Abstract
Bat lyssaviruses were identified in Taiwan’s bat population during 2016–2017. The lyssavirus surveillance system was continuously conducted to understand the epidemiology. Through this system, the found dead bats were collected for lyssavirus detection by direct fluorescent antibody test and reverse transcription polymerase chain reaction. Three bats were identified as positive during 2018–2021. A novel lyssavirus, designated as Taiwan bat lyssavirus 2, was detected in a Nyctalus plancyi velutinus. This lyssavirus had less than 80% nucleotide identity in the nucleoprotein (N) gene with other lyssavirus species, forming a separate branch in the phylogenetic analysis. The other two cases were identified in Pipistrellus abramus (Japanese pipistrelles); they were identified to be similar to the former lyssavirus identified in 2016–2017, which was renominated as Taiwan bat lyssavirus 1 (TWBLV-1) in this study. Even though one of the TWBLV-1 isolates showed high genetic diversity in the N gene compared with other TWBLV-1 isolates, it may be a TWBLV-1 variant but not a new species based on its high amino acid identities in the nucleoprotein, same host species, and same geographic location as the other TWBLV-1.
1. Introduction
To date, 17 species of lyssavirus have been officially recognized by the International Committee on Taxonomy of Viruses (ICTV) [1,2,3,4]. The other two putative new lyssaviruses were published [5,6]. Over the past five years, more lyssavirus species were identified worldwide [4,5,6,7]. These novel lyssaviruses fulfilled the lyssavirus species demarcation criteria of the ICTV [8]: having less than 78–80% or 80% nucleotide identities on whole nucleoprotein (N) gene or concatenated coding regions compared to known lyssaviruses, respectively, and forming a monophyletic group in phylogenetic analysis. Although lyssaviruses are genetically various, all lyssaviruses cause the fatal disease rabies [9]. Among the lyssaviruses, they are divided into at least three phylogroups by their immunogenic properties and genetic diversity [2,10]. Current rabies vaccines confer partial or complete protection against phylogroup I lyssaviruses, but do not provide efficacious protection against lyssaviruses of phylogroups II and III [10,11,12,13,14].
Most studies currently indicate that bats are the reservoir host of lyssaviruses, except for Mokola lyssavirus (MOKV) and Ikoma lyssavirus (IKOV) [1,2,15,16]. In Asia, rabies lyssavirus (RABV) circulates in dogs and many wildlife [17,18]; in comparison, the other five lyssaviruses, Aravan lyssavirus (ARAV) [19], Khujand lyssavirus (KHUV) [19], Irkut lyssavirus (IRKV) [20], Gannoruwa bat lyssavirus (GBLV) [7], and Taiwan bat lyssavirus (TWBLV) [4], were merely found in bats. All circulating lyssaviruses in Asia belong to phylogroup I. There were two lyssaviruses discovered in Taiwan. The rabies lyssavirus was identified in Melogale moschata subaurantiaca (Formosan ferret-badger) in 2013 [21,22] and the other, Taiwan bat lyssavirus, was identified in Pipistrellus abramus (Japanese pipistrelle) in 2016 [4]. To monitor the activities of lyssavirus in Taiwan’s bat populations, a passive lyssavirus surveillance system has been continuously implemented in Taiwan for years. Thus, another novel bat lyssavirus was discovered and characterized between 2018 and 2021.
2. Materials and Methods
2.1. Sample Collection, Lyssavirus Diagnosis, and Virus Isolation
The lyssavirus surveillance system and the diagnostic methods performed in this study were described previously [23]. Briefly, the found dead bats were collected by nongovernmental organizations and local animal disease inspection authorities and were submitted to the Animal Health Research Institute in 2018–2021. The bats were necropsied in a biosafety level 2 laboratory, and their brain tissues were assayed by direct fluorescent antibody test (FAT) and reverse transcription polymerase chain reaction (RT-PCR).
Detecting lyssavirus antigen using the FAT, the brain smear was stained with each of the two commercially available FITC-conjugated anti-rabies antibodies (Catalog No. 800-092, Fujirebio Diagnostic Inc., Malvern, PA, USA; Catalog No. 5100, EMD Millipore Corporation, Temecula, CA, USA).
