Off-Target Effect of Activation of NF-κB by HIV Latency Reversal Agents on Transposable Elements Expression
Abstract
:1. Introduction
2. Materials and Methods
2.1. RNA-seq Datasets
2.2. Bioinformatic Analyses
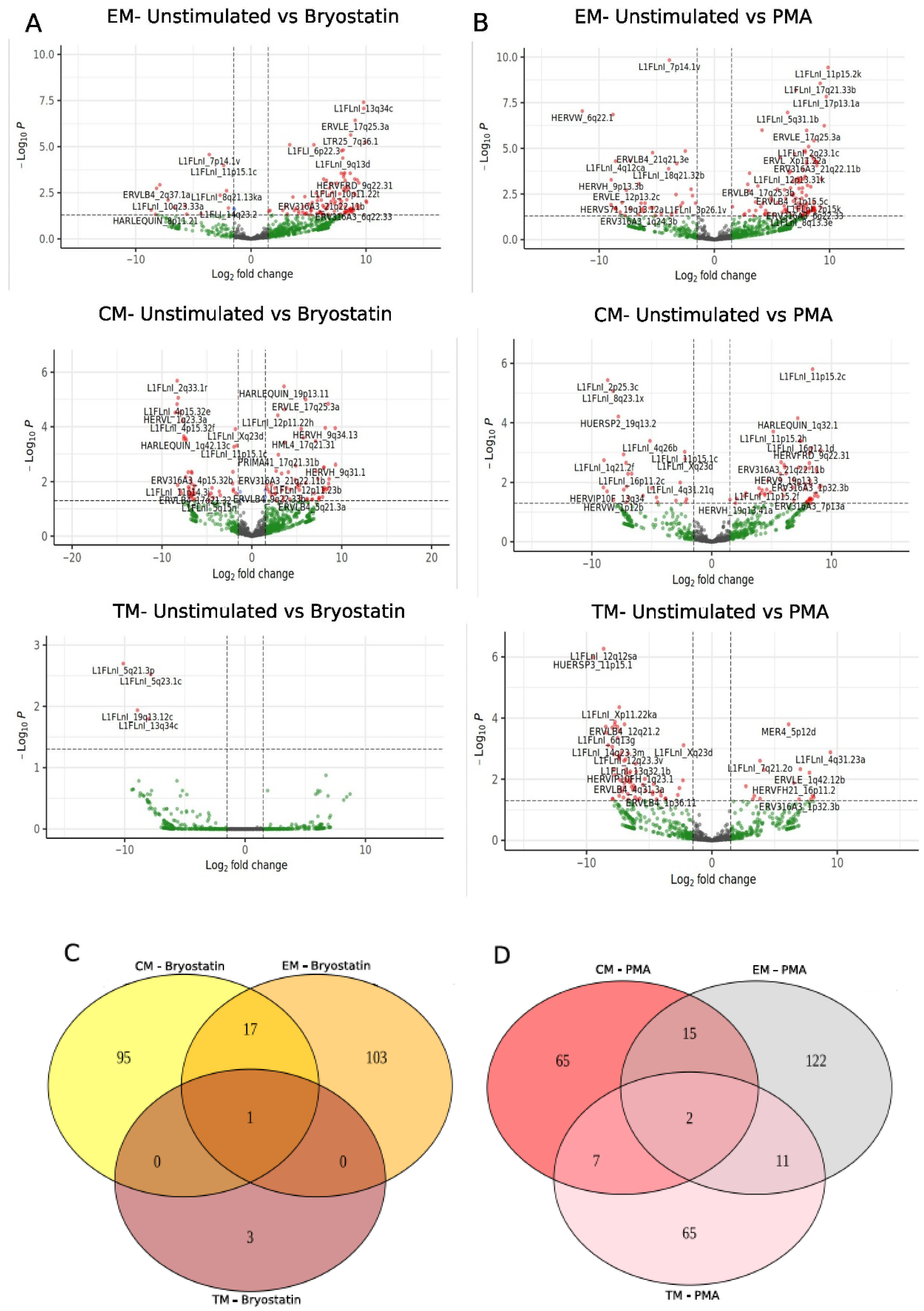
