A TaqMan Probe-Based Multiplex Real-Time PCR for Simultaneous Detection of Porcine Epidemic Diarrhea Virus Subtypes G1 and G2, and Porcine Rotavirus Groups A and C
Abstract
:1. Introduction
2. Materials and Methods
2.1. Design of the Primers and Probes
2.2. Development and Optimization of the Multiplex Real-Time PCR
2.3. Verification of the Multiplex Real-Time PCR
3. Results
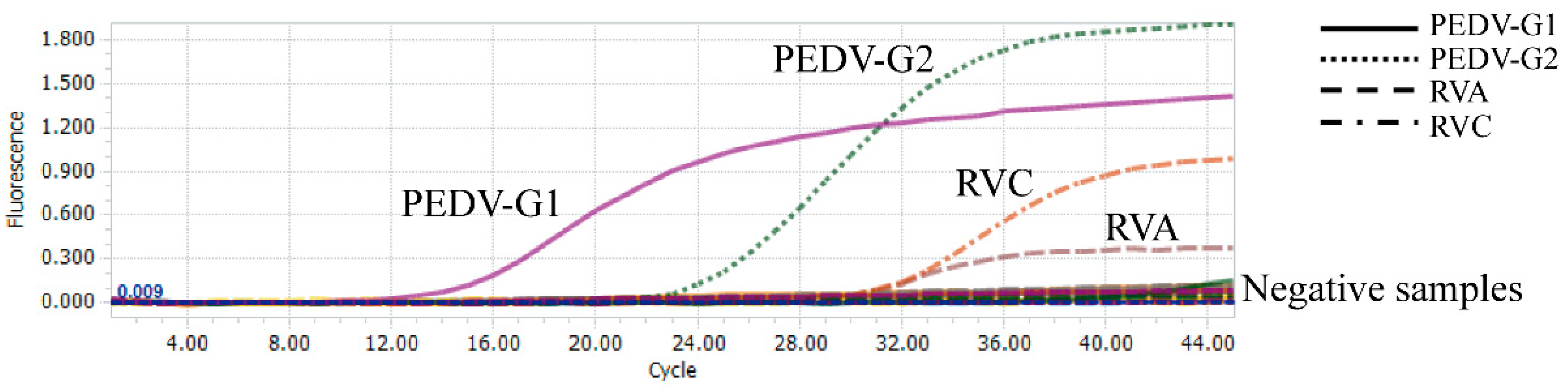
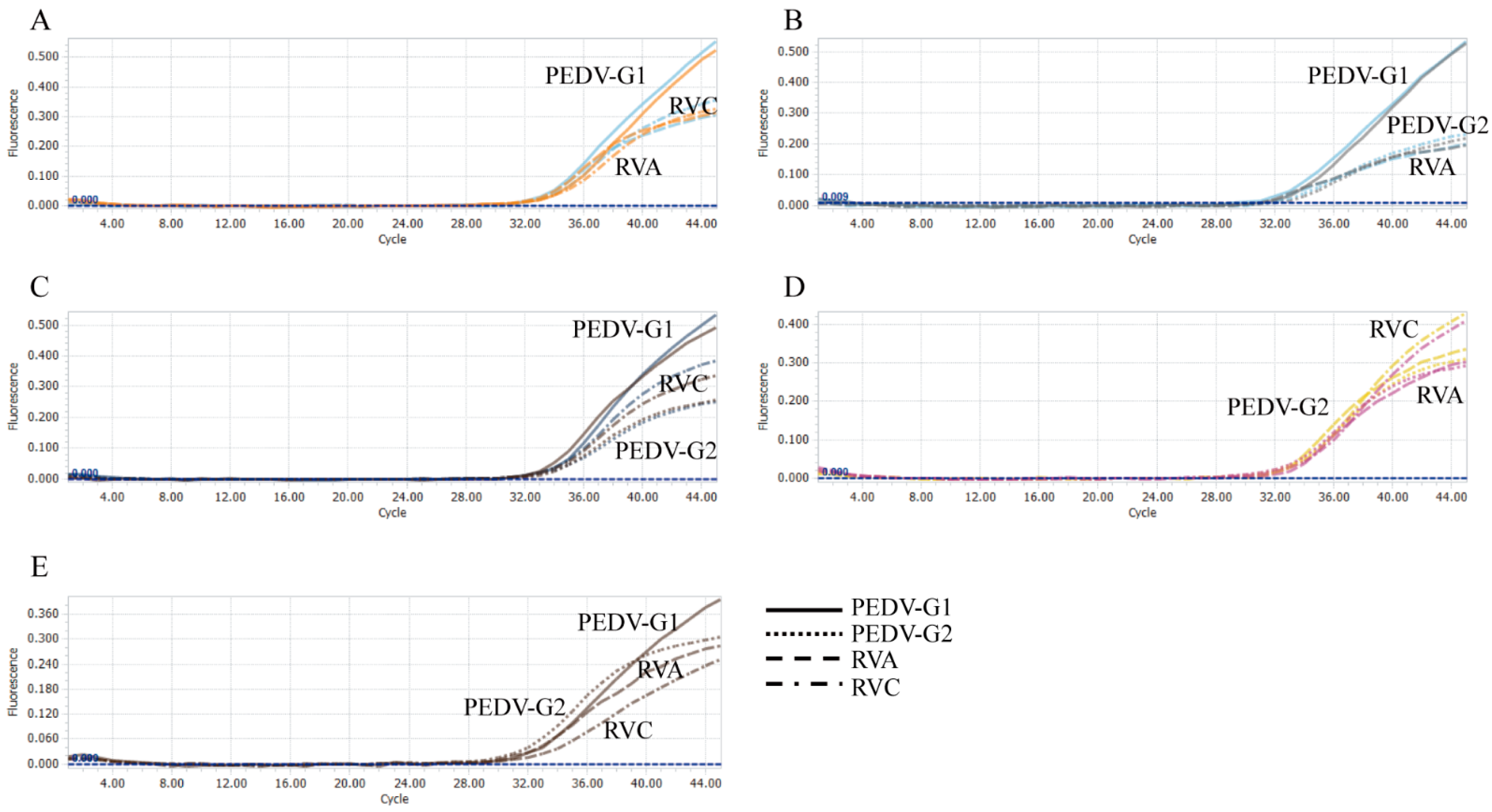
3.1. Evaluation of Sensitivity and Cut-Off Values
3.2. Evaluation of the Specificity and Reproducibility
3.3. Evaluation of the Simulated Co-Infections
3.4. Evaluation Using the Clinical Samples
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Li, Z.; Ma, Z.; Li, Y.; Gao, S.; Xiao, S. Porcine epidemic diarrhea virus: Molecular mechanisms of attenuation and vaccines. Microb. Pathog. 2020, 149, 104553. [Google Scholar] [CrossRef]
- Park, J.; Lee, C. Emergence and evolution of novel G2b-like porcine epidemic diarrhea virus inter-subgroup G1b recombinants. Arch. Virol. 2020, 165, 2471–2478. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.; Saif, L.J.; Wang, Q. Porcine epidemic diarrhea virus (PEDV): An update on etiology, transmission, pathogenesis, and prevention and control. Virus Res. 2020, 286, 198045. [Google Scholar] [CrossRef]
- Yu, J.; Chai, X.; Cheng, Y.; Xing, G.; Liao, A.; Du, L.; Wang, Y.; Lei, J.; Gu, J.; Zhou, J. Molecular characteristics of the spike gene of porcine epidemic diarrhoea virus strains in Eastern China in 2016. Virus Res. 2018, 247, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Li, Y.; Liu, Y.; Chen, Y.; Jiao, W.; Feng, H.; Wei, Q.; Wang, J.; Zhang, Y.; Zhang, G. Isolation and Identification of a Recombinant Porcine Epidemic Diarrhea Virus With a Novel Insertion in S1 Domain. Front. Microbiol. 2021, 12, 667084. [Google Scholar] [CrossRef]
- Ji, Z.; Shi, D.; Shi, H.; Wang, X.; Chen, J.; Liu, J.; Ye, D.; Liu, Q.; Fan, Q.; Li, M.; et al. A porcine epidemic diarrhea virus strain with distinct characteristics of four amino acid insertion in the COE region of spike protein. Vet. Microbiol. 2020, 253, 108955. [Google Scholar] [CrossRef]
