, J.; van Steenis, G.; Osterhaus, A.D.M.E. Isolation and characterization of a papovavirus from cynomolgus macaque kidney cells. Virology 1984, 134, 254–257. [Google Scholar] [CrossRef]
- Parry, J.V.; Richmond, J.E.; Gardner, S.D. Polyomavirus in fetal rhesus monkey kidney cell lines used to grow hepatitis A virus. Lancet 1983, 321, 994. [Google Scholar] [CrossRef]
- Parry, J.V.; Lucas, M.H.; Richmond, J.E.; Gardner, S.D. Evidence for a bovine origin of the polyomaviurs detected in foetal rhesus monkey cells, FRhK-4 and-6. Arch. Virol. 1983, 78, 151–165. [Google Scholar] [CrossRef] [PubMed]
- Schuurman, R.; van Steenis, B.; van Strien, A.; van der Noordaa, J.; Sol, C. Frequent detection of bovine polyomavirus in commercial batches of calf serum by using the polymerase chain reaction. J. Gen. Virol. 1991, 72, 2739–2745. [Google Scholar] [CrossRef] [PubMed]
- van der Noordaa, J.; Sol, C.J.; Schuurman, R. Bovine polyomavirus, a frequent contaminant of calf sera. Dev. Biol. Stand. 1999, 99, 45–47. [Google Scholar] [CrossRef] [PubMed]
- Nairn, C.; Lovatt, A.; Galbraith, D.N. Detection of infectious bovine polyomavirus. Biologicals 2003, 31, 303–306. [Google Scholar] [CrossRef]
- Wang, J.; Horner, G.W.; O’Keefe, J.S. Detection and molecular characterisation of bovine polyomavirus in bovine sera in New Zealand. N. Z. Vet. J. 2005, 53, 26–30. [Google Scholar] [CrossRef]
- Paim, W.P.; Maggioli, M.F.; Falkenberg, S.M.; Ramachandran, A.; Weber, M.N.; Canal, C.W.; Bauermann, F.V. Virome characterization in commercial bovine serum batches—A potentially needed testing strategy for biological products. Viruses 2021, 13, 2425. [Google Scholar] [CrossRef]
- Coackley, W.; Maker, D.; Smith, V.W. A possible bovine polyomavirus. Arch. Virol. 1980, 66, 161–166. [Google Scholar] [CrossRef]
- Parry, J.V.; Gardner, S.D. Human exposure to bovine polyomavirus: A zoonosis? Arch. Virol. 1986, 87, 287–296. [Google Scholar] [CrossRef]
- Schuurman, R.; Sol, C.; van der Noordaa, J. The complete nucleotide sequence of bovine polyomavirus. J. Gen. Virol. 1990, 71, 1723–1735. [Google Scholar] [CrossRef]
- Peretti, A.; FitzGerald, P.C.; Bliskovsky, V.; Buck, C.B.; Pastrana, D.V. Hamburger polyomaviruses. J. Gen. Virol. 2015, 96, 833–839. [Google Scholar] [CrossRef]
- Hierweger, M.M.; Koch, M.C.; Seuberlich, T. Bovine polyomavirus 2 is a probable cause of non-suppurative encephalitis in cattle. Pathogens. 2020, 9, 620. [Google Scholar] [CrossRef]
- Moens, U.; Van Ghelue, M.; Song, X.; Ehlers, B. Serological cross-reactivity between human polyomaviruses. Rev. Med. Virol. 2013, 23, 250–264. [Google Scholar] [CrossRef] [PubMed]
- Viscidi, R.P.; Clayman, B. Serological cross reactivity between polyomavirus capsids. Adv. Exp. Med. Biol. 2006, 577, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Clothier KAnderson, M. Evaluation of bovine abortion cases and tissue suitability for identification of infectious agents in California diagnostic laboratory cases from 2007 to 2012. Theriogenology 2016, 85, 933–938. [Google Scholar] [CrossRef] [PubMed]
- Wolf-Jäckel, G.A.; Hansen, M.S.; Larsen, G.; Holm, E.; Agerholm, J.S.; Jensen, T.K. Diagnostic studies of abortion in Danish cattle 2015–2017. Acta Vet. Scand. 2020, 62, 1. [Google Scholar] [CrossRef]
- Dorsch, M.A.; Cantón, G.J.; Driemeier, D.; Anderson, M.L.; Moeller, R.B.; Giannitti, F. Bacterial, protozoal and viral abortions in sheep and goats in South America: A review. Small Rumin. Res. 2021, 205, 106547. [Google Scholar] [CrossRef]