To test the viral RNA by RT-PCR, the brain tissue was homogenized into 10% (w/v) brain homogenate in minimum essential medium, and the supernatant was used for nucleic acid extraction after centrifugation. Detecting lyssavirus RNA by the RT-PCR, two sets of RT-PCR primers, JW12/N165-146 [24] and N113F/N304R [25,26], were used. The reagent preparation and thermal cycling conditions were followed as per the corresponding references.
Virus isolation was performed when a positive result was given by either the FAT or the RT-PCR [23]. Briefly, the supernatant of 10% (w/v) brain homogenate was mixed with a suspension of 3 × 106 mouse neuroblastoma cell/mL, and the mixture was incubated at 37 °C in a 1% CO2 atmosphere for 1 h. After incubation, the mixed brain homogenate cell suspension was then transferred to a flask and a few control slides and cultivated for three to four days. One of the control slides was fixed and stained with FITC-conjugated anti-rabies antibodies each day for examining the possible fluorescence. Several blind passages were performed when no fluorescence was observed after the four-day incubation.
2.2. Identification of Bat Species
Bat species were identified by external morphological characteristics as previously described [27,28]. For the lyssavirus positive case, the bat’s species was further confirmed through the DNA barcoding [29,30,31].
2.3. Complete Genome Sequencing of the Lyssavirus Isolate
For the whole genome amplification of lyssavirus, twelve sets of RT-PCR primers were used [4], and a few of the primers were modified according to the sequences of the isolate (Table S1). The RT-PCRs were performed using the SuperScript III One-Step RT-PCR System with Platinum Taq polymerase High Fidelity kit (Invitrogen, Life Technologies, Carlsbad, CA, USA) following the manufacturer’s instruction. The reaction was initiated at 42 °C for 40 min and then at 95 °C for 2 min, followed by 35 cycles of 95 °C for 40 s, 50 °C for 50 s, and 72 °C for 80 s, and the reaction ended after a final extension of 72 °C for 7 min.
The termini sequences at the 3′ and 5′ ends of viral genome were obtained using the SMARTer RACE kit (Clontech Laboratories, TaKaRa Bio Company, Mountain View, CA, USA) following the manufacturer’s instructions. Due to the fact that the amount of bat brain tissue was limited, the nucleic acid extracted from the cell culture supernatant of the virus isolation was used for the amplification of the termini sequences.
The RT-PCR products were sequenced with the 3700XL DNA analyzer (Applied Biosystems, Waltham, MA, USA) by a commercial sequencing service (Mission Biotech Co., Taipei, Taiwan).
2.4. Phylogenetic Analysis
The obtained sequences of the N gene in this study were aligned with those of representative lyssavirus strains retrieved from GenBank using the MUSCLE program implemented in the software Molecular Evolutionary Genetic Analysis version X [32]. The best-fit model of nucleotide substitution and partition scheme was evaluated using ModelFinder implemented in the phylogenomic inference software IQ-TREE [33,34,35] The maximum likelihood phylogenetic tree was constructed with IQ-TREE2 v2.1.3 [36], and the branch supports were estimated using 1000 ultrafast bootstrap replicates [37]. The phylogenetic tree produced by the IQ-TREE was further edited and visualized by the Figtree v1.4.4 program (http://tree.bio.ed.ac.uk/software/figtree/ accessed on 16 September 2019).
The genome sequences of all representative lyssaviruses used in the analyses were Rabies lyssavirus (RABV; GenBank accession number: NC001542), Lagos bat lyssavirus (LBV; NC020807), Mokola lyssavirus (MOKV; NC006429), Duvenhage lyssavirus (DUVV; NC020810), European bat lyssavirus 1 (EBLV-1; NC009527), European bat lyssavirus 2 (EBLV-2; NC009528), Australian bat lyssavirus (ABLV; NC003243), Aravan lyssavirus (ARAV; NC020808), Khujand lyssavirus (KHUV; NC025385), Irkut lyssavirus (IRKV; NC020809), West Caucasian bat lyssavirus (WCBV; NC025377), Shimoni bat lyssavirus (SHIBV; NC025365), Ikoma lyssavirus (IKOV; NC018629), Bokeloh bat lyssavirus (BBLV; NC025251), Lleida bat lyssavirus (LLEBV; NC031955), Gannoruwa bat lyssavirus (GBLV; NC031988), Taiwan bat lyssavirus (TWBLV; NC055474), Kotalahti bat lyssavirus (KBLV; LR994545), and Matlo bat lyssavirus (MBLV; MW653808). In addition to the aforementioned representative sequences, more lyssavirus sequences of each species in GenBank were employed, and a total of 233 sequences were used in this study (Table S2). The phylogroup III lyssaviruses were used as an outgroup in the phylogenetic tree.