- Lee, C. Porcine epidemic diarrhea virus: An emerging and re-emerging epizootic swine virus. Virol. J. 2015, 12, 193. [Google Scholar] [CrossRef]
- He, W.-T.; Bollen, N.; Xu, Y.; Zhao, J.; Dellicour, S.; Yan, Z.; Gong, W.; Zhang, C.; Zhang, L.; Lu, M.; et al. Phylogeography Reveals Association between Swine Trade and the Spread of Porcine Epidemic Diarrhea Virus in China and across the World. Mol. Biol. Evol. 2021, 39, msab364. [Google Scholar] [CrossRef]
- Tian, J.; Sun, J.; Li, D.; Wang, N.; Wang, L.; Zhang, C.; Meng, X.; Ji, X.; Suchard, M.A.; Zhang, X.; et al. Emerging viruses: Cross-species transmission of coronaviruses, filoviruses, henipaviruses, and rotaviruses from bats. Cell Rep. 2022, 39, 110969. [Google Scholar] [CrossRef]
- Banyai, K.; Kemenesi, G.; Budinski, I.; Foldes, F.; Zana, B.; Marton, S.; Varga-Kugler, R.; Oldal, M.; Kurucz, K.; Jakab, F. Candidate new rotavirus species in Schreiber’s bats, Serbia. Infect. Genet. Evol. 2017, 48, 19–26. [Google Scholar] [CrossRef]
- Mihalov-Kovács, E.; Gellért, Á.; Marton, S.; Farkas, S.L.; Fehér, E.; Oldal, M.; Jakab, F.; Martella, V.; Bányai, K. Candidate New Rotavirus Species in Sheltered Dogs, Hungary. Emerg. Infect. Dis. 2015, 21, 660–663. [Google Scholar] [CrossRef] [PubMed]
- Matthijnssens, J.; Otto, P.H.; Ciarlet, M.; Desselberger, U.; Van Ranst, M.; Johne, R. VP6-sequence-based cutoff values as a criterion for rotavirus species demarcation. Arch. Virol. 2012, 157, 1177–1182. [Google Scholar] [CrossRef]
- Hull, J.J.A.; Qi, M.; Montmayeur, A.M.; Kumar, D.; Velasquez, D.E.; Moon, S.-S.; Magaña, L.C.; Betrapally, N.; Ng, T.F.F.; Jiang, B.; et al. Metagenomic sequencing generates the whole genomes of porcine rotavirus A, C, and H from the United States. PLoS ONE 2020, 15, e0244498. [Google Scholar] [CrossRef] [PubMed]
- Martella, V.; Bányai, K.; Ciarlet, M.; Iturriza-Gómara, M.; Lorusso, E.; De Grazia, S.; Arista, S.; Decaro, N.; Elia, G.; Cavalli, A.; et al. Relationships among porcine and human P [6] rotaviruses: Evidence that the different human P [6] lineages have originated from multiple interspecies transmission events. Virology 2006, 344, 509–519. [Google Scholar] [CrossRef]
