PEDV: Insights and Advances into Types, Function, Structure, and Receptor Recognition
Abstract
:1. Introduction
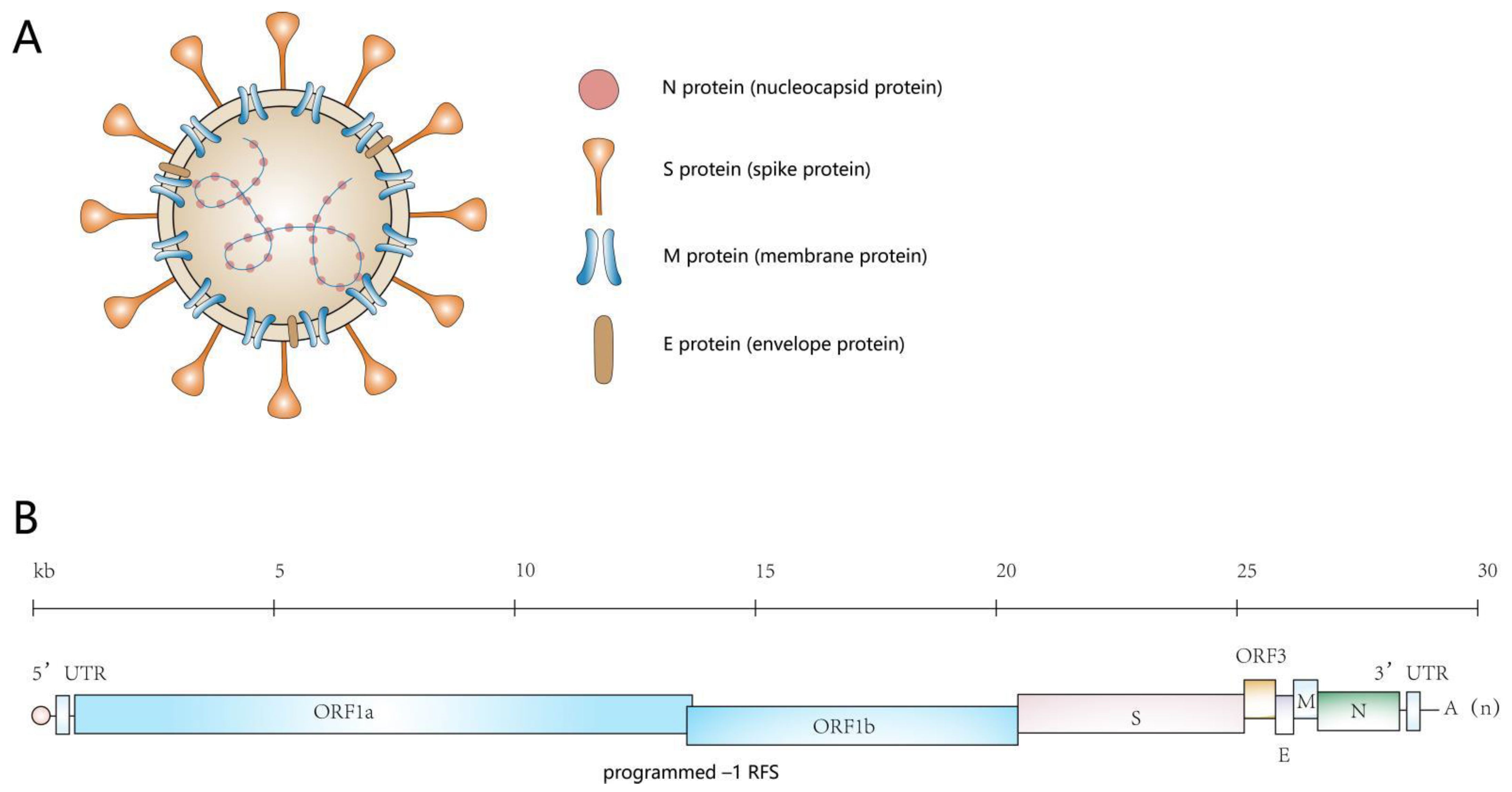
2. An Overview of the Genome of PEDV and the Proteins It Encodes
3. Types of PEDV Strains
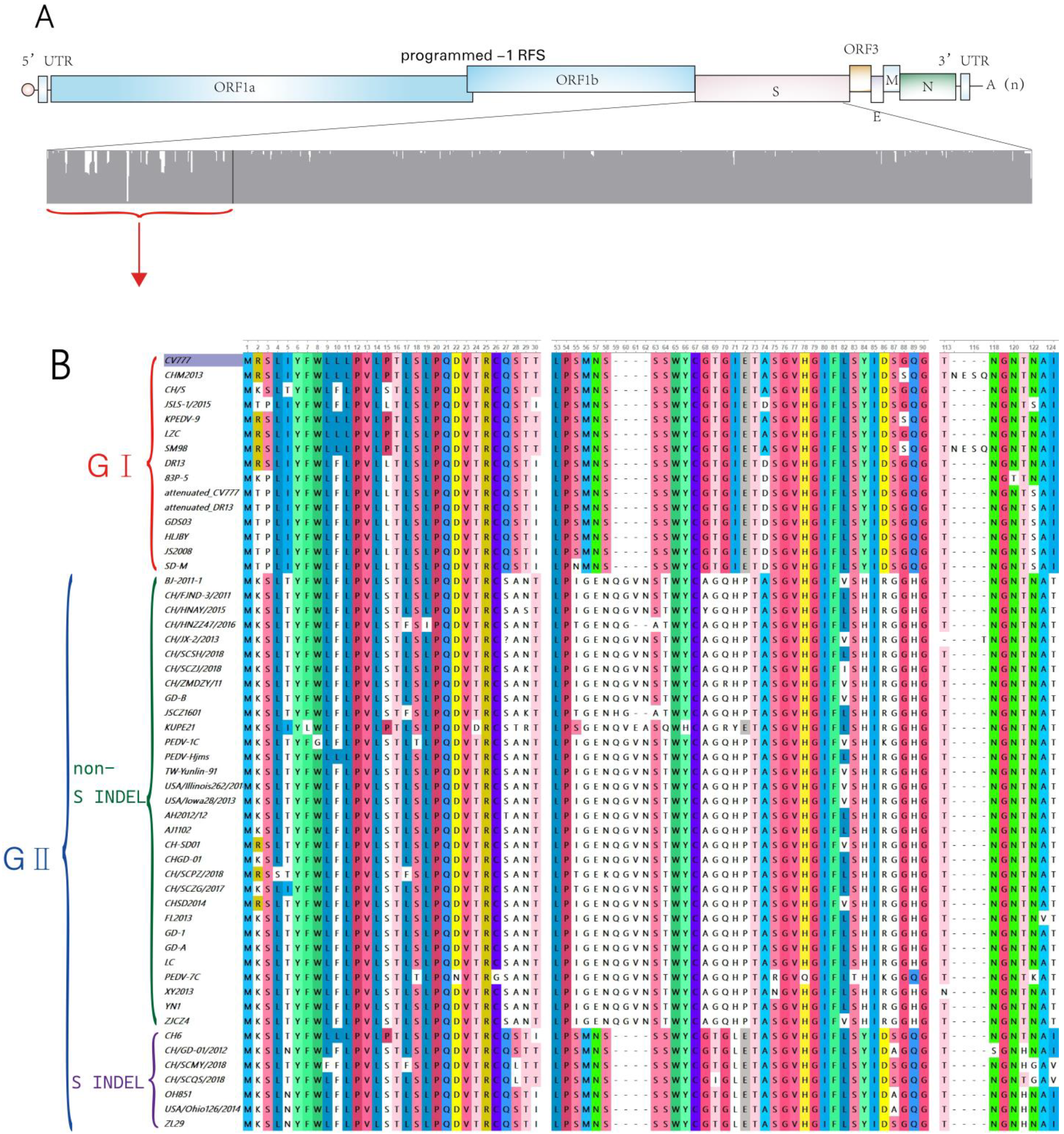
3.1. Classic Strains (GⅠ)
3.2. Mutant Strains (GII)
3.2.1. Non-S INDEL
3.2.2. S INDEL
4. Life Cycle of PEDV
5. Key to the Entry of PEDV into Target Cells—The S Protein
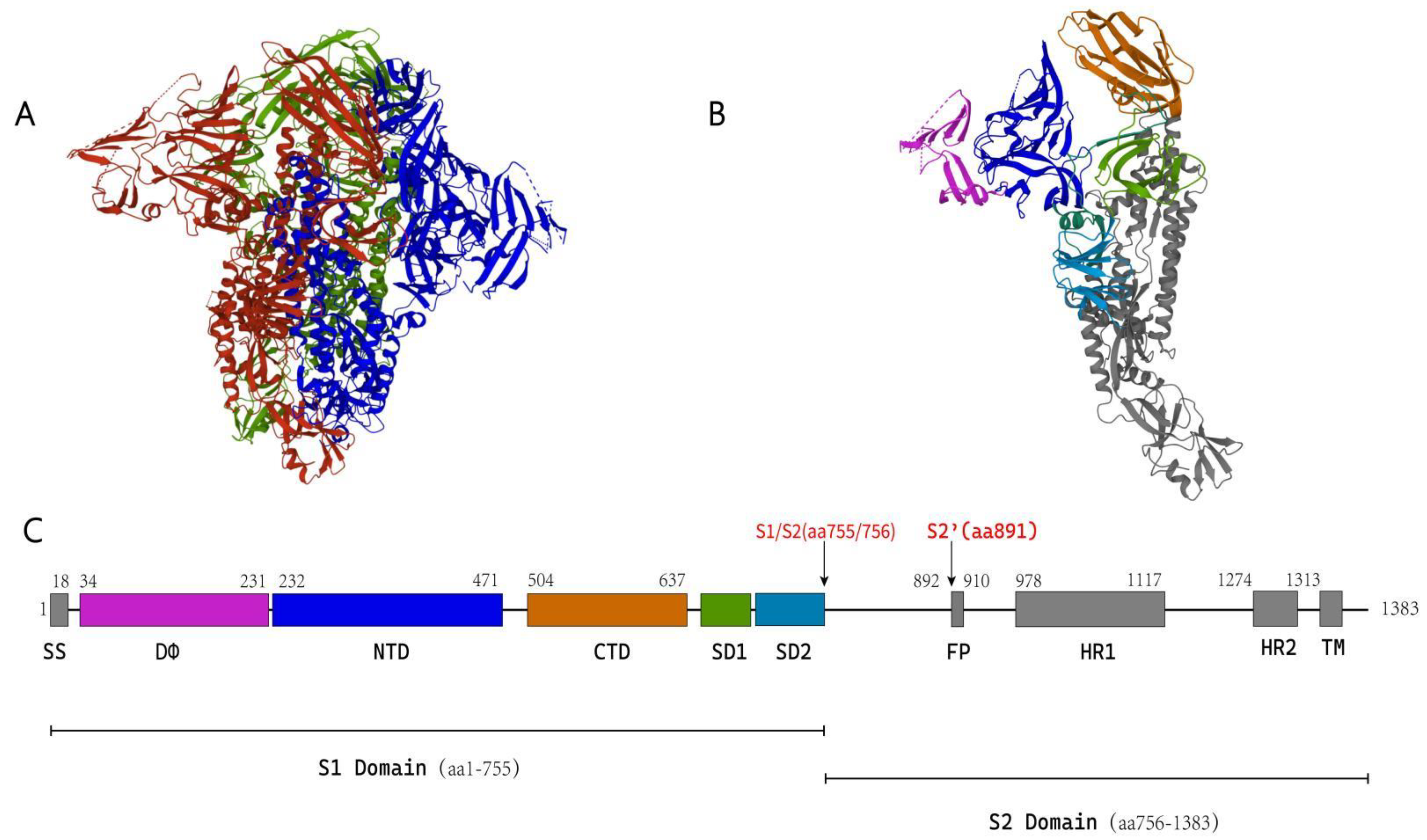
5.1. Structure of the Coronavirus S Protein
5.1.1. S1 Subunit
5.1.2. S2 Subunit
5.2. Protease-Mediated Fusion Activation of Coronavirus S Protein
5.3. S Protein-Mediated PEDV Entry into Host Cells
5.4. The S Protein Is the Key Determinant of Viral Host Range
6. Advances in PEDV Receptor Research
6.1. Aminopeptidase N
6.2. Sialic Acid
6.3. DC-SIGN/L-SIGN
6.4. Heparan Sulfate
6.5. Other Auxiliary Receptors in PEDV Infection
6.5.1. Occludin
6.5.2. Integrin αvβ3
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pensaert, M.B.; de Bouck, P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978, 58, 243–247. [Google Scholar] [CrossRef] [Green Version]
- Cavanagh, D. Nidovirales: A new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 1997, 142, 629–633. [Google Scholar] [PubMed]
- Wood, E. An apparently new syndrome of porcine epidemic diarrhoea. Veter. Rec. 1977, 100, 243–244. [Google Scholar] [CrossRef] [PubMed]
- Kocherhans, R.; Bridgen, A.; Ackermann, M.; Tobler, K. Completion of the porcine epidemic diarrhoea coronavirus (PEDV) genome sequence. Virus Genes 2001, 23, 137–144. [Google Scholar] [CrossRef] [Green Version]
- Duarte, M.; Tobler, K.; Bridgen, A.; Rasschaert, D.; Ackermann, M.; Laude, H. Sequence analysis of the porcine epidemic diarrhea virus genome between the nucleocapsid and spike protein genes reveals a polymorphic ORF. Virology 1994, 198, 466–476. [Google Scholar] [CrossRef]
- Bosch, B.J.; van der Zee, R.; de Haan, C.A.; Rottier, P.J.M. The Coronavirus Spike Protein Is a Class I Virus Fusion Protein: Structural and Functional Characterization of the Fusion Core Complex. J. Virol. 2003, 77, 8801–8811. [Google Scholar] [CrossRef] [Green Version]
- Cavanagh, D. The Coronavirus Surface Glycoprotein: The Coronaviridae; Springer: Boston, MA, USA, 1995. [Google Scholar]
- Klumperman, J.; Locker, J.K.; Meijer, A.; Horzinek, M.C.; Geuze, H.J.; Rottier, P.J. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J. Virol. 1994, 68, 6523–6534. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, V.P.; Hogue, B.G. Protein interactions during coronavirus assembly. J. Virol. 1997, 71, 9278–9284. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Yu, R.; Chen, B.; Si, F.; Wang, J.; Xie, C.; Men, C.; Dong, S.; Li, Z. Identification of host cell proteins that interact with the M protein of porcine epidemic diarrhea virus. Veter. Microbiol. 2020, 246, 108729. [Google Scholar] [CrossRef]
- Tan, Y.W.; Fang, S.; Fan, H.; Lescar, J.; Liu, D. Amino acid residues critical for RNA-binding in the N-terminal domain of the nucleocapsid protein are essential determinants for the infectivity of coronavirus in cultured cells. Nucleic Acids Res. 2006, 34, 4816–4825. [Google Scholar] [CrossRef]
- Baric, R.S.; Nelson, G.W.; Fleming, J.O.; Deans, R.J.; Keck, J.G.; Casteel, N.; Stohlman, S.A. Interactions between coronavirus nucleocapsid protein and viral RNAs: Implications for viral transcription. J. Virol. 1988, 62, 4280–4287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corse, E.; Machamer, C.E. Infectious Bronchitis Virus E Protein Is Targeted to the Golgi Complex and Directs Release of Virus-Like Particles. J. Virol. 2000, 74, 4319–4326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.; Zhang, H.; Zhang, Q.; Dong, J.; Liang, Y.; Huang, Y.; Liu, H.-J.; Tong, D. Porcine epidemic diarrhea virus E protein causes endoplasmic reticulum stress and up-regulates interleukin-8 expression. Virol. J. 2013, 10, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, L.; Wang, X.; Guo, D.; Cao, J.; Cheng, L.; Li, X.; Zou, D.; Zhang, Y.; Xu, J.; Wu, X.; et al. Porcine epidemic diarrhea virus E protein suppresses RIG-I signaling-mediated interferon-β production. Veter. Microbiol. 2021, 254, 108994. [Google Scholar] [CrossRef]
- Wang, K.; Lu, W.; Chen, J.; Xie, S.; Shi, H.; Hsu, H.; Yu, W.; Xu, K.; Bian, C.; Fischer, W.B.; et al. PEDV ORF3 encodes an ion channel protein and regulates virus production. FEBS Lett. 2012, 586, 384–391. [Google Scholar] [CrossRef] [Green Version]
- Ye, S.; Li, Z.; Chen, F.; Li, W.; Guo, X.; Hu, H.; He, Q. Porcine epidemic diarrhea virus ORF3 gene prolongs S-phase, facilitates formation of vesicles and promotes the proliferation of attenuated PEDV. Virus Genes 2015, 51, 385–392. [Google Scholar] [CrossRef]