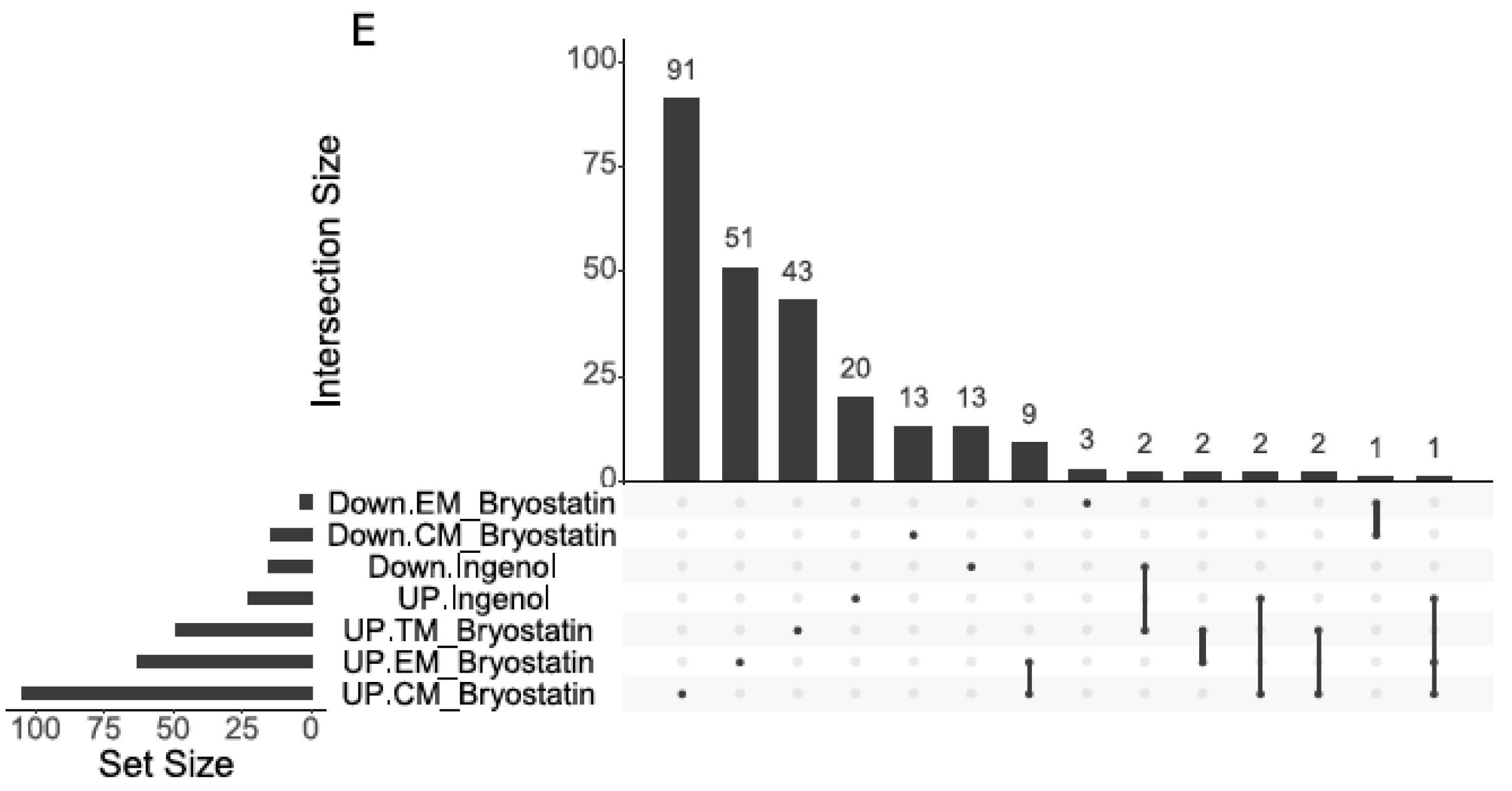
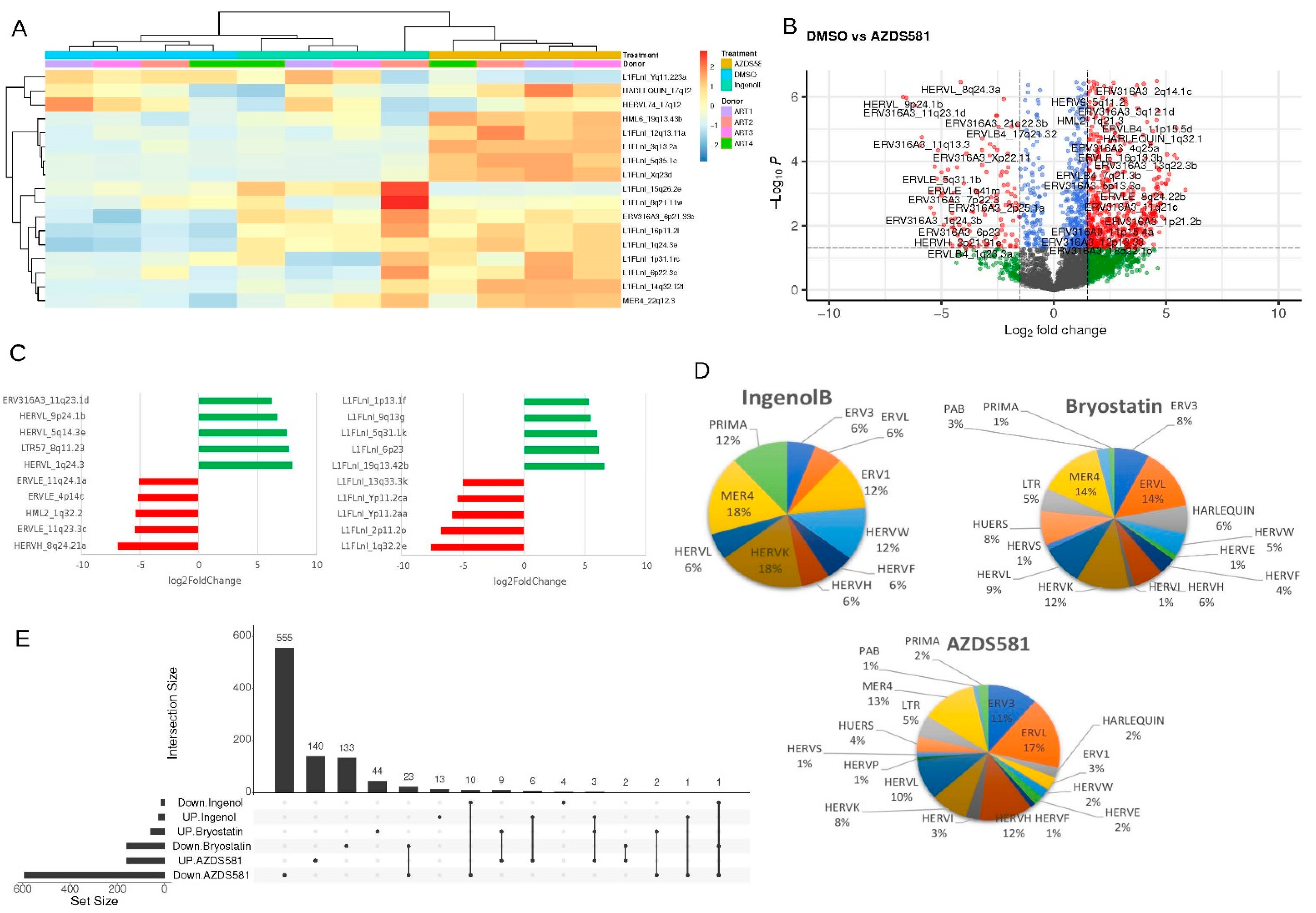
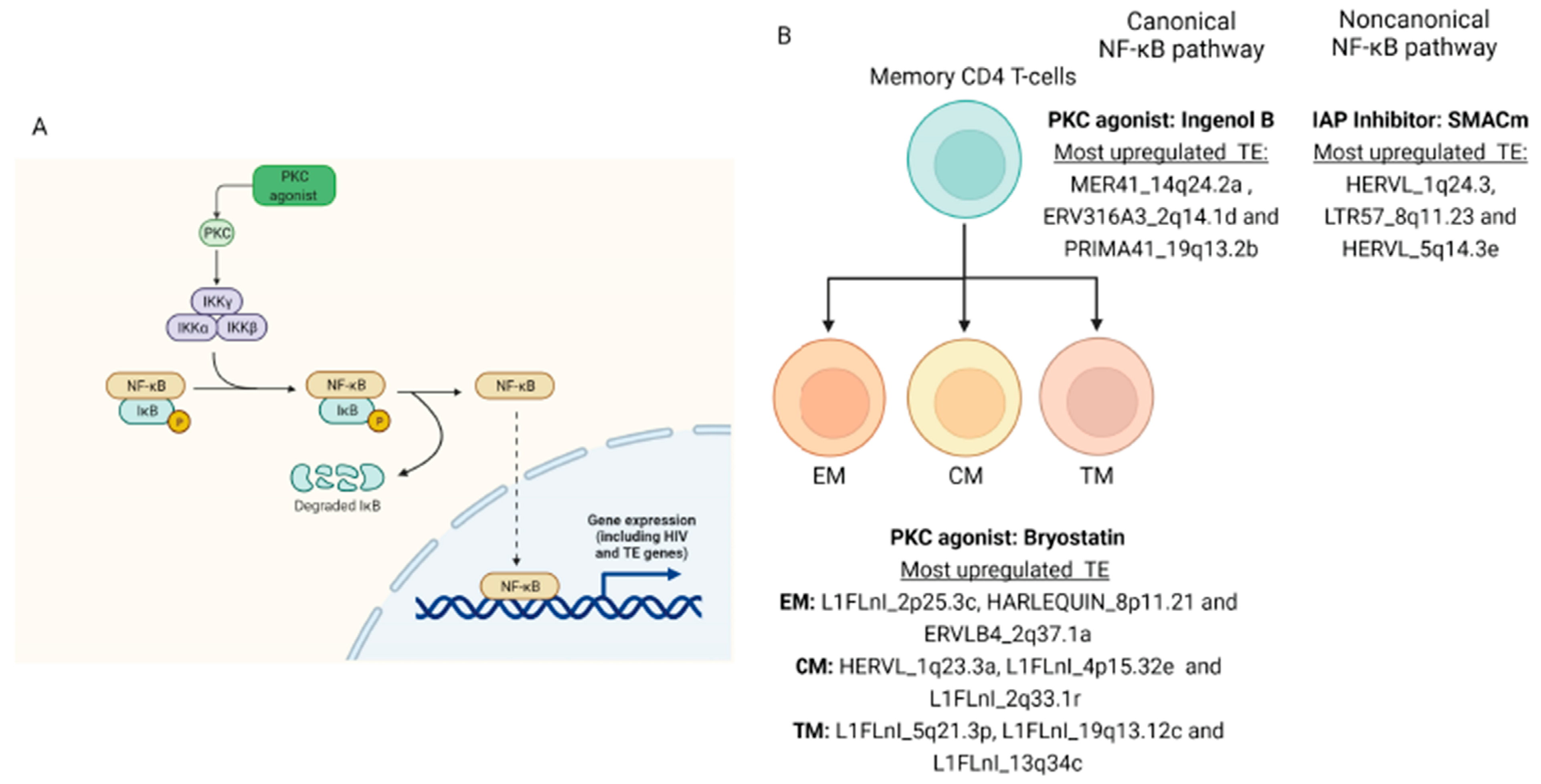
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- UNAIDS Epidemiological Estimates. Available online: https://aidsinfo.unaids.org (accessed on 17 April 2022).
- Saag, M.S.; Gandhi, R.T.; Hoy, J.F.; Landovitz, R.J.; Thompson, M.A.; Sax, P.E.; Smith, D.M.; Benson, C.A.; Buchbinder, S.P.; del Rio, C.; et al. Antiretroviral Drugs for Treatment and Prevention of HIV Infection in Adults. JAMA 2020, 324, 1651. [Google Scholar] [CrossRef] [PubMed]
- Eggleton, J.S.; Nagalli, S. Highly Active Antiretroviral Therapy (HAART). Available online: www.ncbi.nlm.nih.gov/books/NBK554533/ (accessed on 17 April 2022).