- Mukherjee, A.; Dutta, D.; Ghosh, S.; Bagchi, P.; Chattopadhyay, S.; Nagashima, S.; Kobayashi, N.; Dutta, P.; Krishnan, T.; Naik, T.N.; et al. Full genomic analysis of a human group A rotavirus G9P [6] strain from Eastern India provides evidence for porcine-to-human interspecies transmission. Arch. Virol. 2009, 154, 733–746. [Google Scholar] [CrossRef] [PubMed]
- Martella, V.; Bányai, K.; Matthijnssens, J.; Buonavoglia, C.; Ciarlet, M. Zoonotic aspects of rotaviruses. Vet. Microbiol. 2010, 140, 246–255. [Google Scholar] [CrossRef]
- Costa, F.B.; Flores, P.S.; Amorim, A.R.; Mendes, G.D.S.; Santos, N. Porcine rotavirus C strains carrying human-like NSP4 and NSP5. Zoonoses Public Health 2020, 67, 849–861. [Google Scholar] [CrossRef]
- Kattoor, J.J.; Saurabh, S.; Malik, Y.S.; Sircar, S.; Dhama, K.; Ghosh, S.; Bányai, K.; Kobayashi, N.; Singh, R.K. Unexpected detection of porcine rotavirus C strains carrying human origin VP6 gene. Vet. Q. 2017, 37, 252–261. [Google Scholar] [CrossRef]
- Molinari, B.L.D.; Possatti, F.; Lorenzetti, E.; Alfieri, A.; Alfieri, A. Unusual outbreak of post-weaning porcine diarrhea caused by single and mixed infections of rotavirus groups A, B, C, and H. Vet. Microbiol. 2016, 193, 125–132. [Google Scholar] [CrossRef]
- Chepngeno, J.; Diaz, A.; Paim, F.C.; Saif, L.J.; Vlasova, A.N. Rotavirus C: Prevalence in suckling piglets and development of virus-like particles to assess the influence of maternal immunity on the disease development. Vet. Res. 2019, 50, 84. [Google Scholar] [CrossRef]
- He, W.-T.; Hou, X.; Zhao, J.; Sun, J.; He, H.; Si, W.; Wang, J.; Jiang, Z.; Yan, Z.; Xing, G.; et al. Virome characterization of game animals in China reveals a spectrum of emerging pathogens. Cell 2022, 185, 1117–1129.e8. [Google Scholar] [CrossRef] [PubMed]