- Sato, T.; Takeyama, N.; Katsumata, A.; Tuchiya, K.; Kodama, T.; Kusanagi, K.-I. Mutations in the spike gene of porcine epidemic diarrhea virus associated with growth adaptation in vitro and attenuation of virulence in vivo. Virus Genes 2011, 43, 72–78. [Google Scholar] [CrossRef]
- LLin, C.-M.; Saif, L.J.; Marthaler, D.; Wang, Q. Evolution, antigenicity and pathogenicity of global porcine epidemic diarrhea virus strains. Virus Res. 2016, 226, 20–39. [Google Scholar] [CrossRef] [Green Version]
- Sun, R.-Q.; Cai, R.-J.; Chen, Y.-Q.; Liang, P.-S.; Chen, D.-K.; Song, C.-X. Outbreak of Porcine Epidemic Diarrhea in Suckling Piglets, China. Emerg. Infect. Dis. 2012, 18, 161–163. [Google Scholar] [CrossRef]
- Stevenson, G.W.; Hoang, H.; Schwartz, K.J.; Burrough, E.R.; Sun, D.; Madson, D.; Cooper, V.L.; Pillatzki, A.; Gauger, P.; Schmitt, B.J.; et al. Emergence of Porcine epidemic diarrhea virus in the United States: Clinical signs, lesions, and viral genomic sequences. J. Veter. Diagn. Investig. 2013, 25, 649–654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, J.; Guo, J.; Fang, L.; Fang, L.; Ye, X.; Ye, X.; Chen, J.; Chen, J.; Xu, S.; Xu, S.; et al. Evolutionary and genotypic analyses of global porcine epidemic diarrhea virus strains. Transbound. Emerg. Dis. 2019, 66, 111–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, C. Porcine epidemic diarrhea virus: An emerging and re-emerging epizootic swine virus. Virol. J. 2015, 12, 193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Li, H.; Liu, Y.; Pan, Y.; Deng, F.; Song, Y.; Tang, X.; He, Q. New Variants of Porcine Epidemic Diarrhea Virus, China, 2011. Emerg. Infect. Dis. 2012, 18, 1350–1353. [Google Scholar] [CrossRef]
- Wang, L.; Byrum, B.; Zhang, Y. New Variant of Porcine Epidemic Diarrhea Virus, United States, 2014. Emerg. Infect. Dis. 2014, 20, 917–919. [Google Scholar] [CrossRef]
- Vlasova, A.N.; Marthaler, D.; Wang, Q.; Culhane, M.R.; Rossow, K.D.; Rovira, A.; Collins, J.; Saif, L.J. Distinct Characteristics and Complex Evolution of PEDV Strains, North America, May 2013–February 2014. Emerg. Infect. Dis. 2014, 20, 1620–1628. [Google Scholar] [CrossRef] [Green Version]
- Stadler, K.; Masignani, V.; Eickmann, M.; Becker, S.; Abrignani, S.; Klenk, H.-D.; Rappuoli, R. SARS—Beginning to understand a new virus. Nat. Rev. Genet. 2003, 1, 209–218. [Google Scholar] [CrossRef]
- Chi, S.; Chen, S.; Jia, W.; He, Y.; Ren, L.; Wang, X. Non-structural proteins of bovine viral diarrhea virus. Virus Genes 2022, 1–10. [Google Scholar] [CrossRef]
- Wei, X.; She, G.; Wu, T.; Xue, C.; Cao, Y. PEDV enters cells through clathrin-, caveolae-, and lipid raft-mediated endocytosis and traffics via the endo-/lysosome pathway. Veter. Res. 2020, 51, 10. [Google Scholar] [CrossRef] [Green Version]
- Sola, I.; Almazán, F.; Zúñiga, S.; Enjuanes, L. Continuous and Discontinuous RNA Synthesis in Coronaviruses. Annu. Rev. Virol. 2015, 2, 265–288. [Google Scholar] [CrossRef] [Green Version]
- Dufour, D.; Mateos-Gomez, P.A.; Enjuanes, L.; Gallego, J.; Sola, I. Structure and Functional Relevance of a Transcription-Regulating Sequence Involved in Coronavirus Discontinuous RNA Synthesis. J. Virol. 2011, 85, 4963–4973. [Google Scholar] [CrossRef] [Green Version]
- Sawicki, S.G.; Sawicki, D.L.; Siddell, S.G. A Contemporary View of Coronavirus Transcription. J. Virol. 2007, 81, 20–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perlman, S.; Netland, J. Coronaviruses post-SARS: Update on replication and pathogenesis. Nat. Rev. Microbiol. 2009, 7, 439–450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walls, A.C.; Tortorici, M.A.; Snijder, J.; Xiong, X.; Bosch, B.-J.; Rey, F.A.; Veesler, D. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc. Natl. Acad. Sci. USA 2017, 114, 11157–11162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, F.; Li, W.; Farzan, M.; Harrison, S.C. Structure of SARS Coronavirus Spike Receptor-Binding Domain Complexed with Receptor. Science 2005, 309, 1864–1868. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Hu, Y.; Wang, Q.; Qi, J.; Gao, F.; Li, Y.; Zhang, Y.; Zhang, W.; Yuan, Y.; Bao, J.; et al. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature 2013, 500, 227–231. [Google Scholar] [CrossRef] [Green Version]
- Kirchdoerfer, R.N.; Cottrell, C.A.; Wang, N.; Pallesen, J.; Yassine, H.M.; Turner, H.L.; Corbett, K.S.; Graham, B.S.; McLellan, J.S.; Ward, A.B. Pre-fusion structure of a human coronavirus spike protein. Nature 2016, 531, 118–121. [Google Scholar] [CrossRef] [Green Version]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef]