Furthermore, the sequences of the corresponding coding regions (i.e., N gene, phosphoprotein gene, matrix protein gene, glycoprotein gene, and RNA-dependent RNA polymerase gene) of the identified lyssavirus and the representative sequences were aligned respectively, as described before. The percentage of nucleotide identities in each gene between the identified lyssavirus and the representative sequences were calculated using the MegAlign program of the Lasergene software version 7 (DNASTAR Inc., Madison, WI, USA).
The nucleotide and amino acid sequences of the N gene of ABLV from GenBank (Table S2) were used to verify the range of the intra-genotypic nucleotide and amino acid identities of lyssavirus. The sequences were aligned as described before and the identities between nucleotide sequences and between amino acid sequences were calculated using the Sequence Identity Matrix tool in BioEdit [38] to obtain the intra-genotypic identities of the analyzed lyssaviruses.
2.5. Histopathological Examination
When a lyssavirus-positive case was diagnosed, its tissues of central nerve system and salivary glands were investigated for the possible lesions. These fresh tissues were fixed in 10% buffered formalin, embedded in paraffin wax, and stained with hematoxylin and eosin for histopathological examination.
3. Results
3.1. Lyssavirus Surveillance
From 2018 to 2021, 407 bat specimens were received and tested by the Animal Health Research Institute. These specimens covered at least 13 bat species and came from 18 cities or counties in Taiwan (Table S3). Of the received specimens, 62.7% (N = 225) were Pipistrellus abramus, which is a bat species distributing throughout Island of Taiwan (Figure S1). Three of the 407 bat specimens were positive for lyssavirus, detected by RT-PCR and FAT. Confirming the bats’ species by DNA barcoding, two in 2018 and 2020 were P. abramus, and the other was identified in 2018 as Nyctalus