- Rodger, S.M.; Murray, J.; Underwood, C.; Buxton, D. Microscopical lesions and antigen distribution in bovine fetal tissues and placentae following experimental infection with bovine herpesvirus-1 during pregnancy. J. Comp. Pathol. 2007, 137, 94–101. [Google Scholar] [CrossRef]
- Woods, L.W.; Schumaker, B.A.; Pesavento, P.A.; Crossley, B.M.; Swift, P.K. Adenoviral hemorrhagic disease in California mule deer, 1990–2014. J. Vet. Diagn Investig. 2018, 30, 530–573. [Google Scholar] [CrossRef]
- Martines, R.B.; Ritter, J.M.; Matkovic, E.; Gary, J.; Bollweg, B.C.; Bullock, H.; Goldsmith, C.S.; Silva-Flannery, L.; Seixas, J.N.; Reagan-Steiner, S.; et al. Pathology and pathogenesis of SARS-CoV-2 associated with fatal coronavirus disease, United States. Emerg Infect. Dis. 2020, 26, 2005–2015. [Google Scholar] [CrossRef]
- Ashbaugh, S.E.; Thompson, K.E.; Belknap, E.B.; Schultheiss, P.C.; Chowdhury, S.; Collins, J.K. Specific detection of shedding and latency of bovine herpesvirus 1 and 5 using a nested polymerase chain reaction. J. Vet. Diagn Investig. 1997, 9, 387–394. [Google Scholar] [CrossRef] [Green Version]
- Campos, F.S.; Franco, A.C.; Oliveira, M.T.; Firpo, R.; Strelczuk, G.; Fontoura, F.E.; Kulmann, M.I.R.; Maidana, S.; Romera, S.A.; Spilki, F.R.; et al. Detection of bovine herpesvirus 2 and bovine herpesvirus 4 DNA in trigeminal ganglia of naturally infected cattle by polymerase chain reaction. Vet. Microbiol. 2014, 171, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Maya, L.; Puentes, R.; Reolón, E.; Acuña, P.; Riet, F.; Rivero, R.; Cristina, J.; Colina, R. Molecular diversity of bovine viral diarrhea virus in Uruguay. Arch. Virol. 2016, 161, 529–535. [Google Scholar] [CrossRef] [PubMed]
- Vilček, S.; Herring, A.J.; Herring, J.A.; Nettleton, P.F.; Lowings, J.P.; Paton, D.J. Pestiviruses isolated from pigs, cattle and sheep can be allocated into at least three genogroups using polymerase chain reaction and restriction endonuclease analysis. Arch. Virol. 1994, 136, 309–323. [Google Scholar] [CrossRef]
- Cabrera, A.; Fresia, P.; Berná, L.; Silveira, C.; Macías-Rioseco, M.; Arevalo, A.P.; Crispo, M.; Pritsch, O.; Riet-Correa, F.; Giannitti, F.; et al. Isolation and molecular characterization of four novel Neospora caninum strains. Parasitol. Res. 2019, 118, 3535–3542. [Google Scholar] [CrossRef] [PubMed]
- Stoddard, R.A.; Gee, J.E.; Wilkins, P.P.; McCaustland, K.; Hoffmaster, A.R. Detection of pathogenic Leptospira spp. through TaqMan polymerase chain reaction targeting the LipL32 gene. Diagn Microbiol Infect. Dis. 2009, 64, 247–255. [Google Scholar] [CrossRef]
- Hundesa, A.; Bofill-Mas, S.; Maluquer de Motes, C.; Rodriguez-Manzano, J.; Bach, A.; Casas, M.; Girones, R. Development of a quantitative PCR assay for the quantitation of bovine polyomavirus as a microbial source-tracking tool. J. Virol. Methods 2010, 163, 385–389. [Google Scholar] [CrossRef]
- De Coster, W.; D’Hert, S.; Schultz, D.T.; Cruts, M.; Van Broeckhoven, C. NanoPack: Visualizing and processing long-read sequencing data. Bioinformatics 2018, 34, 2666–2669. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [Green Version]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [PubMed]
- Anisimova, M.; Gascuel, O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst. Biol. 2006, 55, 539–552. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Van Borm, S.; Rosseel, T.; Behaeghel, I.; Saulmont, M.; Delooz, L.; Petitjean, T.; Mathijs, E.; Vandenbussche, F. Complete genome sequence of Bovine polyomavirus type 1 from aborted cattle, isolated in Belgium in 2014. Genome Announc. 2016, 4, e01646-15. [Google Scholar] [CrossRef] [PubMed]