- Vanhamel, J.; Bruggemans, A.; Debyser, Z. Establishment of latent HIV-1 reservoirs: What do we really know? J. Virus Erad. 2019, 5, 3–9. [Google Scholar] [CrossRef]
- Cohn, L.B.; Chomont, N.; Deeks, S.G. The Biology of the HIV-1 Latent Reservoir and Implications for Cure Strategies. Cell Host Microbe 2020, 27, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Deeks, S.G.; Archin, N.; Cannon, P.; Collins, S.; Jones, R.B.; de Jong, M.A.W.P.; Lambotte, O.; Lamplough, R.; Ndung’u, T.; Sugarman, J.; et al. Research priorities for an HIV cure: International AIDS Society Global Scientific Strategy 2021. Nat. Med. 2021, 27, 2085–2098. [Google Scholar] [CrossRef]
- Kim, Y.; Anderson, J.L.; Lewin, S.R. Getting the “Kill” into “Shock and Kill”: Strategies to Eliminate Latent HIV. Cell Host Microbe 2018, 23, 14–26. [Google Scholar] [CrossRef] [Green Version]
- Sadowski, I.; Hashemi, F.B. Strategies to eradicate HIV from infected patients: Elimination of latent provirus reservoirs. Cell. Mol. Life Sci. 2019, 76, 3583–3600. [Google Scholar] [CrossRef] [Green Version]
- Curty, G.; Iñiguez, L.P.; Nixon, D.F.; Soares, M.A.; de Mulder Rougvie, M. Hallmarks of Retroelement Expression in T-Cells Treated With HDAC Inhibitors. Front. Virol. 2021, 1, 756635. [Google Scholar] [CrossRef]
- Díaz-Carballo, D.; Saka, S.; Acikelli, A.H.; Homp, E.; Erwes, J.; Demmig, R.; Klein, J.; Schröer, K.; Malak, S.; D’Souza, F.; et al. Enhanced antitumoral activity of TLR7 agonists via activation of human endogenous retroviruses by HDAC inhibitors. Commun. Biol. 2021, 4, 276. [Google Scholar] [CrossRef]
- White, C.H.; Beliakova-Bethell, N.; Lada, S.M.; Breen, M.S.; Hurst, T.P.; Spina, C.A.; Richman, D.D.; Frater, J.; Magiorkinis, G.; Woelk, C.H. Transcriptional Modulation of Human Endogenous Retroviruses in Primary CD4+ T Cells Following Vorinostat Treatment. Front. Immunol. 2018, 9, 603. [Google Scholar] [CrossRef]
- Bannert, N.; Kurth, R. Retroelements and the human genome: New perspectives on an old relation. Proc. Natl. Acad. Sci. USA 2004, 101, 14572–14579. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Montojo, M.; Doucet-O’Hare, T.; Henderson, L.; Nath, A. Human endogenous retrovirus-K (HML-2): A comprehensive review. Crit. Rev. Microbiol. 2018, 44, 715–738. [Google Scholar] [CrossRef] [PubMed]
- van der Kuyl, A.C. HIV infection and HERV expression: A review. Retrovirology 2012, 9, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Curty, G.; Marston, J.L.; de Mulder Rougvie, M.; Leal, F.E.; Nixon, D.F.; Soares, M.A. Human Endogenous Retrovirus K in Cancer: A Potential Biomarker and Immunotherapeutic Target. Viruses 2020, 12, 726. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.B.; Garrison, K.E.; Mujib, S.; Mihajlovic, V.; Aidarus, N.; Hunter, D.V.; Martin, E.; John, V.M.; Zhan, W.; Faruk, N.F.; et al. HERV-K-specific T cells eliminate diverse HIV-1/2 and SIV primary isolates. J. Clin. Investig. 2012, 122, 4473–4489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johanning, G.L.; Malouf, G.G.; Zheng, X.; Esteva, F.J.; Weinstein, J.N.; Wang-Johanning, F.; Su, X. Expression of human endogenous retrovirus-K is strongly associated with the basal-like breast cancer phenotype. Sci. Rep. 2017, 7, 41960. [Google Scholar] [CrossRef] [Green Version]