plancyi velutinus, which was the first case in this species found infected in Taiwan. The lyssaviruses of the three positive cases were recovered by virus isolation and named as TWBLV-1/YiL/2018, TWBLV-1/KL/2020, and TWBLV-2/NT/2018, respectively. The results of the FAT, RT-PCR, and virus isolation of the positive cases were listed in Table S4. The isolates, bat species, location, and year of the bat lyssavirus found in Taiwan were listed in Table 1.

Table 1.
The Lyssaviruses isolated in bats in Taiwan during passive surveillance.
3.2. Complete Genome Sequencing and Phylogenetic Analysis
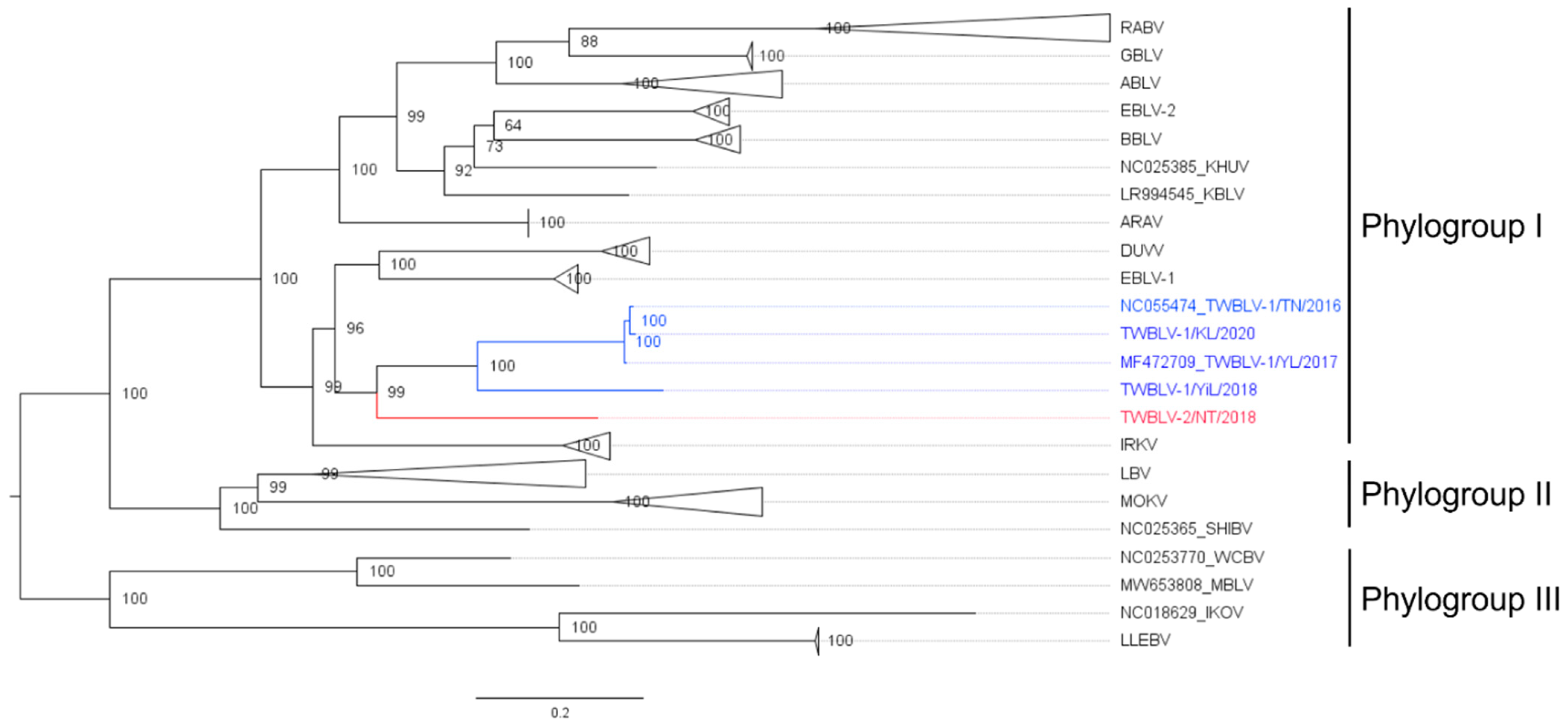
The length of the complete genome of the isolate TWBLV-2/NT/2018 was 11,990 nucleotides with the 43.17% G + C content. The nucleotide similarity between TWBLV-2/NT/2018 and the other lyssaviruses ranged 70.7–79.6% in the N gene and 63.3–76.2% in the concatenated coding genes (Table 2). Among the lyssaviruses, the N gene of TWBLV-2/NT/2018 shared the highest nucleotide identity with those of TWBLV (79.1~79.6%), EBLV-1 (79.1%), IRKV (78.1%), and DUVV (78%). The phylogenetic analysis demonstrated that TWBLV-2/NT/2018 was grouped into phylogroup I and most closely related to, but separate from, TWBLV (Figure 1). Following the species criteria of lyssavirus by the ICTV, TWBLV-2/NT/2018 was suggested to be a novel species of lyssavirus, designated as Taiwan bat lyssavirus 2 (TWBLV-2). As a new lyssavirus species discovered in a bat in Taiwan, the original TWBLV, which was identified in P. abramus, was renominated as Taiwan bat lyssavirus 1 (TWBLV-1) in this article.

Table 2.
The nucleotide identities (%) for the nucleoprotein (N), phosphoprotein (P), matrix protein (M), glycoprotein (G), RNA-dependent RNA polymerase (L) genes, and the concatenated coding genes (N + P + M + G + L) among Taiwan bat lyssavirus 2 and other lyssaviruses.
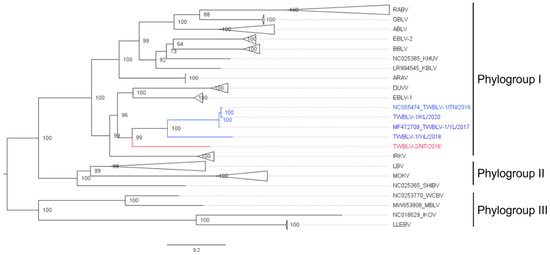
Figure 1.
