Hypovirulence of Colletotrichum gloesporioides Associated with dsRNA Mycovirus Isolated from a Mango Orchard in Thailand
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Isolates, Growth Conditions, and Taxonomic Analysis
2.2. Extraction of dsRNA from Fungal Mycelium
2.3. Full-Length Analysis by Next-Generation Sequencing (NGS) Approach and RACE
2.4. Bioinformatic and Phylogenetic Analysis
2.5. Elimination of C. gloeosporioides Strain Ssa-44.1 dsRNA
2.6. Virus Transmission
Horizontal Transmission
2.7. Effect of Mycovirus on Fungal Colony Morphology, Growth, and Pathogenicity
2.8. Statistical Analyses
3. Results
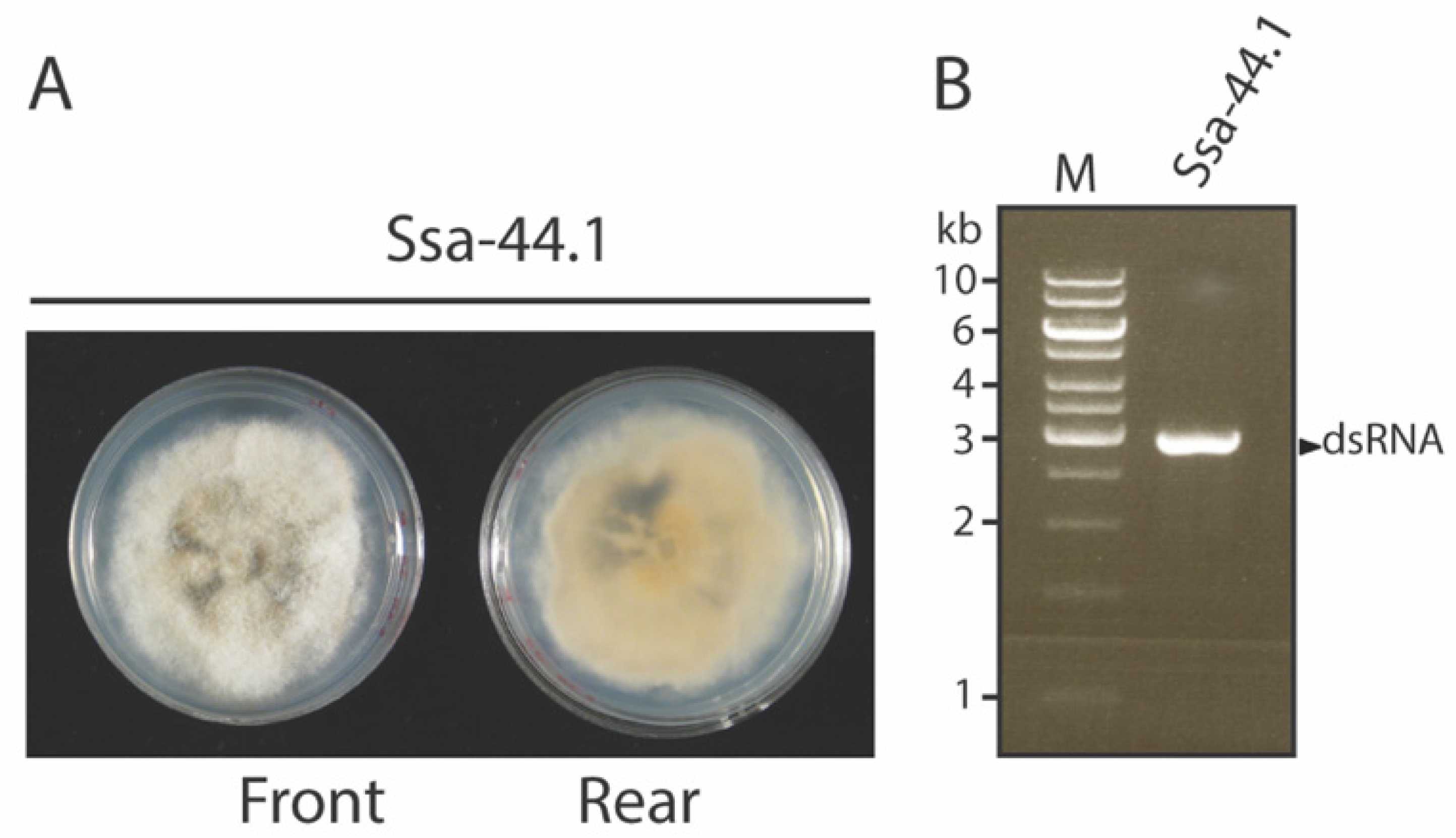
3.1. Identification and Biological Characteristic of Virus-Infected Isolate Ssa-44.1
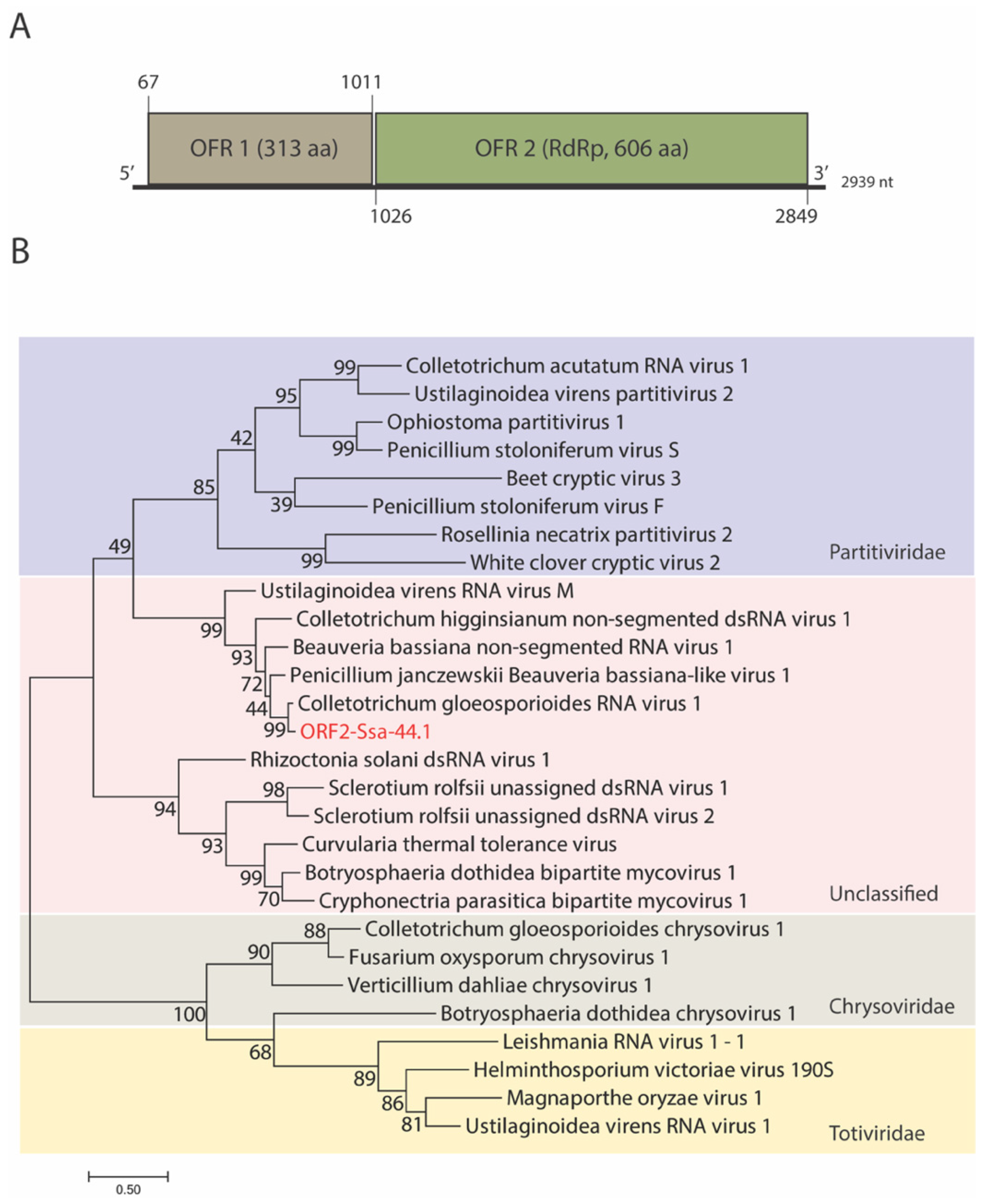
3.2. Genome Organization and Phylogenetic Analysis of Mycovirus Ssa-44.1
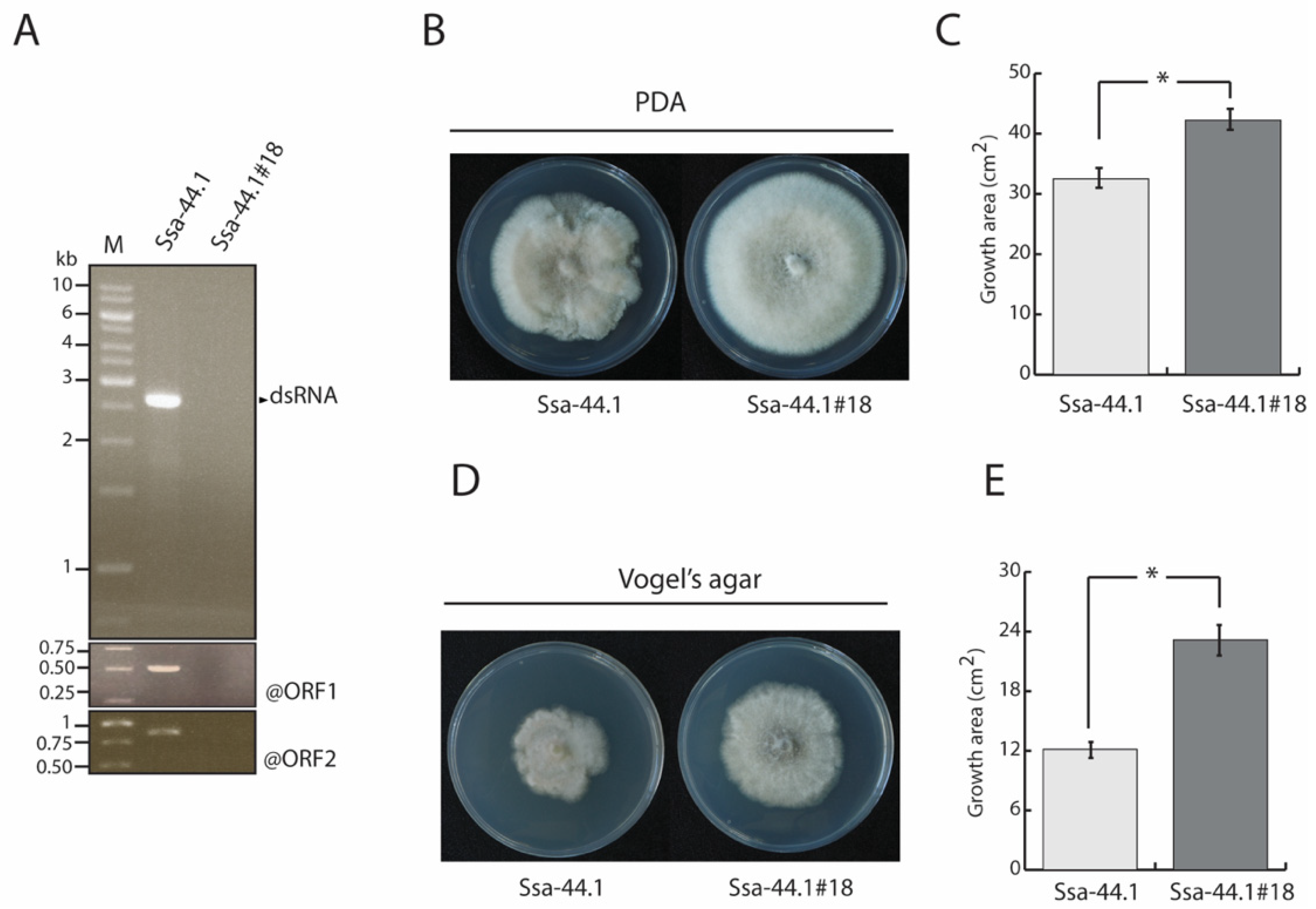
3.3. Biological Properties of CgRV1-Ssa-44.1 on C. gloeosporioides
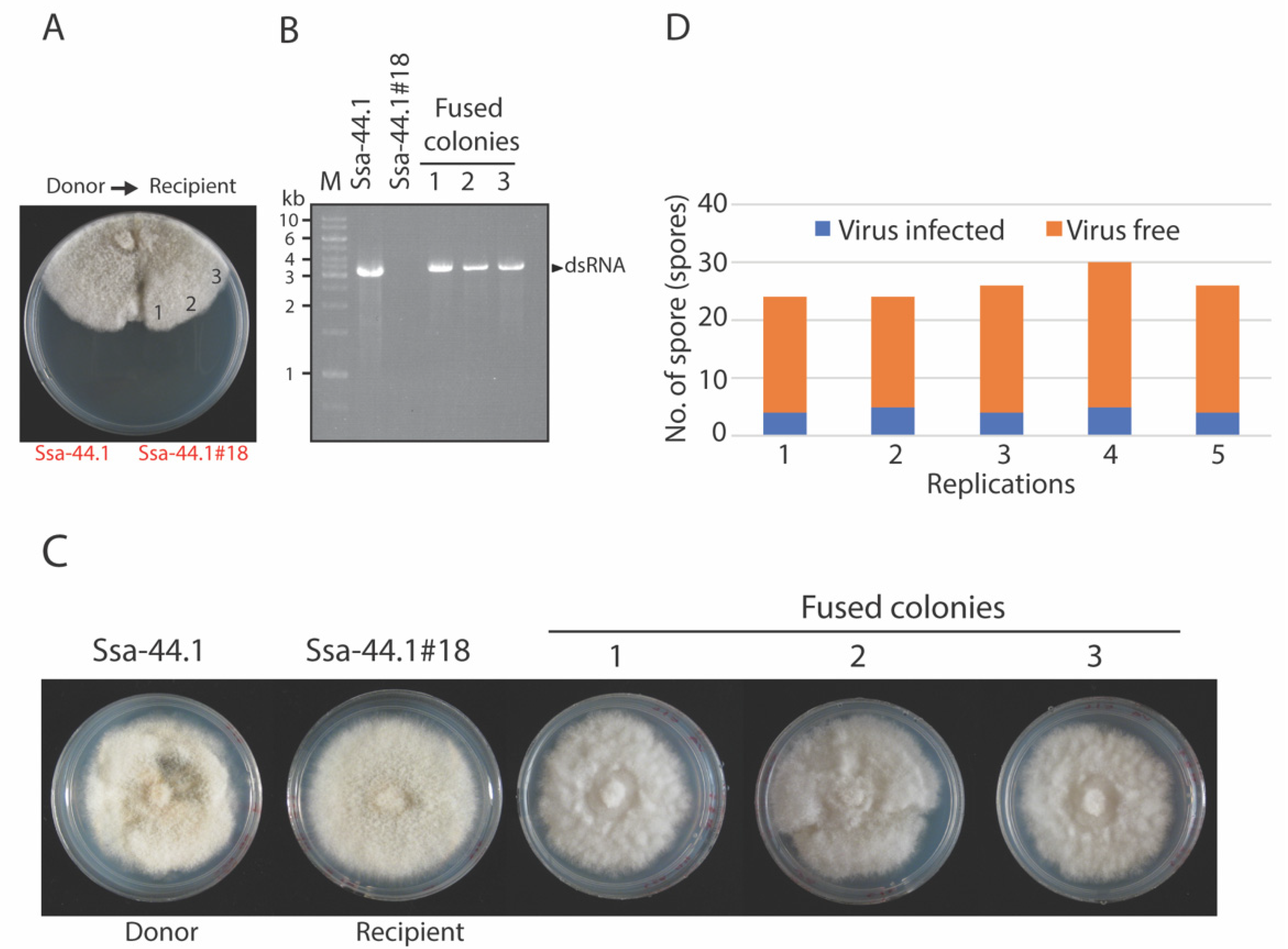
3.4. Transmission of CgRV1-Ssa-44.1 in C. gloeosporioides
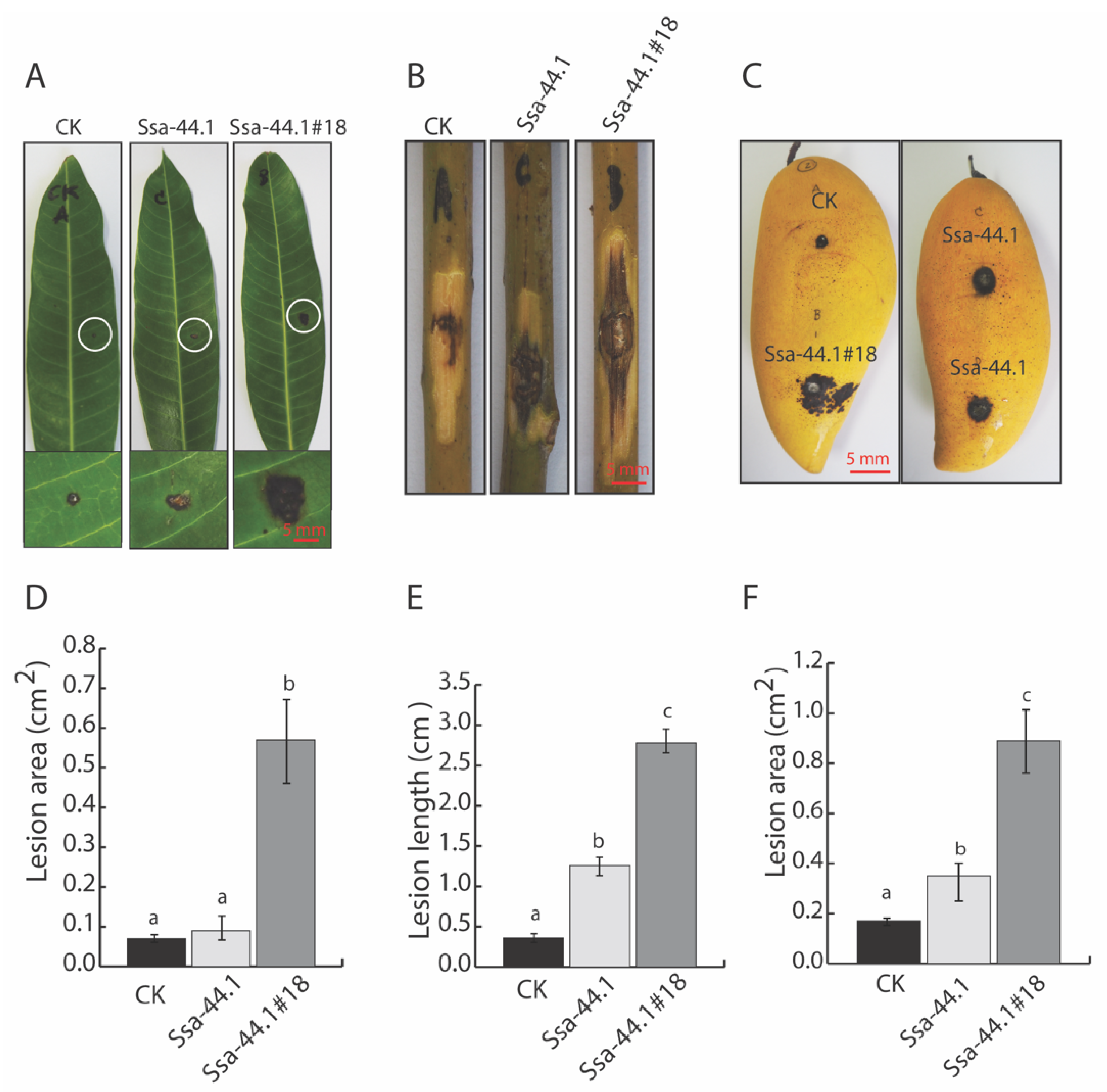
3.5. Effect of CgRV1-Ssa-44.1 on Virulence of C. gloeosporioides
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arauz, L.F. Mango Anthracnose: Economic Impact and Current Options for Integrated Management. Plant Dis. 2000, 84, 600–611. [Google Scholar] [CrossRef] [PubMed]
- Cannon, P.F.; Damm, U.; Johnston, P.R.; Weir, B.S. Colletotrichum—Current status and future directions. Stud. Mycol. 2012, 73, 181–213. [Google Scholar] [CrossRef] [PubMed]
- Gautam, A.K. The Genera Colletotrichum: An Incitant of Numerous New Plant Diseases in India. J. New Biol. Rep. 2014, 3, 9–21. [Google Scholar]