- Bannert, N.; Hofmann, H.; Block, A.; Hohn, O. HERVs New Role in Cancer: From Accused Perpetrators to Cheerful Protectors. Front. Microbiol. 2018, 9, 178. [Google Scholar] [CrossRef]
- Monde, K.; Contreras-Galindo, R.; Kaplan, M.H.; Markovitz, D.M.; Ono, A. Human Endogenous Retrovirus K Gag Coassembles with HIV-1 Gag and Reduces the Release Efficiency and Infectivity of HIV-1. J. Virol. 2012, 86, 11194–11208. [Google Scholar] [CrossRef] [Green Version]
- Okoye, A.A.; Fromentin, R.; Takata, H.; Brehm, J.H.; Fukazawa, Y.; Randall, B.; Pardons, M.; Tai, V.; Tang, J.; Smedley, J.; et al. The ingenol-based protein kinase C agonist GSK445A is a potent inducer of HIV and SIV RNA transcription. PLOS Pathog. 2022, 18, e1010245. [Google Scholar] [CrossRef]
- Wong, L.M.; Jiang, G. NF-κB sub-pathways and HIV cure: A revisit. EBioMedicine 2021, 63, 103159. [Google Scholar] [CrossRef]
- Díaz, L.; Martínez-Bonet, M.; Sánchez, J.; Fernández-Pineda, A.; Jiménez, J.L.; Muñoz, E.; Moreno, S.; Álvarez, S.; Muñoz-Fernández, M.Á. Bryostatin activates HIV-1 latent expression in human astrocytes through a PKC and NF-ĸB-dependent mechanism. Sci. Rep. 2015, 5, 12442. [Google Scholar] [CrossRef] [Green Version]
- Tong-Starkesen, S.E.; Luciw, P.A.; Peterlin, B.M. Signaling through T lymphocyte surface proteins, TCR/CD3 and CD28, activates the HIV-1 long terminal repeat. J. Immunol. 1989, 142, 702–707. [Google Scholar] [PubMed]
- Steffan, N.M.; Bren, G.D.; Frantz, B.; Tocci, M.J.; O’Neill, E.A.; Paya, C. V Regulation of IkB alpha phosphorylation by PKC- and Ca(2+)-dependent signal transduction pathways. J. Immunol. 1995, 155, 4685–4691. [Google Scholar] [PubMed]
- Nixon, C.C.; Mavigner, M.; Sampey, G.C.; Brooks, A.D.; Spagnuolo, R.A.; Irlbeck, D.M.; Mattingly, C.; Ho, P.T.; Schoof, N.; Cammon, C.G.; et al. Systemic HIV and SIV latency reversal via non-canonical NF-κB signalling in vivo. Nature 2020, 578, 160–165. [Google Scholar] [CrossRef] [PubMed]
- Falcinelli, S.D.; Peterson, J.J.; Turner, A.-M.W.; Irlbeck, D.; Read, J.; Raines, S.L.M.; James, K.S.; Sutton, C.; Sanchez, A.; Emery, A.; et al. Combined noncanonical NF-κB agonism and targeted BET bromodomain inhibition reverse HIV latency ex vivo. J. Clin. Investig. 2022, 132, e157281. [Google Scholar] [CrossRef]
- Hennessy, E.J.; Adam, A.; Aquila, B.M.; Castriotta, L.M.; Cook, D.; Hattersley, M.; Hird, A.W.; Huntington, C.; Kamhi, V.M.; Laing, N.M.; et al. Discovery of a Novel Class of Dimeric Smac Mimetics as Potent IAP Antagonists Resulting in a Clinical Candidate for the Treatment of Cancer (AZD5582). J. Med. Chem. 2013, 56, 9897–9919. [Google Scholar] [CrossRef]
- Kulpa, D.A.; Talla, A.; Brehm, J.H.; Ribeiro, S.P.; Yuan, S.; Bebin-Blackwell, A.-G.; Miller, M.; Barnard, R.; Deeks, S.G.; Hazuda, D.; et al. Differentiation into an Effector Memory Phenotype Potentiates HIV-1 Latency Reversal in CD4 + T Cells. J. Virol. 2019, 93, e00969-19. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Bendall, M.L.; de Mulder, M.; Iñiguez, L.P.; Lecanda-Sánchez, A.; Pérez-Losada, M.; Ostrowski, M.A.; Jones, R.B.; Mulder, L.C.F.; Reyes-Terán, G.; Crandall, K.A.; et al. Telescope: Characterization of the retrotranscriptome by accurate estimation of transposable element expression. PLoS Comput. Biol. 2019, 15, e1006453. [Google Scholar] [CrossRef] [Green Version]