- Song, X.; Shi, Y.; Ding, W.; Niu, T.; Sun, L.; Tan, Y.; Chen, Y.; Shi, J.; Xiong, Q.; Huang, X.; et al. Cryo-EM analysis of the HCoV-229E spike glycoprotein reveals dynamic prefusion conformational changes. Nat. Commun. 2021, 12, 141. [Google Scholar] [CrossRef]
- Walls, A.; Tortorici, M.A.; Frenz, B.; Snijder, J.; Li, W.; Rey, F.; DiMaio, F.; Bosch, B.-J.; Veesler, D. Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat. Struct. Mol. Biol. 2016, 23, 899–905. [Google Scholar] [CrossRef]
- Wrapp, D.; McLellan, J.S. The 3.1-Angstrom Cryo-electron Microscopy Structure of the Porcine Epidemic Diarrhea Virus Spike Protein in the Prefusion Conformation. J. Virol. 2019, 93, e00923-19. [Google Scholar] [CrossRef] [Green Version]
- Walls, A.C.; Tortorici, M.A.; Bosch, B.-J.; Frenz, B.; Rottier, P.J.M.; DiMaio, F.; Rey, F.A.; Veesler, D. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature 2016, 531, 114–117. [Google Scholar] [CrossRef] [Green Version]
- Gui, M.; Song, W.; Zhou, H.; Xu, J.; Chen, S.; Xiang, Y.; Wang, X. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017, 27, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Cao, D.; Zhang, Y.; Ma, J.; Qi, J.; Wang, Q.; Lu, G.; Wu, Y.; Yan, J.; Shi, Y.; et al. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 2017, 8, 15092. [Google Scholar] [CrossRef] [PubMed]
- Li, F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016, 3, 237–261. [Google Scholar] [CrossRef] [Green Version]
- Li, F. Evidence for a Common Evolutionary Origin of Coronavirus Spike Protein Receptor-Binding Subunits. J. Virol. 2012, 86, 2856–2858. [Google Scholar] [CrossRef] [Green Version]
- Vlasak, R.; Luytjes, W.; Spaan, W.; Palese, P. Human and bovine coronaviruses recognize sialic acid-containing receptors similar to those of influenza C viruses. Proc. Natl. Acad. Sci. USA 1988, 85, 4526–4529. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Dong, W.; Milewska, A.; Golda, A.; Qi, Y.; Zhu, Q.K.; Marasco, W.A.; Baric, R.S.; Sims, A.C.; Pyrc, K.; et al. Human Coronavirus HKU1 Spike Protein Uses O -Acetylated Sialic Acid as an Attachment Receptor Determinant and Employs Hemagglutinin-Esterase Protein as a Receptor-Destroying Enzyme. J. Virol. 2015, 89, 7202–7213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hulswit, R.J.G.; Lang, Y.; Bakkers, M.J.G.; Li, W.; Li, Z.; Schouten, A.; Ophorst, B.; van Kuppeveld, F.J.M.; Boons, G.-J.; Bosch, B.-J.; et al. Human coronaviruses OC43 and HKU1 bind to 9- O -acetylated sialic acids via a conserved receptor-binding site in spike protein domain A. Proc. Natl. Acad. Sci. USA 2019, 116, 2681–2690. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Hulswit, R.J.G.; Widjaja, I.; Raj, V.S.; McBride, R.; Stalin Raj, V.; McBride, R.; Peng, W.; Widagdo, W.; Alejandra Tortoric, M.; et al. Identification of sialic acid-binding function for the Middle East respiratory syndrome coronavirus spike glycoprotein. Proc. Natl. Acad. Sci. USA 2017, 114, E8508–E8517. [Google Scholar] [CrossRef] [Green Version]
- Krempl, C.; Schultze, B.; Laude, H.; Herrler, G. Point mutations in the S protein connect the sialic acid binding activity with the enteropathogenicity of transmissible gastroenteritis coronavirus. J. Virol. 1997, 71, 3285–3287. [Google Scholar] [CrossRef] [Green Version]
- Wong, S.K.; Li, W.; Moore, M.J.; Choe, H.; Farzan, M. A 193-Amino Acid Fragment of the SARS Coronavirus S Protein Efficiently Binds Angiotensin-converting Enzyme 2. J. Biol. Chem. 2004, 279, 3197–3201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, H.-X.; Feng, Y.; Wong, G.; Wang, L.; Li, B.; Zhao, X.; Li, Y.; Smaill, F.; Zhang, C. Identification of residues in the receptor-binding domain (RBD) of the spike protein of human coronavirus NL63 that are critical for the RBD–ACE2 receptor interaction. J. Gen. Virol. 2008, 89, 1015–1024. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, H.; Simmons, G.; Rennekamp, A.J.; Chaipan, C.; Gramberg, T.; Heck, E.; Geier, M.; Wegele, A.; Marzi, A.; Bates, P.; et al. Highly Conserved Regions within the Spike Proteins of Human Coronaviruses 229E and NL63 Determine Recognition of Their Respective Cellular Receptors. J. Virol. 2006, 80, 8639–8652. [Google Scholar] [CrossRef] [Green Version]
- Godet, M.; Grosclaude, J.; Delmas, B.; Laude, H. Major receptor-binding and neutralization determinants are located within the same domain of the transmissible gastroenteritis virus (coronavirus) spike protein. J. Virol. 1994, 68, 8008–8016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, L.; Zhao, G.; Kou, Z.; Ma, C.; Sun, S.; Poon, V.K.M.; Lu, L.; Wang, L.; Debnath, A.K.; Zheng, B.-J.; et al. Identification of a Receptor-Binding Domain in the S Protein of the Novel Human Coronavirus Middle East Respiratory Syndrome Coronavirus as an Essential Target for Vaccine Development. J. Virol. 2013, 87, 9939–9942. [Google Scholar] [CrossRef] [Green Version]