- Food and Agriculture Organization of the United Nations. Major Tropical Fruits: Preliminary Results 2020. Available online: http://www.fao.org/economic/est/est-commodities/tropical-fruits/en/ (accessed on 25 May 2022).
- Paull, R.E.; Duarte, O. Tropical Fruits, 2nd ed.; CABI Publishing: London, UK, 2011; Volume 1, pp. 252–290. [Google Scholar]
- Weir, B.S.; Johnston, P.R.; Damm, U. The Colletotrichum gloeosporioides Species Complex. Stud. Mycol. 2012, 73, 115–180. [Google Scholar] [CrossRef]
- Vidyalakshni, A.; Divya, C.V. New report of Colletotrichum gloeosporioides causing anthracnose of Pisonia alba in India. Arch. Phytopathol. Plant. Prot. 2013, 46, 201–204. [Google Scholar] [CrossRef]
- da Silva, L.L.; Moreno, H.L.A.; Correia, H.L.N.; Santana, M.F.; de Queiroz, M.V. Colletotrichum: Species Complexes, Lifestyle and Peculiarities of Some Sources of Genetic Variability. Appl. Microbiol. Biotechnol. 2020, 104, 1891–1904. [Google Scholar] [CrossRef]
- Yakoby, N.; Beno-Moualem, D.; Keen, N.T.; Dinoor, A.; Pines, O.; Prusky, D. Colletotrichum gloeosporioides PelB Is an Important Virulence Factor in Avocado Fruit-Fungus Interaction. Mol. Plant Microbe Interact. 2001, 14, 988–995. [Google Scholar] [CrossRef]
- Chudasama, K.S.; Monpara, J.K.; Thaker, V.S. Identification and Characterization of Pectin Lyase Gene as a Virulence Factor in Colletotrichum gloeosporioides. Physiol. Mol. Plant. Pathol. 2021, 116, 101706. [Google Scholar] [CrossRef]
- Liang, C.; Zhang, B.; Zhou, Y.; Yin, H.; An, B.; Lin, D.; He, C.; Luo, H. CgNPG1 as a Novel Pathogenic Gene of Colletotrichum Gloeosporioides from Hevea Brasiliensis in Mycelial Growth, Conidiation and the Invasive Structures Development. Front. Microbiol. 2021, 12, 629387. [Google Scholar] [CrossRef]
