HIV-1 Circulating Recombinant Forms (CRFs) and Unique Recombinant Forms (URFs) in Israel, 2010–2018
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of CRFs and URFs Using RIP, RDP5, and Phylogenetic Analysis
2.2. Comparison between Patients Carrying Recombinant viruses and Patients Infected with Non CRFs
2.3. Statistical Analysis and Ethical Approval
3. Results
3.1. Characteristics of HIV-1 Recombinants
3.2. Comparison between PLHIV Carrying Recombinant Viruses to those Infected with Classic Subtypes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hemelaar, J.; Elangovan, R.; Yun, J.; Dickson-Tetteh, L.; Kirtley, S.; Gouws-Williams, E.; Ghys, P.D.; Alash’le G, A.; Agwale, S.; Archibald, C.; et al. Global and regional epidemiology of HIV-1 recombinants in 1990–2015: A systematic review and global survey. Lancet HIV 2020, 7, e772–e781. [Google Scholar] [CrossRef]
- Elangovan, R.; Jenks, M.; Yun, J.; Dickson-Tetteh, L.; Kirtley, S.; Hemelaar, J.; Abimiku, A.L.G.; Agwale, S.; Archibald, C.; Avidor, B.; et al. Global and regional estimates for subtype-specific therapeutic and prophylactic HIV-1 vaccines: A modeling study. Front. Microbiol. 2021, 12, 690647. [Google Scholar] [CrossRef] [PubMed]
- Hemelaar, J. The origin and diversity of the HIV-1 pandemic. Trends Mol. Med. 2012, 18, 182–192. [Google Scholar] [CrossRef] [PubMed]
- Grossman, Z.; Avidor, B.; Mor, Z.; Chowers, M.; Levy, I.; Shahar, E.; Riesenberg, K.; Sthoeger, Z.; Maayan, S.; Shao, W.; et al. A Population-Structured HIV Epidemic in Israel: Roles of Risk and Ethnicity. PLoS ONE 2015, 10, e0135061. [Google Scholar]
- Wagner, T.; Zuckerman, N.S.; Halperin, T.; Chemtob, D.; Levy, I.; Elbirt, D.; Shachar, E.; Olshtain-Pops, K.; Elinav, H.; Chowers, M.; et al. Epidemiology and Transmitted HIV-1 Drug Resistance among Treatment-Naïve Individuals in Israel, 2010–2018. Viruses 2021, 14, 71. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Foley, B.; Schultz, A.-K.; Macke, J.P.; Bulla, I.; Stanke, M.; Morgenstern, B.; Korber, B.; Leitner, T. The role of recombination in the emergence of a complex and dynamic HIV epidemic. Retrovirology 2010, 7, 25. [Google Scholar] [CrossRef] [PubMed]
- Bbosa, N.; Kaleebu, P.; Ssemwanga, D. HIV subtype diversity worldwide. Curr. Opin. HIV AIDS 2019, 14, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Sivay, M.V.; Totmenin, A.V.; Zyryanova, D.P.; Osipova, I.P.; Nalimova, T.M.; Gashnikova, M.P.; Ivlev, V.V.; Meshkov, I.O.; Chokmorova, U.Z.; Narmatova, E.; et al. Characterization of HIV-1 Epidemic in Kyrgyzstan. Front. Microbiol. 2021, 12, 753675. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; An, M.; Zhao, B.; Duan, S.; Yang, S.; Xu, J.; Zhang, M.; McGoogan, J.M.; Takebe, Y.; Shang, H. High Prevalence of HIV-1 Intersubtype B′/C Recombinants among Injecting Drug Users in Dehong, China. PLoS ONE 2013, 8, e65337. [Google Scholar]
- Larsson, A. AliView: A fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.P.; Varsani, A.; Roumagnac, P.; Botha, G.; Maslamoney, S.; Schwab, T.; Kelz, Z.; Kumar, V.; Murrell, B. RDP5: A computer program for analyzing recombination in, and removing signals of recombination from, nucleotide sequence datasets. Virus Evol. 2020, 7, veaa087. [Google Scholar] [CrossRef] [PubMed]
- Posada, D. jModelTest: Phylogenetic Model Averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef] [PubMed]
- HIVDB. HIV Drug Resistance Database. Available online: https://hivdb.stanford.edu/ (accessed on 12 December 2021).