- Deng, F.; Ye, G.; Liu, Q.; Navid, M.T.; Zhong, X.; Li, Y.; Wan, C.; Xiao, S.; He, Q.; Fu, Z.F.; et al. Identification and Comparison of Receptor Binding Characteristics of the Spike Protein of Two Porcine Epidemic Diarrhea Virus Strains. Viruses 2016, 8, 55. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Tomlinson, A.C.; Wong, A.H.; Zhou, D.; Desforges, M.; Talbot, P.J.; Benlekbir, S.; Rubinstein, J.L.; Rini, J.M. The human coronavirus HCoV-229E S-protein structure and receptor binding. eLife 2019, 8, e51230. [Google Scholar] [CrossRef]
- Peng, G.; Sun, D.; Rajashankar, K.R.; Qian, Z.; Holmes, K.V.; Li, F. Crystal structure of mouse coronavirus receptor-binding domain complexed with its murine receptor. Proc. Natl. Acad. Sci. USA 2011, 108, 10696–10701. [Google Scholar] [CrossRef] [Green Version]
- Qian, Z.; Ou, X.; Góes, L.G.B.; Osborne, C.; Castano, A.; Holmes, K.V.; Dominguez, S.R. Identification of the Receptor-Binding Domain of the Spike Glycoprotein of Human Betacoronavirus HKU1. J. Virol. 2015, 89, 8816–8827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mou, H.; Raj, V.S.; van Kuppeveld, F.J.M.; Rottier, P.J.M.; Haagmans, B.L.; Bosch, B.J. The Receptor Binding Domain of the New Middle East Respiratory Syndrome Coronavirus Maps to a 231-Residue Region in the Spike Protein That Efficiently Elicits Neutralizing Antibodies. J. Virol. 2013, 87, 9379–9383. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Wang, B.; Liang, Q.-Z.; Shi, F.-S.; Ji, C.-M.; Yang, X.-L.; Yang, Y.-L.; Qin, P.; Chen, R.; Huang, Y.-W. Roles of Two Major Domains of the Porcine Deltacoronavirus S1 Subunit in Receptor Binding and Neutralization. J. Virol. 2021, 95, e0111821. [Google Scholar] [CrossRef]
- Madu, I.G.; Roth, S.L.; Belouzard, S.; Whittaker, G.R. Characterization of a Highly Conserved Domain within the Severe Acute Respiratory Syndrome Coronavirus Spike Protein S2 Domain with Characteristics of a Viral Fusion Peptide. J. Virol. 2009, 83, 7411–7421. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; van Kuppeveld, F.J.M.; He, Q.; Rottier, P.J.M.; Bosch, B.-J. Cellular entry of the porcine epidemic diarrhea virus. Virus Res. 2016, 226, 117–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belouzard, S.; Millet, J.K.; Licitra, B.N.; Whittaker, G.R. Mechanisms of Coronavirus Cell Entry Mediated by the Viral Spike Protein. Viruses 2012, 4, 1011–1033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belouzard, S.; Chu, V.C.; Whittaker, G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. USA 2009, 106, 5871–5876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Millet, J.K.; Whittaker, G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. USA 2014, 111, 15214–15219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, F. Receptor Recognition Mechanisms of Coronaviruses: A Decade of Structural Studies. J. Virol. 2015, 89, 1954–1964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, G. Furin at the cutting edge: From protein traffic to embryogenesis and disease. Nat. Rev. Mol. Cell Biol. 2002, 3, 753–766. [Google Scholar] [CrossRef] [Green Version]
- Cheng, J.; Zhao, Y.; Hu, Y.; Zhao, J.; Xue, J.; Zhang, G. The Furin-S2′ Site in Avian Coronavirus Plays a Key Role in Central Nervous System Damage Progression. J. Virol. 2021, 95, e02447-20. [Google Scholar] [CrossRef]
- Liu, X.; Wu, Q.; Zhang, Z. Global Diversification and Distribution of Coronaviruses with Furin Cleavage Sites. Front. Microbiol. 2021, 12, 649314. [Google Scholar] [CrossRef]
- Millet, J.K.; Whittaker, G.R. Host cell proteases: Critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2014, 202, 120–134. [Google Scholar] [CrossRef]
- Cheng, Y.-W.; Chao, T.-L.; Li, C.-L.; Chiu, M.-F.; Kao, H.-C.; Wang, S.-H.; Pang, Y.-H.; Lin, C.-H.; Tsai, Y.-M.; Lee, W.-H.; et al. Furin Inhibitors Block SARS-CoV-2 Spike Protein Cleavage to Suppress Virus Production and Cytopathic Effects. Cell Rep. 2020, 33, 108254. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Pöhlmann, S. A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol. Cell 2020, 78, 779–784. [Google Scholar] [CrossRef] [PubMed]
- Papa, G.; Mallery, D.L.; Albecka, A.; Welch, L.G.; Cattin-Ortolá, J.; Luptak, J.; Paul, D.; McMahon, H.T.; Goodfellow, I.G.; Carter, A.; et al. Furin cleavage of SARS-CoV-2 Spike promotes but is not essential for infection and cell-cell fusion. PLOS Pathog. 2021, 17, e1009246. [Google Scholar] [CrossRef] [PubMed]