- Jakhar, M.S.; Pathak, S. Effect of Pre-harvest Nutrients Application and Bagging on Quality and Shelf Life of Mango (Mangifera indica L.). J. Agric. Sci. Technol. 2016, 18, 717–729. [Google Scholar]
- Jamalizadeh, M.; Etebarian, H.R.; Aminia, H.; Alizadeh, A. A review of mechanisms of action of biological control organisms against post-harvest fruit spoilage. Bull. OEPP 2011, 41, 65–71. [Google Scholar] [CrossRef]
- Droby, S.; Wisniewski, M.; Macarisin, D.; Wilson, C. Twenty years of postharvest biocontrol research: Is it time for a new paradigm? Postharvest Biol. Technol. 2009, 5, 137–145. [Google Scholar] [CrossRef]
- Tariq, M.; Khan, A.; Asif, M.; Khan, F.; Ansari, T.; Shariq, M.; Siddiqui, M.A. Biological control: A sustainable and practical approach for plant disease management, Acta Agric. Scand. B Soil Plant Sci. 2020, 70, 507–524. [Google Scholar] [CrossRef]
- Anagnostakis, S.L. Biological Control of Chestnut Blight. Science 1982, 215, 466–471. [Google Scholar] [CrossRef]
- Heiniger, U.; Rigling, D. Biological control of chestnut blight in Europe. Annu. Rev. Phytopathol. 1994, 32, 581–599. [Google Scholar] [CrossRef]
- Chiba, S.; Salaipeth, L.; Lin, Y.S.; Sasaki, A.; Kanematsu, S.; Suzuki, N. A Novel Bipartite Double-Stranded RNA Mycovirus from the White Root Rot Fungus Rosellinia necatrix: Molecular and Biological Characterization, Taxonomic Considerations and Potential for Biological Control. J. Virol. 2009, 83, 12801–12812. [Google Scholar] [CrossRef]
- Yu, X.; Li, B.; Fu, Y.; Xie, J.; Cheng, J.; Ghabrial, S.A.; Li, G.; Yi, X.; Jiang, D. Extracellular transmission of a DNA mycovirus and its use as a natural fungicide. Proc. Natl. Acad. Sci. USA 2013, 110, 1452–1457. [Google Scholar] [CrossRef]