Phylogenetic relationship of a novel lyssavirus (red font) identified in Nyctalus plancyi velutinus in 2018, Taiwan. The nucleotide sequences of representatives of all lyssavirus species and their isolates from GenBank were analyzed. The Taiwan bat lyssavirus 1 isolates discovered in Taiwan were marked in blue font. The maximum likelihood phylogeny was constructed with full-length nucleoprotein sequences by IQ-TREE using the best fit model of nucleotide substitution with 1000 ultrafast bootstrap. Numbers at the nodes indicate ultrafast bootstrap supports. The phylogroup III lyssavirus were used as the outgroup. A clade composed of the same lyssavirus species was collapsed, and the species names are labeled on the right. The scale bar indicates the number of substitutions per site. The detailed phylogenetic tree with accession numbers of all genomes is shown in Figure S2. ABLV, Australia bat lyssavirus; ARAV, Aravan lyssavirus; BBLV, Bokeloh bat lyssavirus; DUVV, Duvenhage lyssavirus; EBLV-1, European bat lyssavirus 1; EBLV-2, European bat lyssavirus 2; IKOV, Ikoma lyssavirus; IRKV, Irkut lyssavirus; KHUV, Khujand lyssavirus; KBLV, Kotalahti bat lyssavirus; LBV, Lagos bat lyssavirus; LLEBV, Lleida bat lyssavirus; MBLV, Matlo Bat Lyssavirus; MOKV, Mokola lyssavirus; SHIBV, Shimoni bat lyssavirus; RABV, Rabies lyssavirus; TWBLV-1, Taiwan bat lyssavirus 1; TWBLV-2, Taiwan bat lyssavirus 2; WCBV, West Caucasian bat lyssavirus.
The complete genome sequences of the isolates TWBLV-1/YiL/2018 and TWBLV-1/KL/2020 were compared to the TWBLV-1 isolates in 2016 and 2017. The nucleotide similarity between TWBLV-1/KL/2020 and TWBLV-1 identified in 2016 and 2017 was 98.5~99.0% in the N gene and 98.6~98.9% in the concatenated coding genes (Table 3). The genomic organization of the identified lyssavirus in this study are listed in Table S5.

Table 3.
(A) The intra-genotypic nucleotide and amino acid identities of the nucleoprotein gene of Taiwan bat lyssavirus 1 (TWBLV-1) isolates and Australian bat lyssavirus (ABLV) isolates. (B) The percent identities of nucleotide and amino acid (underlined) between the nucleoprotein gene of Taiwan bat lyssavirus 1 (TWBLV-1) and Taiwan bat lyssavirus 2 (TWBLV-2).
The nucleotide similarities between TWBLV-1/YiL/2018 and the other three TWBLV-1 isolates were 81.2~81.4% in the N gene and 79.7~79.8% in the concatenated coding genes, which fulfilled the criteria in 2018 (80–82% for the complete N gene or 80% for the concatenated coding regions). Therefore, the TWBLV-1/YiL/2018 could be a novel lyssavirus. In order to verify the range of intra-genotypic nucleotide and amino acid identities of the lyssavirus, the ABLV sequences were collected from GenBank, and further analysis was performed in this study. The intra-genotypic nucleotide and amino acid identities of the N gene were 83.6~100% and 95.5~100% among ABLV (Table 3), respectively. It was assumed that the isolate TWBLV-1/YiL/2018 belonged to TWBLV-1, and the intra-genotypic nucleotide and amino acid identities of the N gene among TWBLV-1 were 81.2~99% and 96.2~100%, which was consistent with those of RABV (93.7~99.0%), EBLV-1 (97.8–100%), EBLV-2 (97.3–99.8%), and ABLV (95.5~100%) [39].