- Su, S.; Wong, G.; Shi, W.; Liu, J.; Lai, A.C.K.; Zhou, J.; Liu, W.; Bi, Y.; Gao, G.F. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends Microbiol. 2016, 24, 490–502. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Hu, Y.; Liu, P.; Zhu, Z.; Liu, P.; Chen, C.; Wu, X. Development and application of a duplex real-time PCR assay for differentiation of genotypes I and II African swine fever viruses. Transbound. Emerg. Dis. 2022. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.; Lu, J.; Wang, N.; He, W.T.; Zhang, L.; Zhao, W.; Su, S. Development of a TaqMan-probe-based multiplex real-time PCR for the simultaneous detection of emerging and reemerging swine coronaviruses. Virulence 2020, 11, 707–718. [Google Scholar] [CrossRef]
- Liu, G.; Jiang, Y.; Opriessnig, T.; Gu, K.; Zhang, H.; Yang, Z. Detection and differentiation of five diarrhea related pig viruses utilizing a multiplex PCR assay. J. Virol. Methods 2018, 263, 32–37. [Google Scholar] [CrossRef]
- Vidal, A.; Martin-Valls, G.; Tello, M.; Mateu, E.; Martín, M.; Darwich, L. Prevalence of enteric pathogens in diarrheic and non-diarrheic samples from pig farms with neonatal diarrhea in the North East of Spain. Vet. Microbiol. 2019, 237, 108419. [Google Scholar] [CrossRef]
- Turlewicz-Podbielska, H.; Pomorska-Mól, M. Porcine Coronaviruses: Overview of the State of the Art. Virol. Sin. 2021, 36, 833–851. [Google Scholar] [CrossRef]
- Monteagudo, L.V.; Benito, A.A.; Lázaro-Gaspar, S.; Arnal, J.L.; Martin-Jurado, D.; Menjon, R.; Quílez, J. Occurrence of Rotavirus A Genotypes and Other Enteric Pathogens in Diarrheic Suckling Piglets from Spanish Swine Farms. Animals 2022, 12, 251. [Google Scholar] [CrossRef]
- Aguilera, E.R.; Pfeiffer, J.K. Strength in numbers: Mechanisms of viral co-infection. Virus Res. 2019, 265, 43–46. [Google Scholar] [CrossRef]
- Zhang, H.; Han, F.; Shu, X.; Li, Q.; Ding, Q.; Hao, C.; Yan, X.; Xu, M.; Hu, H. Co-infection of porcine epidemic diarrhoea virus and porcine deltacoronavirus enhances the disease severity in piglets. Transbound. Emerg. Dis. 2021, 69, 1715–1726. [Google Scholar] [CrossRef]