- Wang, X.; Lai, J.; Hu, H.; Yang, J.; Zang, K.; Zhao, F.; Zeng, G.; Liao, Q.; Gu, Z.; Du, Z. Infection of Nigrospora nonsegmented RNA Virus 1 Has Important Biological Impacts on a Fungal Host. Viruses 2022, 14, 795. [Google Scholar] [CrossRef]
- Kotta-Loizou, I.; Coutts, R.H.A. Mycoviruses in Aspergilli: A Comprehensive Review. Front. Microbiol. 2017, 8, 1699. [Google Scholar] [CrossRef]
- Ghabrial, S.A.; Suzuki, N. Viruses of plant pathogenic fungi. Annu. Rev. Phytopathol. 2009, 47, 353–384. [Google Scholar] [CrossRef]
- Lemus-Minor, C.G.; Cañizares, M.C.; García-Pedrajas, M.D.; Pérez-Artés, E. Horizontal and Vertical Transmission of the Hypovirulence-Associated Mycovirus Fusarium Oxysporum f. Sp. Dianthi Virus 1. Eur. J. Plant Pathol. 2019, 153, 645–650. [Google Scholar] [CrossRef]
- Pearson, M.N.; Beever, R.E.; Boine, B.; Arthur, K. Mycoviruses of Filamentous Fungi and Their Relevance to Plant Pathology. Mol. Plant. Pathol. 2009, 10, 115–128. [Google Scholar] [CrossRef] [PubMed]
- Figueirêdo, L.C.; Figueirêdo, G.S.; Giancoli, Á.C.H.; Tanaka, F.A.O.; Silva, L.A.O.; Kitajima, E.W.; Filho, S.A.; Azevedo, J.L. Detection of isometric, dsRNA-containing viral particles in Colletotrichum gloeosporioides isolated from cashew tree. Trop. Plant Pathol. 2012, 37, 142–145. [Google Scholar] [CrossRef]
- Zhong, J.; Pang, X.D.; Zhu, H.J.; Gao, B.D.; Huang, W.K.; Zhou, Q. Molecular Characterization of a Trisegmented Mycovirus from the Plant Pathogenic Fungus Colletotrichum gloeosporioides. Viruses 2016, 8, 268. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Zhu, J.Z.; Zhou, X.Y.; Zhong, J.; Li, C.H.; Zhang, Z.G.; Zhu, H.J. A novel ourmia-like mycovirus isolated from the plant pathogenic fungus Colletotrichum gloeosporioides. Arch. Virol. 