- Bennett, D.E.; Camacho, R.J.; Otelea, D.; Kuritzkes, D.R.; Fleury, H.; Kiuchi, M.; Heneine, W.; Kantor, R.; Jordan, M.R.; Schapiro, J.M.; et al. Drug Resistance Mutations for Surveillance of Transmitted HIV-1 Drug-Resistance: 2009 Update. PLoS ONE 2009, 4, e4724. [Google Scholar] [CrossRef] [PubMed]
- van de Laar, M.J.; Bosman, A.; Pharris, A.; Andersson, E.; Assoumou, L.; Ay, E.; Bannert, N.; Bartmeyer, B.; Brady, M.; Chaix, M.-L.; et al. Piloting a surveillance system for HIV drug resistance in the European Union. Eurosurveillance 2019, 24, 1800390. [Google Scholar] [CrossRef] [PubMed]
- Beloukas, A.; Psarris, A.; Giannelou, P.; Kostaki, E.; Hatzakis, A.; Paraskevis, D. Molecular epidemiology of HIV-1 infection in Europe: An overview. Infect. Genet. Evol. 2016, 46, 180–189. [Google Scholar] [CrossRef] [PubMed]
- Lau, K.A.; Wong, J.J. Current trends of HIV recombination worldwide. Infect. Dis. Rep. 2013, 5 (Suppl. 1), e4. [Google Scholar] [CrossRef] [PubMed]
- Grossman, Z.; Avidor, B.; Girshengoren, S.; Katchman, E.; Maldarelli, F.; Turner, D. Transmission Dynamics of HIV Subtype A in Tel Aviv, Israel: Implications for HIV Spread and Eradication. Open Forum Infect. Dis. 2019, 6, ofz304. [Google Scholar] [CrossRef] [PubMed]
- Zuckerman, N.S.; Mor, Z.; Bucris, E.; Wax, M.; Mendelson, E.; Mor, O. Sexual intermingling of Arab and Jewish MSM in Israel: Results of a molecular epidemiology study. AIDS 2019, 33, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Battistini Garcia, S.A.; Guzman, N. Acquired Immune Deficiency Syndrome CD4+ Count. In Treasure Island; StatPearls Publishing: Tampa, FL, USA, 2022. [Google Scholar]