- Dalianis, T.; Hirsch, H.H. Human polyomaviruses in disease and cancer. Virology 2013, 437, 63–72. [Google Scholar] [CrossRef]
- Ramos, E.; Drachenberg, C.B.; Wali, R.; Hirsch, H.H. The decade of polyomavirus BK-associated nephropathy: State of affairs. Transplantation 2009, 87, 621–630. [Google Scholar] [CrossRef]
- Alcendor, D.J. BK polyomavirus virus glomerular tropism: Implications for virus reactivation from latency and amplification during immunosuppression. J. Clin. Med. 2019, 8, 1477. [Google Scholar] [CrossRef]
- Costa, C.; Cavallo, R. Polyomavirus-associated nephropathy. World J. Transplant. 2012, 2, 84–94. [Google Scholar] [CrossRef]
- Dubey, J.P.; Hemphill, A.; Calero-Bernal, R.; Schares, G. Neosporosis in Animals, 1st ed.; CRC Press: Boca Raton, FL, USA, 2017. [Google Scholar] [CrossRef]
- Venturini, M.C.; Venturini, L.; Bacigalupe, D.; Machuca, M.; Echaide, I.; Basso, W.; Unzaga, J.M.; Di Lorenzo, C.; Guglielmone, A.; Jenkins, M.C.; et al. Neospora caninum infections in bovine foetuses and dairy cows with abortions in Argentina. Int. J. Parasitol. 1999, 29, 1705–1708. [Google Scholar] [CrossRef]
- Bacigalupe, D.; Basso, W.; Caspe, S.G.; Moré, G.; Lischinsky, L.; Gos, M.L.; Leunda, M.; Campero, L.; Moore, D.P.; Schares, G.; et al. Neospora caninum NC-6 Argentina induces fetopathy in both serologically positive and negative experimentally inoculated pregnant dams. Parasitol. Res. 2013, 112, 2585–2592. [Google Scholar] [CrossRef]
- Giakoumelou, S.; Wheelhouse, N.; Cuschieri, K.; Entrican, G.; Howie, S.E.M.; Horne, A.W. The role of infection in miscarriage. Hum. Reprod. Update 2016, 22, 116–133. [Google Scholar] [CrossRef] [PubMed]
- Gräfe, D.; Ehlers, B.; Mäde, D.; Ellerbroek, L.; Seidler, T.; Johne, R. Detection and genome characterization of bovine polyomaviruses in beef muscle and ground beef samples from Germany. Int. J. Food Microbiol. 2017, 241, 168–172. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Li, L.; Deng, X.; Kapusinszky, B.; Delwart, E. What is for dinner? Viral metagenomics of US store bought beef, pork, and chicken. Virology 2004, 468–470, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Ben Salem, N.; Pérez de Val, B.; Martin, M.; Moens, U.; Ehlers, B. Genome sequence of Bovine polyomavirus 1 detected in a Salers cow (Bos taurus) from Catalonia, Spain. Genome Announc. 2016, 4, e01658-15. [Google Scholar] [CrossRef]
- Rusiñol, M.; Fernandez-Cassi, X.; Hundesa, A.; Vieira, C.; Kern, A.; Eriksson, I.; Ziros, P.; Kay, D.; Miagostovich, M.; Vargha, M.; et al. Application of human and animal viral microbial source tracking tools in fresh and marine waters from five different geographical areas. Water Res. 2014, 59, 119–129. [Google Scholar] [CrossRef]
- Hundesa, A.; Maluquer de Motes, C.; Bofill-Mas, S.; Albinana-Gimenez, N.; Girones, R. Identification of human and animal adenoviruses and polyomaviruses for determination of sources of fecal contamination in the environment. Appl. Environ. Microbiol. 2006, 72, 7886–7893. [Google Scholar] [CrossRef] [Green Version]
- Byrne, P. The Role of the Large T Antigen in the In Vitro Replication of BPyV. Master’s Thesis, University of Glasgow, Scotland, UK, 2009. Available online: https://theses.gla.ac.uk/576/ (accessed on 13 September 2022).