- Zheng, L.L.; Cui, J.T.; Han, H.Y.; Hou, H.L.; Wang, L.; Liu, F.; Chen, H.Y. Development of a duplex SYBR Green based real-time PCR assay for detection of porcine epidemic diarrhea virus and porcine bocavirus3/4/5. Mol. Cell Probes. 2020, 51, 101544. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Liu, X.; Shi, D.; Shi, H.; Zhang, X.; Li, C.; Chi, Y.; Feng, L. Detection and Molecular Diversity of Spike Gene of Porcine Epidemic Diarrhea Virus in China. Viruses 2013, 5, 2601–2613. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.-D.; Bai, J.; Jiang, P.; Tang, T.-S.; Li, Y.; Tan, C.; Shi, X. Development of a multiplex TaqMan probe-based real-time PCR for discrimination of variant and classical porcine epidemic diarrhea virus. J. Virol. Methods 2014, 206, 150–155. [Google Scholar] [CrossRef]
- Su, Y.; Liu, Y.; Chen, Y.; Xing, G.; Hao, H.; Wei, Q.; Liang, Y.; Xie, W.; Li, D.; Huang, H.; et al. A novel duplex TaqMan probe-based real-time RT-qPCR for detecting and differentiating classical and variant porcine epidemic diarrhea viruses. Mol. Cell Probes. 2018, 37, 6–11. [Google Scholar] [CrossRef]
- Marthaler, D.; Homwong, N.; Rossow, K.; Culhane, M.; Goyal, S.; Collins, J.; Matthijnssens, J.; Ciarlet, M. Rapid detection and high occurrence of porcine rotavirus A, B, and C by RT-qPCR in diagnostic samples. J. Virol. Methods 2014, 209, 30–34. [Google Scholar] [CrossRef] [PubMed]
- Vlasova, A.N.; Amimo, J.O.; Saif, L.J. Porcine Rotaviruses: Epidemiology, Immune Responses and Control Strategies. Viruses 2017, 9, 48. [Google Scholar] [CrossRef]


| Primer/Probe Name | Sequence 5′-3′ | Gene | Length (bp) |
|---|---|---|---|
| PEDV-G1-F | TGTTTTGGGTGGTTATCTACCTA | S | 168 a |
| PEDV-G1-R | AGCTGGTAACCACTAGGAT | ||
| PEDV-G1-Probe | FAM-TGTGCCACAGTACCAGCTAGAAGA-MGB | ||
| PEDV-G2-F | CCAGTACTTTCAACACTTAGCCTA | S | 195 b |
| PEDV-G2-R | GCCACTAGCAGTTGGATG | ||
| PEDV-G2-Probe | Texas Red-CAAGTTGAATTGACACCCTGGTTT-BHQ2 | ||
| RVA-F | CAACGAAACGGAATAGCACC | VP6 | 123 c |
| RVA-R | CCGCCTATTCTGTAGATTCCAA | ||
| RVA-Probe | CY5-ACCCGACAGCTTTCTTAGTGCTT-BHQ3 | ||
| RVC-F | GTGAAGAGAATGGTGHTGTAG | VP6 | 121 d |
| RVC-R | CATGCGCATTTGCCCCTACGC | ||
| RVC-Probe | VIC-CATGATTCACGAATGGGTTTAG-BHQ1 |
| Pathogen | Concentration (Copies/μL) | Repeat Times | Positive Number | Positive Rate | 95% Positive Rate |
|---|---|---|---|---|---|
| PEDV-G1 | 2 × 101 | 23 | 23 | 100% | >95% |
| 1 × 101 | 23 | 0 | 0% | <95% | |