- Huang, I.-C.; Bosch, B.J.; Li, F.; Li, W.; Lee, K.H.; Ghiran, S.; Vasilieva, N.; Dermody, T.S.; Harrison, S.C.; Dormitzer, P.R.; et al. SARS Coronavirus, but Not Human Coronavirus NL63, Utilizes Cathepsin L to Infect ACE2-expressing Cells. J. Biol. Chem. 2006, 281, 3198–3203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, Z.; Dominguez, S.R.; Holmes, K.V. Role of the Spike Glycoprotein of Human Middle East Respiratory Syndrome Coronavirus (MERS-CoV) in Virus Entry and Syncytia Formation. PLoS ONE 2013, 8, e76469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, Y.X.; Ren, J.; Heinen, P.; Zambon, M.; Jones, I.M. Cleavage and Serum Reactivity of the Severe Acute Respiratory Syndrome Coronavirus Spike Protein. J. Infect. Dis. 2004, 190, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Matsuyama, S.; Ujike, M.; Morikawa, S.; Tashiro, M.; Taguchi, F. Protease-mediated enhancement of severe acute respiratory syndrome coronavirus infection. Proc. Natl. Acad. Sci. USA 2005, 102, 12543–12547. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Ma, Y.; Yang, Y.; Zheng, Y.; Shang, J.; Zhou, Y.; Jiang, S.; Du, L.; Li, J.; Li, F. Cell Entry of Porcine Epidemic Diarrhea Coronavirus Is Activated by Lysosomal Proteases. J. Biol. Chem. 2016, 291, 24779–24786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Wicht, O.; van Kuppeveld, F.J.M.; He, Q.; Rottier, P.J.M.; Bosch, B.-J. A Single Point Mutation Creating a Furin Cleavage Site in the Spike Protein Renders Porcine Epidemic Diarrhea Coronavirus Trypsin Independent for Cell Entry and Fusion. J. Virol. 2015, 89, 8077–8081. [Google Scholar] [CrossRef] [Green Version]
- Wicht, O.; Li, W.; Willems, L.; Meuleman, T.J.; Wubbolts, R.W.; van Kuppeveld, F.J.M.; Rottier, P.J.M.; Bosch, B.J. Proteolytic Activation of the Porcine Epidemic Diarrhea Coronavirus Spike Fusion Protein by Trypsin in Cell Culture. J. Virol. 2014, 88, 7952–7961. [Google Scholar] [CrossRef] [Green Version]
- Shirato, K.; Matsuyama, S.; Ujike, M.; Taguchi, F. Role of Proteases in the Release of Porcine Epidemic Diarrhea Virus from Infected Cells. J. Virol. 2011, 85, 7872–7880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.-E.; Cruz, D.J.M.; Shin, H.-J. Receptor-bound porcine epidemic diarrhea virus spike protein cleaved by trypsin induces membrane fusion. Arch. Virol. 2011, 156, 1749–1756. [Google Scholar] [CrossRef] [PubMed]
- Zamolodchikova, T.S. Serine proteases of small intestine mucosa—Localization, functional properties, and physiological role. Biochemistry 2012, 77, 820–829. [Google Scholar] [CrossRef] [PubMed]
- Nam, E.; Lee, C. Contribution of the porcine aminopeptidase N (CD13) receptor density to porcine epidemic diarrhea virus infection. Veter. Microbiol. 2010, 144, 41–50. [Google Scholar] [CrossRef]
- Pallesen, J.; Wang, N.; Corbett, K.S.; Wrapp, D.; Kirchdoerfer, R.N.; Turner, H.L.; Cottrell, C.A.; Becker, M.M.; Wang, L.; Shi, W.; et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. USA 2017, 114, E7348–E7357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Wong, S.-K.; Li, F.; Kuhn, J.H.; Huang, I.-C.; Choe, H.; Farzan, M. Animal Origins of the Severe Acute Respiratory Syndrome Coronavirus: Insight from ACE2-S-Protein Interactions. J. Virol. 2006, 80, 4211–4219. [Google Scholar] [CrossRef] [Green Version]
- Graham, R.L.; Baric, R.S. Recombination, Reservoirs, and the Modular Spike: Mechanisms of Coronavirus Cross-Species Transmission. J. Virol. 2010, 84, 3134–3146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Zhang, C.; Sui, J.; Kuhn, J.H.; Moore, M.J.; Luo, S.; Wong, S.-K.; Huang, I.-C.; Xu, K.; Vasilieva, N.; et al. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 2005, 24, 1634–1643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qu, X.-X.; Hao, P.; Song, X.-J.; Jiang, S.-M.; Liu, Y.-X.; Wang, P.-G.; Rao, X.; Song, H.-D.; Wang, S.-Y.; Zuo, Y.; et al. Identification of Two Critical Amino Acid Residues of the Severe Acute Respiratory Syndrome Coronavirus Spike Protein for Its Variation in Zoonotic Tropism Transition via a Double Substitution Strategy. J. Biol. Chem. 2005, 280, 29588–29595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Shi, Z.; Yu, M.; Ren, W.; Smith, C.; Epstein, J.H.; Wang, H.; Crameri, G.; Hu, Z.; Zhang, H.; et al. Bats Are Natural Reservoirs of SARS-Like Coronaviruses. Science 2005, 310, 676–679. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Ma, Z.; Dong, L.; Yang, T.; Li, Y.; Jiao, D.; Han, W.; Zheng, H.; Xiao, S. Molecular Mechanism of Porcine Epidemic Diarrhea Virus Cell Tropism. mBio 2022, 13, e0373921. [Google Scholar] [CrossRef] [PubMed]