| Epsilonpolyomavirus bovis (BoPyV-1) | BoPyV-2 | BoPyV-3 | |||||
|---|---|---|---|---|---|---|---|
| Nucleotide | Amino Acid | Nucleotide | Amino Acid | Nucleotide | Amino Acid | ||
| BoPyV-1/Faber/2021/Uy | Complete genome | 93.6–99.3 | - | 40.0–41.0 | - | 37.6 | - |
| Large T antigen | 93.8–99.3 | 94.1–99.6 | 36.1–37.5 | 30.3–31.6 | 48.3 | 36.9 | |
| Small T antigen | 99.2–99.7 | 98.3–99.1 | 26.5–28.9 | 15.7–17.1 | 26.7 | 18.2 | |
| VP1 | 97.1–99.3 | 98.6–99.7 | 53.7–55.0 | 50.5–52.2 | 39.7 | 27.5 | |
| VP2 | 97.0–99.3 | 96.8–99.7 | 31.0–32.1 | 23.7–25.1 | 31.8 | 16.4 | |
| VP3 | 96.1–99.2 | 96.1–100 | 36.1–37.7 | 20.6–22.1 | 30.1 | 13.0 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giannitti, F.; da Silva Silveira, C.; Bullock, H.; Berón, M.; Fernández-Ciganda, S.; Benítez-Galeano, M.J.; Rodríguez-Osorio, N.; Silva-Flannery, L.; Perdomo, Y.; Cabrera, A.; et al. Bovine Polyomavirus-1 (Epsilonpolyomavirus bovis): An Emerging Fetal Pathogen of Cattle That Causes Renal Lesions Resembling Polyomavirus-Associated Nephropathy of Humans. Viruses 2022, 14, 2042. https://doi.org/10.3390/v14092042
Giannitti F, da Silva Silveira C, Bullock H, Berón M, Fernández-Ciganda S, Benítez-Galeano MJ, Rodríguez-Osorio N, Silva-Flannery L, Perdomo Y, Cabrera A, et al. Bovine Polyomavirus-1 (Epsilonpolyomavirus bovis): An Emerging Fetal Pathogen of Cattle That Causes Renal Lesions Resembling Polyomavirus-Associated Nephropathy of Humans. Viruses. 2022; 14(9):2042. https://doi.org/10.3390/v14092042
Chicago/Turabian StyleGiannitti, Federico, Caroline da Silva Silveira, Hannah Bullock, Marina Berón, Sofía Fernández-Ciganda, María José Benítez-Galeano, Nélida Rodríguez-Osorio, Luciana Silva-Flannery, Yisell Perdomo, Andrés Cabrera, and et al. 2022. "Bovine Polyomavirus-1 (Epsilonpolyomavirus bovis): An Emerging Fetal Pathogen of Cattle That Causes Renal Lesions Resembling Polyomavirus-Associated Nephropathy of Humans" Viruses 14, no. 9: 2042. https://doi.org/10.3390/v14092042
APA StyleGiannitti, F., da Silva Silveira, C., Bullock, H., Berón, M., Fernández-Ciganda, S., Benítez-Galeano, M. J., Rodríguez-Osorio, N., Silva-Flannery, L., Perdomo, Y., Cabrera, A., Puentes, R., Colina, R., Ritter, J. M., & Castells, M. (2022). Bovine Polyomavirus-1 (Epsilonpolyomavirus bovis): An Emerging Fetal Pathogen of Cattle That Causes Renal Lesions Resembling Polyomavirus-Associated Nephropathy of Humans. Viruses, 14(9), 2042. https://doi.org/10.3390/v14092042