| PEDV-G2 | 1 × 102 | 23 | 23 | 100% | >95% |
| 1 × 101 | 23 | 0 | 0% | <95% | |
| RVA | 5 × 101 | 23 | 23 | 100% | >95% |
| 1 × 101 | 23 | 0 | 0% | <95% | |
| RVC | 5 × 101 | 23 | 23 | 100% | >95% |
| 1 × 101 | 23 | 0 | 0% | <95% |
| Sample Type | Controls | Multiplex Real-Time PCR in This Study (Ct Value) | |||
|---|---|---|---|---|---|
| PEDV-G1 | PEDV-G2 | RVA | RVC | ||
| Fecal | PEDV-G1 | 12.57 | - | - | - |
| Anal swab | PEDV-G2 | - | 21.49 | - | - |
| Fecal | RVA | - | - | 29.04 | - |
| Anal swab | RVC | - | - | - | 30.99 |
| Anal swab | TGEV | - | - | - | - |
| Fecal | PDCoV | - | - | - | - |
| Lung | PRRSV | - | - | - | - |
| Lung | CSFV | - | - | - | - |
| Nasal swab | SIV | - | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Jiang, Z.; Zhou, Z.; Sun, J.; Yan, S.; Gao, W.; Shao, Y.; Bai, Y.; Wu, Y.; Yan, Z.; et al. A TaqMan Probe-Based Multiplex Real-Time PCR for Simultaneous Detection of Porcine Epidemic Diarrhea Virus Subtypes G1 and G2, and Porcine Rotavirus Groups A and C. Viruses 2022, 14, 1819. https://doi.org/10.3390/v14081819
Zhang L, Jiang Z, Zhou Z, Sun J, Yan S, Gao W, Shao Y, Bai Y, Wu Y, Yan Z, et al. A TaqMan Probe-Based Multiplex Real-Time PCR for Simultaneous Detection of Porcine Epidemic Diarrhea Virus Subtypes G1 and G2, and Porcine Rotavirus Groups A and C. Viruses. 2022; 14(8):1819. https://doi.org/10.3390/v14081819
Chicago/Turabian StyleZhang, Letian, Zhiwen Jiang, Zitong Zhou, Jiumeng Sun, Shiyu Yan, Wenting Gao, Yuekun Shao, Yuhe Bai, Yifan Wu, Zefei Yan, and et al. 2022. "A TaqMan Probe-Based Multiplex Real-Time PCR for Simultaneous Detection of Porcine Epidemic Diarrhea Virus Subtypes G1 and G2, and Porcine Rotavirus Groups A and C" Viruses 14, no. 8: 1819. https://doi.org/10.3390/v14081819
APA StyleZhang, L., Jiang, Z., Zhou, Z., Sun, J., Yan, S., Gao, W., Shao, Y., Bai, Y., Wu, Y., Yan, Z., Sheng, S., Lai, A., & Su, S. (2022). A TaqMan Probe-Based Multiplex Real-Time PCR for Simultaneous Detection of Porcine Epidemic Diarrhea Virus Subtypes G1 and G2, and Porcine Rotavirus Groups A and C. Viruses, 14(8), 1819. https://doi.org/10.3390/v14081819