- Barnieh, F.M.; Loadman, P.M.; Falconer, R.A. Is tumour-expressed aminopeptidase N (APN/CD13) structurally and functionally unique? Biochim. Biophys. Acta 2021, 1876, 188641. [Google Scholar] [CrossRef] [PubMed]
- Delmas, B.; Gelfi, J.; L’Haridon, R.; Vogel, L.K.; Sjöström, H.; Norén, O.; Laude, H. Aminopeptidase N is a major receptor for the enteropathogenic coronavirus TGEV. Nature 1992, 357, 417–420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tresnan, D.B.; Levis, R.; Holmes, K.V. Feline aminopeptidase N serves as a receptor for feline, canine, porcine, and human coronaviruses in serogroup I. J. Virol. 1996, 70, 8669–8674. [Google Scholar] [CrossRef] [Green Version]
- Yeager, C.L.; Ashmun, R.A.; Williams, R.K.; Cardellichio, C.B.; Shapiro, L.H.; Look, A.T.; Holmes, K.V. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature 1992, 357, 420–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sánchez, C.M.; Gebauer, F.; Suñé, C.; Mendez, A.; Dopazo, J.; Enjuanes, L. Genetic evolution and tropism of transmissible gastroenteritis coronaviruses. Virology 1992, 190, 92–105. [Google Scholar] [CrossRef]
- Sueyoshi, M.; Tsuda, T.; Yamazaki, K.; Yoshida, K.; Nakazawa, M.; Sato, K.; Minami, T.; Iwashita, K.; Watanabe, M.; Suzuki, Y.; et al. An immunohistochemical investigation of porcine epidemic diarrhoea. J. Comp. Pathol. 1995, 113, 59–67. [Google Scholar] [CrossRef]
- Oh, J.S.; Song, D.S.; Park, B.K. Identification of a putative cellular receptor 150 kDa polypeptide for porcine epidemic diarrhea virus in porcine enterocytes. J. Veter. Sci. 2003, 4, 269–275. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Ge, J.; Li, Y. Porcine aminopeptidase N is a functional receptor for the PEDV coronavirus. Virology 2007, 365, 166–172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Yang, Y.; Chen, L.; Lin, Y.-L.; Li, F. A Unified Mechanism for Aminopeptidase N-based Tumor Cell Motility and Tumor-homing Therapy. J. Biol. Chem. 2014, 289, 34520–34529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Lin, Y.-L.; Peng, G.; Li, F. Structural basis for multifunctional roles of mammalian aminopeptidase N. Proc. Natl. Acad. Sci. USA 2012, 109, 17966–17971. [Google Scholar] [CrossRef] [Green Version]
- Kamau, A.N.; Park, J.-E.; Park, E.-S.; Yu, J.-E.; Rho, J.; Shin, H.-J. Porcine amino peptidase N domain VII has critical role in binding and entry of porcine epidemic diarrhea virus. Virus Res. 2016, 227, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Luo, R.; He, Q.; van Kuppeveld, F.J.; Rottier, P.J.; Bosch, B.-J. Aminopeptidase N is not required for porcine epidemic diarrhea virus cell entry. Virus Res. 2017, 235, 6–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, C.-M.; Wang, B.; Zhou, J.; Huang, Y.-W. Aminopeptidase-N-independent entry of porcine epidemic diarrhea virus into Vero or porcine small intestine epithelial cells. Virology 2018, 517, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Whitworth, K.M.; Rowland, R.R.; Ewen, C.L.; Trible, B.R.; Kerrigan, M.A.; Cino-Ozuna, A.G.; Samuel, M.S.; Lightner, J.E.; McLaren, D.G.; Mileham, A.J.; et al. Gene-edited pigs are protected from porcine reproductive and respiratory syndrome virus. Nat. Biotechnol. 2016, 34, 20–22. [Google Scholar] [CrossRef]
- Whitworth, K.M.; Rowland, R.R.R.; Petrovan, V.; Sheahan, M.; Cino-Ozuna, A.G.; Fang, Y.; Hesse, R.; Mileham, A.; Samuel, M.S.; Wells, K.D.; et al. Resistance to coronavirus infection in amino peptidase N-deficient pigs. Transgen. Res. 2018, 28, 21–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whitworth, K.M.; Lee, K.; Benne, J.A.; Beaton, B.P.; Spate, L.D.; Murphy, S.L.; Samuel, M.S.; Mao, J.; O’Gorman, C.; Walters, E.M.; et al. Use of the CRISPR/Cas9 System to Produce Genetically Engineered Pigs from In Vitro-Derived Oocytes and Embryos1. Biol. Reprod. 2014, 91, 78. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Wu, Z.; Yang, H. Aminopeptidase N Knockout Pigs Are Not Resistant to Porcine Epidemic Diarrhea Virus Infection. Virol. Sin. 2019, 34, 592–595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shirato, K.; Maejima, M.; Islam, T.; Miyazaki, A.; Kawase, M.; Matsuyama, S.; Taguchi, F. Porcine aminopeptidase N is not a cellular receptor of porcine epidemic diarrhea virus, but promotes its infectivity via aminopeptidase activity. J. Gen. Virol. 2016, 97, 2528–2539. [Google Scholar] [CrossRef]
- Stencel-Baerenwald, J.E.; Reiss, K.; Reiter, D.M.; Stehle, T.; Dermody, T.S. The sweet spot: Defining virus–sialic acid interactions. Nat. Rev. Genet. 2014, 12, 739–749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, X.; Tan, M.; Xia, M.; Huang, P.; Kennedy, M.A. Intra-species sialic acid polymorphism in humans: A common niche for influenza and coronavirus pandemics? Emerg. Microbes Infect. 2021, 10, 1191–1199. [Google Scholar] [CrossRef]