- Kovaka, S.; Zimin, A.V.; Pertea, G.M.; Razaghi, R.; Salzberg, S.L.; Pertea, M. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 2019, 20, 278. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, e02804-20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beliakova-Bethell, N.; Mukim, A.; White, C.H.; Deshmukh, S.; Abewe, H.; Richman, D.D.; Spina, C.A. Histone deacetylase inhibitors induce complex host responses that contribute to differential potencies of these compounds in HIV reactivation. J. Biol. Chem. 2019, 294, 5576–5589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maxwell, J.W.; Falcinelli, S.D.; Nefedov, A.; Dorfmeier, C.; Wu, G.; Dewey, M.; Webber, A.L.; Archin, N.M.; Margolis, D.M.; Hazuda, D.J.; et al. Cellular Gene Modulation of HIV-Infected CD4 T Cells in Response to Serial Treatment with the Histone Deacetylase Inhibitor Vorinostat. J. Virol. 2020, 94, e00351-20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, T.J.; Sun, M.-K.; Lim, C.; Sen, A.; Khan, T.; Chirila, F.V.; Alkon, D.L. Bryostatin Effects on Cognitive Function and PKCɛ in Alzheimer’s Disease Phase IIa and Expanded Access Trials. J. Alzheimer’s Dis. 2017, 58, 521–535. [Google Scholar] [CrossRef] [Green Version]
- Kollár, P.; Rajchard, J.; Balounová, Z.; Pazourek, J. Marine natural products: Bryostatins in preclinical and clinical studies. Pharm. Biol. 2014, 52, 237–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Zhang, R.; Yu, J. New Understanding of the Relevant Role of LINE-1 Retrotransposition in Human Disease and Immune Modulation. Front. Cell Dev. Biol. 2020, 8, 657. [Google Scholar] [CrossRef]
- Garrison, K.E.; Jones, R.B.; Meiklejohn, D.A.; Anwar, N.; Ndhlovu, L.C.; Chapman, J.M.; Erickson, A.L.; Agrawal, A.; Spotts, G.; Hecht, F.M.; et al. T Cell Responses to Human Endogenous Retroviruses in HIV-1 Infection. PLoS Pathog. 2007, 3, e165. [Google Scholar] [CrossRef]
- Jameson, S.C.; Masopust, D. Understanding Subset Diversity in T Cell Memory. Immunity 2018, 48, 214–226. [Google Scholar] [CrossRef] [Green Version]
- Fromentin, R.; Chomont, N. HIV persistence in subsets of CD4+ T cells: 50 shades of reservoirs. Semin. Immunol. 2021, 51, 101438. [Google Scholar] [CrossRef]
- Kwon, K.J.; Timmons, A.E.; Sengupta, S.; Simonetti, F.R.; Zhang, H.; Hoh, R.; Deeks, S.G.; Siliciano, J.D.; Siliciano, R.F. Different human resting memory CD4+ T cell subsets show similar low inducibility of latent HIV-1 proviruses. Sci. Transl. Med. 2020, 12, eaax6795. [Google Scholar] [CrossRef]
- Hiener, B.; Horsburgh, B.A.; Eden, J.-S.; Barton, K.; Schlub, T.E.; Lee, E.; von Stockenstrom, S.; Odevall, L.; Milush, J.M.; Liegler, T.; et al. Identification of Genetically Intact HIV-1 Proviruses in Specific CD4+ T Cells from Effectively Treated Participants. Cell Rep. 2017, 21, 813–822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, J.A.; Carrel, L.; Chakravarti, A.; Eichler, E.E. Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: The Lyon repeat hypothesis. Proc. Natl. Acad. Sci. USA 2000, 97, 6634–6639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chow, J.C.; Ciaudo, C.; Fazzari, M.J.; Mise, N.; Servant, N.; Glass, J.L.; Attreed, M.; Avner, P.; Wutz, A.; Barillot, E.; et al. LINE-1 Activity in Facultative Heterochromatin Formation during X Chromosome Inactivation. Cell 2010, 141, 956–969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lyon, M.F. LINE-1 elements and X chromosome inactivation: A function for “junk” DNA? Proc. Natl. Acad. Sci. USA 2000, 97, 6248–6249. [Google Scholar] [CrossRef] [Green Version]