2019, 164, 2631–2635. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, S.; Zhu, H.J.; Zhong, J. Molecular characterization of a novel mycovirus from the plant pathogenic fungus Colletotrichum gloeosporioides. Arch. Virol. 2019, 164, 2859–2863. [Google Scholar] [CrossRef] [PubMed]
- Balijja, A.; Kvarnheden, A.; Turchetti, T. A non-phenol–chloroform extraction of double-stranded RNA from plant and fungal tissues. J. Virol. Methods 2008, 152, 32–37. [Google Scholar] [CrossRef]
- Chiba, S.; Lin, Y.H.; Kondo, H.; Kanematsu, S.; Suzuki, N. Effects of defective interfering RNA on symptom induction by and replication of, a novel partitivirus from a phytopathogenic fungus, Rosellinia necatrix. J. Virol. 2013, 87, 2330–2341. [Google Scholar] [CrossRef]
- Eusebio-Cope, A.; Sun, L.; Hillman, B.I.; Suzuki, N. Mycoreovirus 1 S4-coded protein is dispensable for viral replication but necessary for efficient vertical transmission and normal symptom induction. Virology 2010, 397, 399–408. [Google Scholar] [CrossRef]
- Vogel, H.J. A Convenient Growth Medium for Neurospora crassa. Microbiol. Genet. Bull. 1956, 13, 42–47. [Google Scholar]
- Yang, S.; Dai, R.; Salaipeth, L.; Huang, L.; Liu, J.; Andika, I.B.; Sun, L. Infection of Two Heterologous Mycoviruses Reduces the Virulence of Valsa mali, a Fungal Agent of Apple Valsa Canker Disease. Front. Microbiol. 2021, 12, 659210. [Google Scholar] [PubMed]
- Gilbert, K.B.; Holcomb, E.E.; Allscheid, R.L.; Carrington, J.C. Hiding in plain sight: New virus genomes discovered via a systematic analysis of fungal public transcriptomes. PLoS ONE 2019, 14, e0219207. [Google Scholar]