3.3. Histopathological Examination
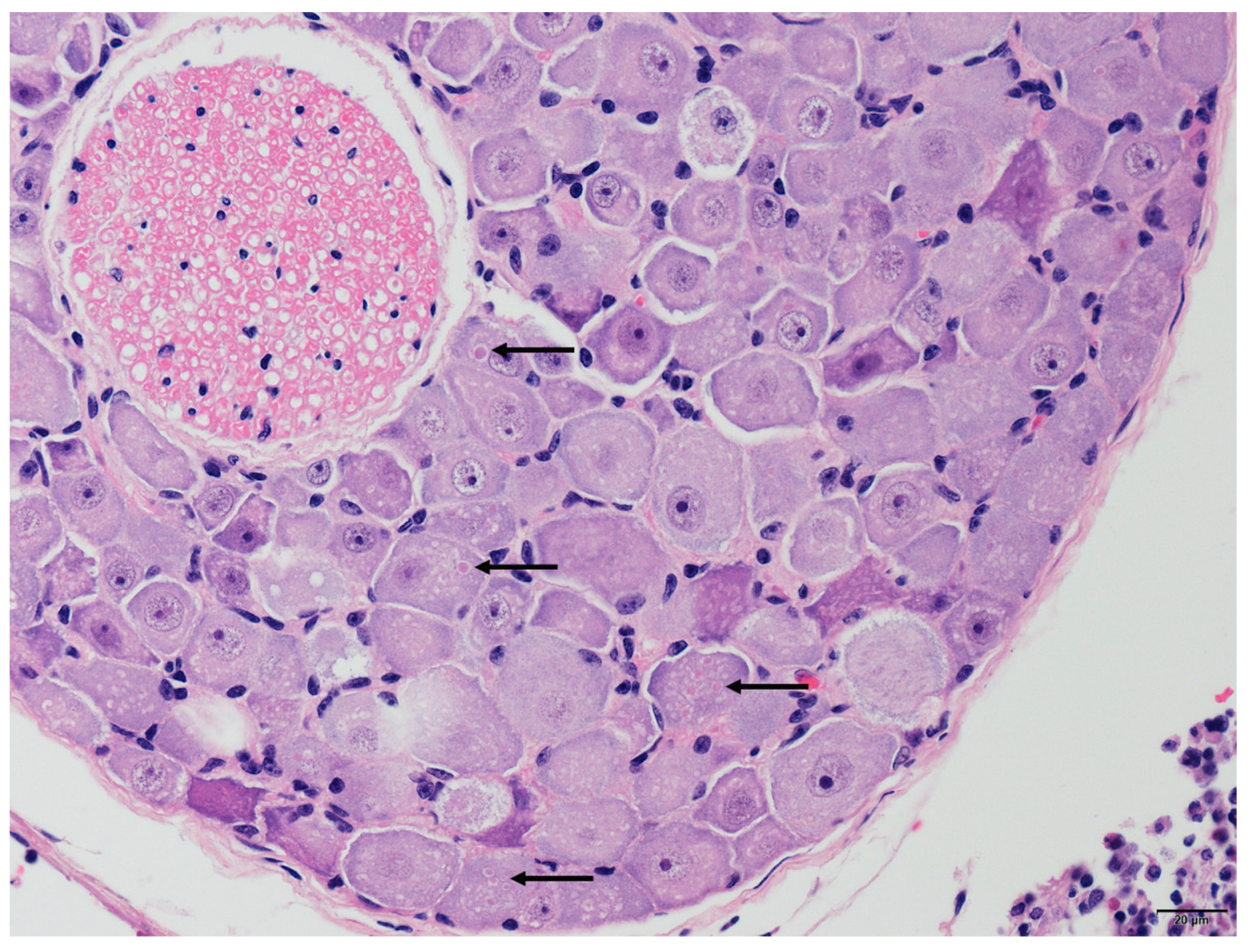
Two of the three positive cases, TWBLV-1/KL/2020 and TWBLV-2/NT/2018, were suitable for histopathological examination. The rest case was ruled out due to the fact of its severe degrees of postmortem changes of specimen. Neuronal degeneration and necrosis and perivascular cuffing with various numbers of mononuclear cells were observed in the cerebrum of N. velutinus from which TWBLV-2/NT/2018 was isolated. The pathogonomonic Negri bodies (ovoid, approximately 2 µm in length) were noted in the cytoplasm of neurons in the cerebrum of the N. velutinus and in the spinal ganglion of P. abramus from which TWBLV-1/KL/2020 was isolated (Figure 2). Lymphocytic infiltrations with varying degrees were also noted in the salivary glands in both cases.
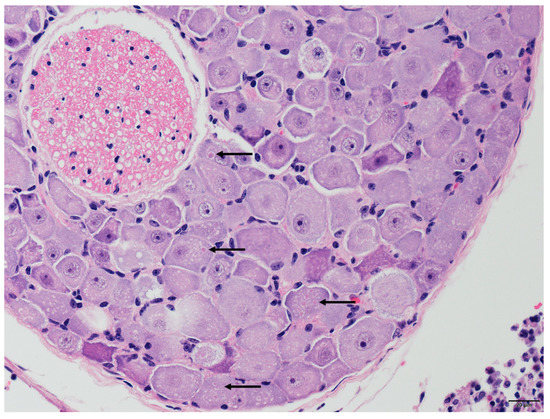
Figure 2.