- Rhee, S.-Y.; Shafer, R.W. Geographically-stratified HIV-1 group M pol subtype and circulating recombinant form sequences. Sci. Data 2018, 5, 180148. [Google Scholar] [CrossRef] [PubMed] [Green Version]

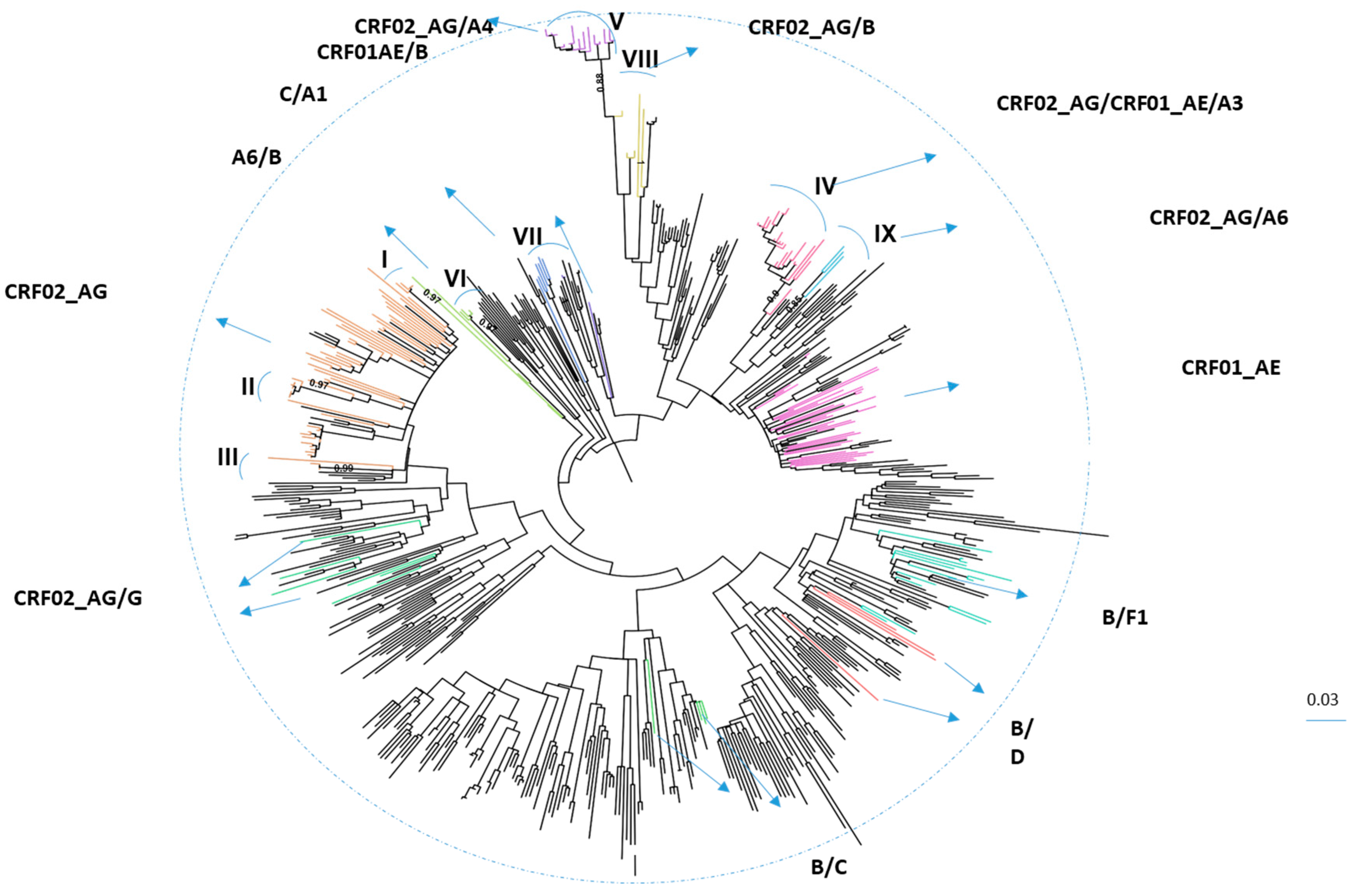
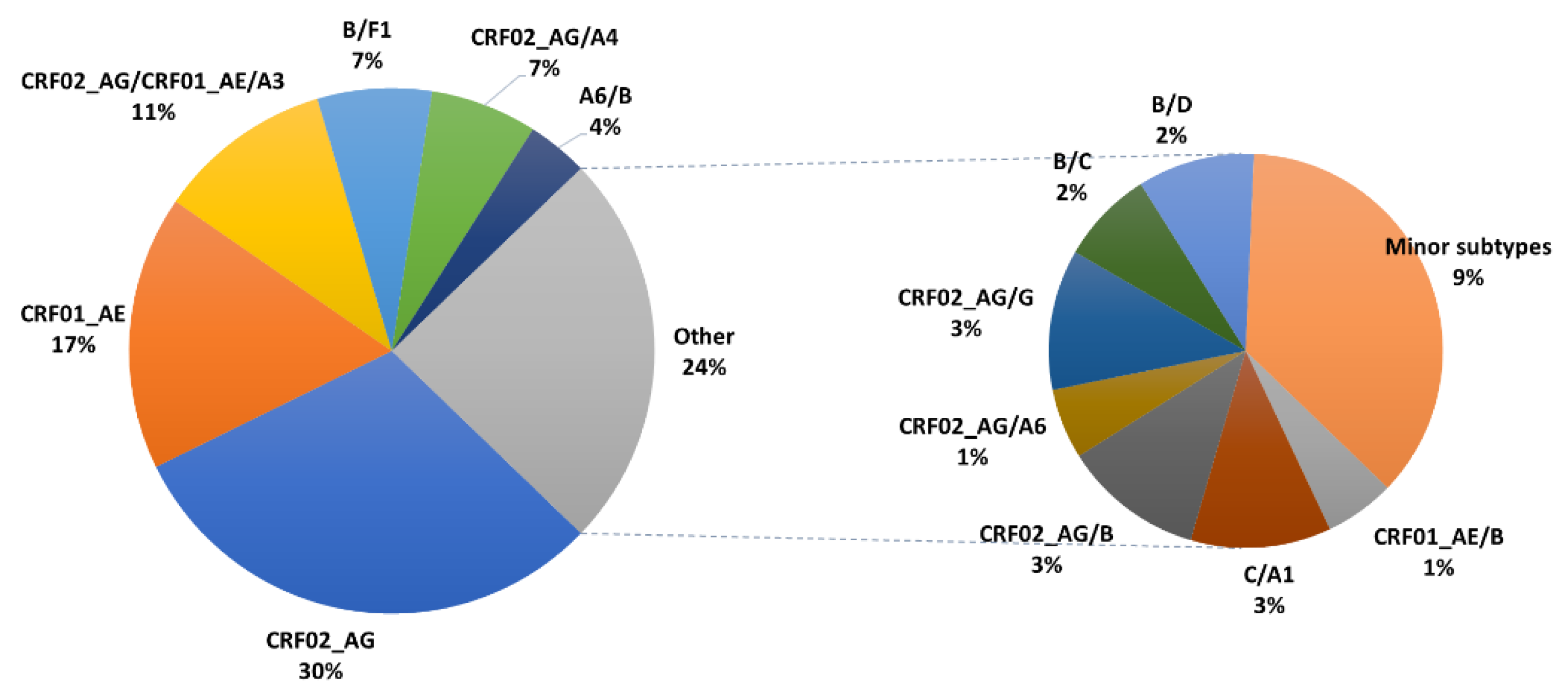

| Recombinants | CRF02_AG | CRF01_AE | CRF02_AG/ CRF01_AE/A3 | B/F1 | CRF02_AG/A4 | A6/B | Other | All |
|---|---|---|---|---|---|---|---|---|
| N (%) | 65 (30.5) | 36 (16.9) | 23 (10.8) | 15 (7) | 14 (6.6) | 8 (3.8) | 52 (24.4) | 213 (100) |
| Age, Year, Median (IQR) | 37 (30–42) | 35 (26–47) | 47 (38–54) | 40 (32–45) | 28 (24–43) | 28 (23–51) | 41 (35–50) | 38 (30–47) |
| Male, N (%) | 45 (69.2) | 31 (86.1) | 22 (95.7) | 12 (80) | 14 (100) | 8 (100) | 43 (82.7) | 175 (82.5) |
| Place of Birth | ||||||||
| SSA | 14/63 (22.2) | 2/23 (8.7) | 2/14 (14.3) | 5/50 (10) | 23 (11) | |||
| EEU/CA | 10/63 (15.9) | 3/36 (8.3) | 2/23 (8.7) | 1/15 (6.7) | 5/14 (35.7) | 1/8 (12.5) | 9/50 (18) | 31 (14.8) |
| WCEU/NA | 6/63 (9.5) | 1/36 (2.8) | 1/23 (4.3) | 1/50 (2) | 9 (4.3) | |||
| Israel | 28/63 (44.4) | 27/36 (75) | 15/23 (65.2) | 10/15 (66.7) | 6/14 (42.9) | 7/8 (87.5) | 34/50 (68) | 127 (60.8) |
| Other | 5/63 (7.9) | 5/36 (13.9) | 3/23 (13) | 4/15 (26.7) | 1/14 (7.1) | 1/50 (2) | 19 (9.1) | |
| Transmission Groups | ||||||||
| MSM | 29/63 (46) | 19/34 (55.9) | 18/23 (78.3) | 10/13 (76.9) | 11/14 (78.6) | 7/8 (87.5) | 26/50 (52) | 120 (58.5) |
| HETERO | 28/63 (44.4) | 14/34 (41.2) | 5/23 (21.7) | 2/13 (15.4) | 3/14 (21.4) | 1/8 (12.5) | 20/50 (40) | 73 (35.6) |