- Nerva, L.; Ciuffo, M.; Vallino, M.; Margaria, P.; Varese, G.C.; Gnavi, G.; Turina, M. Multiple approaches for the detection and characterization of viral and plasmid symbionts from a collection of marine fungi. Virus Res. 2016, 219, 22–38. [Google Scholar] [PubMed]
- Campo, S.; Gilbert, K.B.; Carrington, J.C. Small RNA-Based Antiviral Defense in the Phytopathogenic Fungus Colletotrichum higginsianum. PLoS Pathog. 2016, 12, e1005640. [Google Scholar]
- Jiang, Y.; Zhang, T.; Luo, C.; Jiang, D.; Li, G.; Li, Q.; Hsiang, T.; Huang, J. Prevalence and diversity of mycoviruses infecting the plant pathogen Ustilaginoidea virens. Virus Res. 2015, 195, 47–56. [Google Scholar]
- Zhang, L.; Yan, C.; Guo, Q.; Zhang, J.; Ruiz-Menjivar, J. The impact of agricultural chemical inputs on environment: Global evidence from informetrics analysis and visualization. Int. J. Low Carbon Technol. 2018, 13, 338–352. [Google Scholar] [CrossRef]
- Neang, S.; Bincader, S.; Rangsuwan, S.; Keawmanee, P.; Rin, S.; Salaipeth, L.; Das, S.; Kondo, H.; Suzuki, N.; Sato, I.; et al. Omnipresence of Partitiviruses in Rice Aggregate Sheath Spot Symptom-Associated Fungal Isolates from Paddies in Thailand. Viruses 2021, 13, 2269. [Google Scholar] [CrossRef]
- van Diepeningen, A.D.; Debets, A.J.; Hoekstra, R.F. Dynamics of dsRNA mycoviruses in black Aspergillus populations. Fungal Genet. Biol. 2006, 43, 446–452. [Google Scholar] [CrossRef]
- Chu, Y.M.; Lim, W.S.; Yea, S.J.; Cho, J.D.; Lee, Y.W.; Kim, K.H. Complexity of dsRNA mycovirus isolated from Fusarium graminearum. Virus Genes 2004, 28, 135–143. [Google Scholar] [CrossRef]
- Nuss, D. Hypovirulence: Mycoviruses at the fungal–plant interface. Nat. Rev. Microbiol. 2005, 3, 632–642. [Google Scholar] [CrossRef]