Various sized, ovoid, intracytoplasmic eosinophilic inclusion bodies (Negri bodies, represented by the arrows) were noted in the neurons of the spinal ganglion infected by the Taiwan bat lyssavirus 1 isolate TWBLV-1/KL/2020.
4. Discussion
To the best of authors’ knowledge, we reported a novel lyssavirus, TWBLV-2, identified in Nyctalus plancyi velutinus. This lyssavirus had less than 80% nucleotide identity in the N gene with other lyssavirus species; thus, forming a separate branch in the phylogenetic analysis. The results revealed that the TWBLV-2 isolate in our study, TWBLV-2/NT/2018, met the new species criteria of lyssavirus by the ICTV. The phylogenetic tree based on the complete N gene showed that TWBLV-2/NT/2018 was clustered with TWBLV-1, EBLV-1, and DUVV and belonged to phylogroup I lyssaviruses.
A TWBLV-1 variant, TWBLV-1/YiL/2018, was found in a Pipistrellus abramus from Yilan County, Taiwan. Based on the high amino acid identity in nucleoprotein between TWBLV-1/YiL/2018 and other TWBLV-1 isolates (96.2%), in addition to the same host species (P. abramus) and same geographic distribution (Taiwan), we suggest that this isolate belongs to TWBLV-1 instead of a new species. Furthermore, the ICTV demarcation criteria of lyssavirus was revised down from 80–82% to 78–80% nucleotide identity for the complete N gene in 2021, thus also supporting that TWBLV-1/YiL/2018 was a variant of TWBLV-1. Unlike the variants of ABLV being identified in different bat species (Pteropus species and Saccolaimus flaviventris) [40], TWBLV-1 was identified only in P. abramus. Geographically, the TWBLV-1/YiL/2018 was the only TWBLV-1 isolate found in the eastern Taiwan (Figure S1). The other TWBLV-1 isolates, which were found in western Taiwan, revealed 98.5~99% nucleotide similarity in the N gene. There are mountains along central Taiwan Island from north to south. The mountainous geographic barrier with a high elevation between the eastern and western regions may contribute to the independent evolution of TWBLV-1 in P. abramus and, thus, results in this genetic diversity. The distinct clades of rabies lyssavirus based on geographical separation has been mentioned [41,42]; likewise, the geographical barrier causes the host genetic discontinuity [43].
In the present study, we reported that Nyctalus plancyi velutinus could be a potential reservoir of the TWBLV-2 by one identified case only. The more surveillance of N. velutinus that is performed, the more evidence will be brought up to clarify whether N. velutinus is a reservoir of TWBLV-2 or just incidentally infected. N. velutinus is the subspecies of Nyctalus plancyi, which is distributed in eastern China, Taiwan, and Luzon of the Philippines [44,45]. In Taiwan, N. velutinus is widely distributed from low to middle altitude areas; it is not a commonly observed bat species, with an unknown population size. A total number of 14 roadkill records of N. velutinus were received from the Taiwan Roadkill Observation Network during 2011~2021 [46]. Similarly, we received only four N. velutinus cases (4/407, 1%) during 2018~2021. This indicates that N. velutinus is a rarely observable species in the country. Even though the possibility of bat–human contact is rare, and only one TWBLV-2 isolate was found in Taiwan, our findings imply that the surrounding East Asian countries may also be exposed to a similar risk. N. velutinus, with similar potential long migration ability as other bats in the Nyctalus genus (N. noctule and N. leisleri) [47,48], may be a competent reservoir and responsible transmitter for international transmission of bat lyssaviruses.
The reservoir of TWBLV-1 was Pipistrellus abramus in which four cases were confirmed in Taiwan. P. abramus is a common insectivorous, non-migratory bat of low-altitude, urban areas in East Asia [49]. The prevalence of TWBLV-1 in P. abramus was (2/225, 0.89%) in 2018~2021. P. abramus was the bat species most commonly discovered as a victim of roadkill in Taiwan [50]. Moreover, 62.7% of received bat’s species were P. abramus in this study, reflecting that this species has high contact possibility with people in Taiwan. The awareness of public health should be promoted to avoid bat–human contact so as to prevent human cases.