- Schwegmann-Wessels, C.; Herrler, G. Sialic acids as receptor determinants for coronaviruses. Glycoconj. J. 2006, 23, 51–58. [Google Scholar] [CrossRef]
- Künkel, F.; Herrler, G. Structural and Functional Analysis of the Surface Protein of Human Coronavirus OC43. Virology 1993, 195, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Schultze, B.; Krempl, C.; Ballesteros, M.L.; Shaw, L.; Schauer, R.; Enjuanes, L.; Herrler, G. Transmissible gastroenteritis coronavirus, but not the related porcine respiratory coronavirus, has a sialic acid (N-glycolylneuraminic acid) binding activity. J. Virol. 1996, 70, 5634–5637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwegmann-Weßels, C.; Bauer, S.; Winter, C.; Enjuanes, L.; Laude, H.; Herrler, G. The sialic acid binding activity of the S protein facilitates infection by porcine transmissible gastroenteritis coronavirus. Virol. J. 2011, 8, 435. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Tang, J.; Ma, Y.; Liang, X.; Yang, Y.; Peng, G.; Qi, Q.; Jiang, S.; Li, J.; Du, L.; et al. Receptor Usage and Cell Entry of Porcine Epidemic Diarrhea Coronavirus. J. Virol. 2015, 89, 6121–6125. [Google Scholar] [CrossRef] [Green Version]
- Peng, G.; Xu, L.; Lin, Y.-L.; Chen, L.; Pasquarella, J.R.; Holmes, K.V.; Li, F. Crystal Structure of Bovine Coronavirus Spike Protein Lectin Domain. J. Biol. Chem. 2012, 287, 41931–41938. [Google Scholar] [CrossRef] [Green Version]
- Pan, Y.; Tian, X.; Li, W.; Zhou, Q.; Wang, D.; Bi, Y.; Chen, F.; Song, Y. Isolation and characterization of a variant porcine epidemic diarrhea virus in China. Virol. J. 2012, 9, 195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, Y.; Yang, X.; Li, H.; Ma, B.; Guan, R.; Yang, J.; Chen, D.; Han, X.; Zhou, L.; Song, Z.; et al. Molecular characterization of porcine epidemic diarrhea virus associated with outbreaks in southwest China during 2014–2018. Transbound. Emerg. Dis. 2020, 68, 3482–3497. [Google Scholar] [CrossRef]
- Lozach, P.-Y.; Burleigh, L.; Staropoli, I.; Amara, A.; Lozach, P.-Y.; Burleigh, L.; Staropoli, I.; Amara, A. The C Type Lectins DC-SIGN and L-SIGN. Glycovirol. Protoc. 2007, 379, 51–68. [Google Scholar] [CrossRef]
- Han, D.P.; Lohani, M.; Cho, M.W. Specific Asparagine-Linked Glycosylation Sites Are Critical for DC-SIGN- and L-SIGN-Mediated Severe Acute Respiratory Syndrome Coronavirus Entry. J. Virol. 2007, 81, 12029–12039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Q.; Liu, J.; Zhao, S.; Castro, M.F.G.; Laurent-Rolle, M.; Dong, J.; Ran, X.; Damani-Yokota, P.; Tang, H.; Karakousi, T.; et al. SARS-CoV-2 exacerbates proinflammatory responses in myeloid cells through C-type lectin receptors and Tweety family member 2. Immunity 2021, 54, 1304–1319. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Xu, L.-D.; Zhang, Y.; Cao, H.; Chen, R.; Wang, B.; Huang, Y.-W. Expression of the human or porcine C-type lectins DC-SIGN/L-SIGN confers susceptibility to porcine epidemic diarrhea virus entry and infection in otherwise refractory cell lines. Microb. Pathog. 2021, 157, 104956. [Google Scholar] [CrossRef]
- Milewska, A.; Zarebski, M.; Nowak, P.; Stozek, K.; Potempa, J.; Pyrc, K. Human Coronavirus NL63 Utilizes Heparan Sulfate Proteoglycans for Attachment to Target Cells. J. Virol. 2014, 88, 13221–13230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clausen, T.M.; Sandoval, D.R.; Spliid, C.B.; Pihl, J.; Perrett, H.R.; Painter, C.D.; Narayanan, A.; Majowicz, S.A.; Kwong, E.M.; McVicar, R.N.; et al. SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2. Cell 2020, 183, 1043–1057. [Google Scholar] [CrossRef]
- Huan, C.-C.; Wang, Y.; Ni, B.; Wang, R.; Huang, L.; Ren, X.-F.; Tong, G.-Z.; Ding, C.; Fan, H.-J.; Mao, X. Porcine epidemic diarrhea virus uses cell-surface heparan sulfate as an attachment factor. Arch. Virol. 2015, 160, 1621–1628. [Google Scholar] [CrossRef]
- Zhou, Y.; Jiang, X.; Tong, T.; Fang, L.; Wu, Y.; Liang, J.; Xiao, S. High antiviral activity of mercaptoethane sulfonate functionalized Te/BSA nanostars against arterivirus and coronavirus. RSC Adv. 2020, 10, 14161–14169. [Google Scholar] [CrossRef]
- Zeisel, M.B.; Fofana, I.; Fafi-Kremer, S.; Baumert, T.F. Hepatitis C virus entry into hepatocytes: Molecular mechanisms and targets for antiviral therapies. J. Hepatol. 2011, 54, 566–576. [Google Scholar] [CrossRef]
- Luo, X.; Guo, L.; Zhang, J.; Xu, Y.; Gu, W.; Feng, L.; Wang, Y. Tight Junction Protein Occludin Is a Porcine Epidemic Diarrhea Virus Entry Factor. J. Virol. 2017, 91, e00202-17. [Google Scholar] [CrossRef] [Green Version]