| IDU | 6/63 (9.5) | 1/34 (2.9) | 1/13 (7.7) | 4/50 (8) | 12 (5.9) | |||
| CD4, n/N(%) | 55/65 (28.4) | 35/36 (18) | 23/23 (11.9) | 15/15 (7.7) | 13/14 (6.7) | 8/8 (4.1) | 45/52 (23.2) | 194 (91.1) |
| CD4(cells/mm3), Median (IQR) | 383 (216–515) | 373 (204–609) | 418 (216–514) | 408 (243–473) | 497 (415–739) | 343 (287–749) | 350 (161–451) | 389 (214–511) |
| HIV-1 VL n/N (%) | 61/65 (29.8) | 36/36 (17.6) | 23/23 (11.2) | 15/15 (7.3) | 13/14 (6.3) | 8/8 (3.9) | 49/52 (23.9) | 205 (96.2) |
| HIV-1 VL, Median (IQR) | 4.9 (2.2–5.4) | 4.9 (4.3–5.3) | 4.9 (4.5–5.3) | 5.5 (3.8–6.1) | 4.5 (4–5.2) | 4.5 (4–4.9) | 4.9 (4.6–5.5) | 4.9 (4.3–5.4) |
| Subtypes (N = 808) | Recombinant Forms (N = 120) | p-Value | |
|---|---|---|---|
| Age at diagnosis (years) | N = 802 (87) | N = 120 (13) | 0.936 |
| median (IQR) (n = 922) | 38 (30–45) | 39 (29–45) | |
| Sex, (n = 923, 99.5%) | N = 803 (87) | N = 120 (13) | 0.038 |
| Female | 196 (24.4) | 19 (15.8) | |
| Male | 607 (75.6) | 101 (84.2) | |
| Place of birth, (n = 893, 96.2%) | N = 775 (86.8) | N = 118 (13.2) | |
| SSA | 125 (16.1) | 14 (11.9) | |
| EEU/CA | 213 (27.5) | 17 (14.4) | |
| WCE/NA | 25 (3.2) | 3 (2.5) | |
| Israel | 389 (50.2) | 78 (66.1) | 0.023 |
| LA | 7 (0.9) | 2 (1.7) | |
| AP | 7 (0.9) | 2 (1.7) | |
| ME/NA | 9 (1.2) | 2 (1.7) | |
| Transmission Groups, (n = 899, 96.9%) | N = 782 (87) | N = 117 (13) | |
| MSM | 378 (48.3) | 72 (61.5) | |
| Hetero | 288 (36.8) | 37 (31.6) | 0.011 |
| IDU | 116 (14.8) | 8 (6.8) | |
| CD4 (cells/mm3), (N = 849, 91.5%) | N = 741 (87.3) | N = 108 (12.7) | |
| median CD4 (IQR) | 323 (175–519) | 418 (209–509) | 0.043 |