- Shafik, K.; Umer, M.; You, H.; Aboushedida, H.; Wang, Z.; Ni, D.; Xu, W. Characterization of a Novel Mitovirus Infecting Melanconiella theae Isolated from Tea Plants. Front. Microbiol. 2021, 12, 757556. [Google Scholar] [CrossRef]
- Wang, M.; Wang, Y.; Sun, X.; Cheng, J.; Fu, Y.; Liu, H.; Jiang, D.; Ghabrial, S.A.; Xie, J. Characterization of a Novel Megabirnavirus from Sclerotinia sclerotiorum Reveals Horizontal Gene Transfer from Single-Stranded RNA Virus to Double-Stranded RNA Virus. J. Virol. 2015, 89, 8567–8579. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, Y.; Wan, X.; She, Y.; Li, M.; Xi, H.; Xie, J.; Wen, C. A Novel Ourmia-Like Mycovirus Confers Hypovirulence-Associated Traits on Fusarium oxysporum. Front. Microbiol. 2020, 11, 569869. [Google Scholar] [CrossRef] [PubMed]
- Glass, N.L.; Jacobson, D.J.; Shiu, P.K. The Genetics of Hyphal Fusion and Vegetative Incompatibility in Filamentous Ascomycete Fungi. Annu. Rev. Genet. 2000, 34, 165–186. [Google Scholar] [CrossRef] [PubMed]
- Garnjobst, L.; Wilson, J.F. Heterocaryosis and Protoplasmic Incompability in Neurospora Crassa. Proc. Natl. Acad. Sci. USA 1956, 42, 613–618. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.M.; Cho, W.K.; Yu, J.; Son, M.; Choi, H.; Min, K. A comparison of transcriptional patterns and mycological phenotypes following infection of Fusarium graminearum by four mycoviruses. PLoS ONE 2014, 9, e100989. [Google Scholar] [CrossRef]
- Suzuki, N.; Maruyama, K.; Moriyama, M.; Nuss, D.L. Hypovirus papain-like protease p29 functions in trans to enhance viral double-stranded RNA accumulation and vertical transmission. J. Virol. 2003, 77, 11697–11707. [Google Scholar] [CrossRef][Green Version]