- Stewart, P.L.; Nemerow, G.R. Cell integrins: Commonly used receptors for diverse viral pathogens. Trends Microbiol. 2007, 15, 500–507. [Google Scholar] [CrossRef] [PubMed]
- Guerrero, C.A.; Méndez, E.; Zárate, S.; Isa, P.; López, S.; Arias, C.F. Integrin α v β 3 mediates rotavirus cell entry. Proc. Natl. Acad. Sci. USA 2000, 97, 14644–14649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, D.; Shi, H.; Guo, D.; Chen, J.; Shi, D.; Zhu, Q.; Zhang, X.; Feng, L. Analysis of protein expression changes of the Vero E6 cells infected with classic PEDV strain CV777 by using quantitative proteomic technique. J. Virol. Methods 2015, 218, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Su, M.; Yin, B.; Guo, D.; Wei, S.; Kong, F.; Feng, L.; Wu, R.; Sun, D. Integrin αvβ3 enhances replication of porcine epidemic diarrhea virus on Vero E6 and porcine intestinal epithelial cells. Veter. Microbiol. 2019, 237, 108400. [Google Scholar] [CrossRef] [PubMed]


| Subgroup | Strain | Number of AA Dissimilarities | AA Similarity (%) | GeneBank | |
|---|---|---|---|---|---|
| GⅠ | CV777 | 0 | 100 | AF353511 | |
| CHM2013 | 25 | 98 | KM887144 | ||
| CH/S | 53 | 96 | JN547228 | ||
| JSLS-1/2015 | 59 | 96 | KX534205 | ||
| KPEDV-9 | 42 | 97 | KF898124 | ||
| LZC | 13 | 99 | EF185992 | ||
| SM98 | 24 | 98 | GU937797 | ||
| DR13 | 28 | 98 | JQ023161 | ||
| 83P-5 | 46 | 97 | AB548618 | ||
| attenuated CV777 | 57 | 96 | JN599150 | ||
| attenuated DR13 | 54 | 96 | JQ023162 | ||
| GDS03 | 59 | 96 | AB857235 | ||
| HLJBY | 58 | 96 | KP403802 | ||
| JS2008 | 55 | 96 | KC210146 | ||
| SD-M | 59 | 96 | JX560761 | ||
| GⅡ | non-S INDEL | BJ-2011-1 | 92 | 93 | JN825712 |
| CH/FJND-3/2011 | 95 | 93 | JQ282909 | ||
| CH/HNAY/2015 | 96 | 93 | KR809885 | ||
| CH/HNZZ47/2016 | 97 | 93 | KX981440 | ||
| CH/JX-2/2013 | 104 | 93 | KJ526096 | ||
| CH/SCSH/2018 | 94 | 93 | MH053413 | ||
| CH/SCZJ/2018 | 101 | 93 | MH061342 | ||
| CH/ZMDZY/11 | 98 | 93 | KC196276 | ||
| GD-B | 95 | 93 | JX088695 | ||
| JSCZ1601 | 97 | 93 | KY070587 | ||
| KUPE21 | 89 | 94 | MF737355 | ||
| PEDV-1C | 142 | 90 | KM609203 | ||
| PEDV-Hjms | 100 | 93 | KY007139 | ||
| TW-Yunlin-91 | 96 | 93 | KP276248 | ||
| USA/Illinois262/2014 | 94 | 93 | KR265796 | ||
| USA/Iowa28/2013 | 95 | 93 | KJ645636 | ||
| AH2012/12 | 100 | 93 | KU646831 | ||
| AJ1102 | 95 | 93 | JX188454 | ||
| CH-SD01 | 96 | 93 | KU380331 | ||
| CHGD-01 | 96 | 93 | JX261936 | ||
| CH/SCPZ/2018 | 98 | 93 | MH593147 | ||
| CH/SCZG/2017 | 95 | 93 | MH061337 | ||
| CHSD2014 | 96 | 93 | KX791060 | ||
| FL2013 | 113 | 93 | KP765609 | ||
| GD-1 | 99 | 93 | JX647847 | ||
| GD-A | 97 | 93 | JX112709 | ||
| LC | 95 | 93 | JX489155 | ||
| PEDV-7C | 135 | 90 | KM609204 | ||
| XY2013 | 102 | 93 | KR818832 | ||
| YN1 | 97 | 93 | KT021227 | ||
| ZJCZ4 | 95 | 93 | JX524137 | ||
| S INDEL | CH6 | 57 | 96 | JQ239434 | |
| CH/GD-01/2012 | 59 | 96 | KP870113 | ||
| CH/SCMY/2018 | 71 | 95 | MH061343 | ||
| CH/SCQS/2018 | 58 | 96 | MH593141 | ||
| OH851 | 53 | 96 | KJ399978 | ||
| USA/Ohio126/2014 | 51 | 96 | KJ645702 | ||
| ZL29 | 55 | 96 | KU847996 | ||
| Virus | Receptor | Genus | RBD Position | Reference |
|---|---|---|---|---|
| PEDV | - | Alphacoronavirus | aa 477–629 | [57] |
| TGEV | pAPN | Alphacoronavirus | aa 506–655 | [53] |
| hCoV-NL63 | ACE2 | Alphacoronavirus | aa 476–616 | [53] |
| hCoV-229E | hAPN | Alphacoronavirus | aa 293–435 | [58] |
| MHV | mCEACAM1a | Betacoronavirus group A | aa 15–268 | [59] |
| hCoV-HKU1 | - | Betacoronavirus group A | aa 535–673 | [60] |
| SARS-CoV | ACE2 | Betacoronavirus group B | aa 318–510 | [52] |
| SARS-CoV-2 | ACE2 | Betacoronavirus group B | aa 331–528 | [38] |
| MERS-CoV | DPP4 | Betacoronavirus group C | aa 318–588 | [61] |
| PDCoV | pAPN | Deltacoronavirus | aa 302–425 | [62] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, F.; Zhang, H.; Li, L.; Yang, Y.; Zou, X.; Chen, J.; Tang, X. PEDV: Insights and Advances into Types, Function, Structure, and Receptor Recognition. Viruses 2022, 14, 1744. https://doi.org/10.3390/v14081744
Lin F, Zhang H, Li L, Yang Y, Zou X, Chen J, Tang X. PEDV: Insights and Advances into Types, Function, Structure, and Receptor Recognition. Viruses. 2022; 14(8):1744. https://doi.org/10.3390/v14081744
Chicago/Turabian StyleLin, Feng, Huanyu Zhang, Linquan Li, Yang Yang, Xiaodong Zou, Jiahuan Chen, and Xiaochun Tang. 2022. "PEDV: Insights and Advances into Types, Function, Structure, and Receptor Recognition" Viruses 14, no. 8: 1744. https://doi.org/10.3390/v14081744
APA StyleLin, F., Zhang, H., Li, L., Yang, Y., Zou, X., Chen, J., & Tang, X. (2022). PEDV: Insights and Advances into Types, Function, Structure, and Receptor Recognition. Viruses, 14(8), 1744. https://doi.org/10.3390/v14081744