| HIV-1—RNA (Log copies/mL), | N = 772 (87.2) | N = 113 (12.8) | |
| (N = 885, 95.4) | |||
| median viral load (IQR) | 4.8 (4.2–5.4) | 4.9 (4.3–5.5) | 0.303 |
| TDRM Bennet (N = 906, 97.6%)TDRM, n (%) | N = 786 (86.8) | N = 120 (13.2) | |
| 112 (14.2) | 10 (8.3) | 0.085 | |
| HIVdrm ECDC (N = 906, 97.6%)HIVdrm, n (%) | N = 786 (86.8) | N = 120 (13.2) | |
| 191 (24.3) | 23 (19.2) | 0.249 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wagner, T.; Zuckerman, N.S.; Wax, M.; Shirazi, R.; Gozlan, Y.; Girshengorn, S.; Marom, R.; Mendelson, E.; Turner, D.; Mor, O. HIV-1 Circulating Recombinant Forms (CRFs) and Unique Recombinant Forms (URFs) in Israel, 2010–2018. Viruses 2022, 14, 1970. https://doi.org/10.3390/v14091970
Wagner T, Zuckerman NS, Wax M, Shirazi R, Gozlan Y, Girshengorn S, Marom R, Mendelson E, Turner D, Mor O. HIV-1 Circulating Recombinant Forms (CRFs) and Unique Recombinant Forms (URFs) in Israel, 2010–2018. Viruses. 2022; 14(9):1970. https://doi.org/10.3390/v14091970
Chicago/Turabian StyleWagner, Tali, Neta S. Zuckerman, Marina Wax, Rachel Shirazi, Yael Gozlan, Shirley Girshengorn, Rotem Marom, Ella Mendelson, Dan Turner, and Orna Mor. 2022. "HIV-1 Circulating Recombinant Forms (CRFs) and Unique Recombinant Forms (URFs) in Israel, 2010–2018" Viruses 14, no. 9: 1970. https://doi.org/10.3390/v14091970
APA StyleWagner, T., Zuckerman, N. S., Wax, M., Shirazi, R., Gozlan, Y., Girshengorn, S., Marom, R., Mendelson, E., Turner, D., & Mor, O. (2022). HIV-1 Circulating Recombinant Forms (CRFs) and Unique Recombinant Forms (URFs) in Israel, 2010–2018. Viruses, 14(9), 1970. https://doi.org/10.3390/v14091970