- Jia, H.; Dong, K.; Zhou, L.; Wang, G.; Hong, N.; Jiang, D.; Xu, W. A dsRNA virus with filamentous viral particles. Nat. Commun. 2017, 8, 168. [Google Scholar] [CrossRef]
- Zhou, L.; Li, X.; Kotta-Loizou, I.; Dong, K.; Li, S.; Ni, D.; Hong, N.; Wang, G.; Xu, W. A mycovirus modulates the endophytic and pathogenic traits of a plant associated fungus. ISME J. 2021, 15, 1893–1906. [Google Scholar] [CrossRef]
| Strains | Spore Counting (Spores/mL) | |||
|---|---|---|---|---|
| 1 | 2 | 3 | Mean ± SD * | |
| Ssa-44.1 | 2.58 × 107 | 1.85 × 107 | 2.38 × 107 | 2.27 ± 0.38 × 107 a |
| Ssa-44.1#18 | 4.25 × 108 | 5.39 × 108 | 4.95 × 108 | 4.86 ± 0.57 × 108 b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suharto, A.R.; Jirakkakul, J.; Eusebio-Cope, A.; Salaipeth, L. Hypovirulence of Colletotrichum gloesporioides Associated with dsRNA Mycovirus Isolated from a Mango Orchard in Thailand. Viruses 2022, 14, 1921. https://doi.org/10.3390/v14091921
Suharto AR, Jirakkakul J, Eusebio-Cope A, Salaipeth L. Hypovirulence of Colletotrichum gloesporioides Associated with dsRNA Mycovirus Isolated from a Mango Orchard in Thailand. Viruses. 2022; 14(9):1921. https://doi.org/10.3390/v14091921
Chicago/Turabian StyleSuharto, Aditya R., Jiraporn Jirakkakul, Ana Eusebio-Cope, and Lakha Salaipeth. 2022. "Hypovirulence of Colletotrichum gloesporioides Associated with dsRNA Mycovirus Isolated from a Mango Orchard in Thailand" Viruses 14, no. 9: 1921. https://doi.org/10.3390/v14091921
APA StyleSuharto, A. R., Jirakkakul, J., Eusebio-Cope, A., & Salaipeth, L. (2022). Hypovirulence of Colletotrichum gloesporioides Associated with dsRNA Mycovirus Isolated from a Mango Orchard in Thailand. Viruses, 14(9), 1921. https://doi.org/10.3390/v14091921