- Lyon, M.F. Do LINEs Have a Role in X-Chromosome Inactivation? J. Biomed. Biotechnol. 2006, 2006, 059746. [Google Scholar] [CrossRef]
- Gunter, C. Get in LINE for silencing. Nat. Rev. Genet. 2010, 11, 528–529. [Google Scholar] [CrossRef]
- Attermann, A.S.; Bjerregaard, A.M.; Saini, S.K.; Grønbæk, K.; Hadrup, S.R. Human endogenous retroviruses and their implication for immunotherapeutics of cancer. Ann. Oncol. 2018, 29, 2183–2191. [Google Scholar] [CrossRef]
- Rycaj, K.; Plummer, J.B.; Yin, B.; Li, M.; Garza, J.; Radvanyi, L.; Ramondetta, L.M.; Lin, K.; Johanning, G.L.; Tang, D.G.; et al. Cytotoxicity of Human Endogenous Retrovirus K-Specific T Cells toward Autologous Ovarian Cancer Cells. Clin. Cancer Res. 2015, 21, 471–483. [Google Scholar] [CrossRef] [Green Version]
- Doorbar, J.; Egawa, N.; Griffin, H.; Kranjec, C.; Murakami, I. Human papillomavirus molecular biology and disease association. Rev. Med. Virol. 2015, 25 (Suppl. 1), 2–23. [Google Scholar] [CrossRef] [Green Version]
- Denne, M.; Sauter, M.; Armbruester, V.; Licht, J.D.; Roemer, K.; Mueller-Lantzsch, N. Physical and Functional Interactions of Human Endogenous Retrovirus Proteins Np9 and Rec with the Promyelocytic Leukemia Zinc Finger Protein. J. Virol. 2007, 81, 5607–5616. [Google Scholar] [CrossRef] [Green Version]
- Fulda, S.; Vucic, D. Targeting IAP proteins for therapeutic intervention in cancer. Nat. Rev. Drug Discov. 2012, 11, 109–124. [Google Scholar] [CrossRef] [PubMed]
- Pache, L.; Dutra, M.S.; Spivak, A.M.; Marlett, J.M.; Murry, J.P.; Hwang, Y.; Maestre, A.M.; Manganaro, L.; Vamos, M.; Teriete, P.; et al. BIRC2/cIAP1 Is a Negative Regulator of HIV-1 Transcription and Can Be Targeted by Smac Mimetics to Promote Reversal of Viral Latency. Cell Host Microbe 2015, 18, 345–353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sampey, G.C.; Irlbeck, D.M.; Browne, E.P.; Kanke, M.; McAllister, A.B.; Ferris, R.G.; Brehm, J.H.; Favre, D.; Routy, J.-P.; Jones, C.D.; et al. The SMAC Mimetic AZD5582 is a Potent HIV Latency Reversing Agent. bioRxiv 2018, 312447. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Curty, G.; Iniguez, L.P.; Soares, M.A.; Nixon, D.F.; de Mulder Rougvie, M. Off-Target Effect of Activation of NF-κB by HIV Latency Reversal Agents on Transposable Elements Expression. Viruses 2022, 14, 1571. https://doi.org/10.3390/v14071571
Curty G, Iniguez LP, Soares MA, Nixon DF, de Mulder Rougvie M. Off-Target Effect of Activation of NF-κB by HIV Latency Reversal Agents on Transposable Elements Expression. Viruses. 2022; 14(7):1571. https://doi.org/10.3390/v14071571
Chicago/Turabian StyleCurty, Gislaine, Luis P. Iniguez, Marcelo A. Soares, Douglas F. Nixon, and Miguel de Mulder Rougvie. 2022. "Off-Target Effect of Activation of NF-κB by HIV Latency Reversal Agents on Transposable Elements Expression" Viruses 14, no. 7: 1571. https://doi.org/10.3390/v14071571
APA StyleCurty, G., Iniguez, L. P., Soares, M. A., Nixon, D. F., & de Mulder Rougvie, M. (2022). Off-Target Effect of Activation of NF-κB by HIV Latency Reversal Agents on Transposable Elements Expression. Viruses, 14(7), 1571. https://doi.org/10.3390/v14071571