Infection with either TWBLV-1 or TWBLV-2 in bats could develop histopathologic lesions in the central nervous system of infected bats. Even though no clear neurological symptoms were recorded in the cases, we observed characteristic Negri bodies within the neurons of the TWBLV-1 and TWBLV-2 infected bats. In the previously studies, aggression and attack behaviors were noted in experimentally inoculated bats, and some of the infected bats became emaciated and then died [51,52,53]. To further investigate clinical symptoms, the incubation period, and transmission route of Taiwan bat lyssaviruses in bats, advanced studies are required to understand the pathogenicity and pathogenesis of these bat lyssaviruses in bats.
This passive surveillance system extensively broadened our understanding of bat lyssaviruses in Taiwan. Meanwhile, it had limitations and/or bias in collecting samples of uncommon species. Though some species had few or had never been received and tested for lyssavirus in our study under the passive surveillance system, this surveillance still provided valuable information. Several surveillance systems implemented in Europe and Asia [7,20,54,55,56,57,58] indicate that passive surveillance is more suitable for identifying lyssavirus-infected bats compared to active surveillance on swabs of healthy bats [54]. Once the target of a bat species can be determined by passive surveillance, further research on pathogen monitoring, virus transmission between colonies, or seroprevalence can be conducted. Furthermore, the associated lyssavirus surveillance in other countries with the specific bat species will be encouraged to fill the gap of lyssavirus distribution in bats around the world.
In conclusion, a bat lyssavirus, designated as TWBLV-2, was newly isolated and identified in N. velutinus. The virus isolate fulfilled the criteria as a putatively novel species of the genus lyssavirus, and its epidemiology and pathogenicity deserve further studies.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/v14071562/s1, Table S1: The modified primer used in the whole genome sequence for the isolates in this study; Table S2: The reference sequence of the lyssavirus species included in the phylogenetic tree construction; Table S3: The details of the number and bat species tested in the surveillance between 2018 and 2021; Table S4: The results of the positive cases, employed with different diagnostic methods; Table S5: The genomic organization of the identified lyssaviruses in Taiwan; Figure S1: Map of the Island of Taiwan, showing the tested number of Pipistrellus abramus during 2018~2021 by counties/cities; Figure S2: A detailed phylogenetic tree of the complete nucleoprotein gene of lyssaviruses.
Author Contributions
Conceptualization, S.-C.H., W.-C.H. and F.L.; methodology, W.-C.H., Y.-W.C., Y.-C.T. and J.-C.C.; validation, S.-C.H., Y.-C.T. and W.-C.H.; formal analysis, S.-C.H.; investigation, S.-C.H. and W.-C.H.; resources, C.-L.H.; data curation, S.-C.H.; writing—original draft preparation, S.-C.H.; writing—review and editing, F.L. and W.-C.H.; visualization, S.-C.H.; supervision, F.L. and W.-C.H.; project administration, W.-C.H.; funding acquisition, W.-C.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by grants from the Bureau of Animal and Plant Health Inspection and Quarantine, Council of Agriculture, Executive Yuan, Taiwan. The funding numbers were as following: 107AS-8.7.1-BQ-B2(1), 108AS-8.6.1-BQ-B2(1), 109AS-8.6.1-BQ-B1(1), and 110-AS-5.1.5-BQ-B1(2).
Institutional Review Board Statement
Not applicable, because only found dead animals were tested in this study.
Informed Consent Statement
Not applicable.
Data Availability Statement
All the data pertaining to the study are available in the manuscript.
Acknowledgments
The acknowledgement is made in great memory of Ming-Shiuh Lee for his help in improving the process on whole genome sequence and wildlife disease surveillance. Special thanks to Tien-Cheng Li for his help in improving the manuscript. The authors thank the colleagues of non-governmental organizations and local animal disease inspection authorities for collecting and delivering samples.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kuzmin, I.V. Chapter One-Basic Facts about Lyssaviruses. In Current Laboratory Techniques in Rabies Diagnosis, Research and Prevention; Rupprecht, C., Nagarajan, T., Eds.; Academic Press: Amsterdam, The Netherlands, 2014; Volume 1, pp. 3–21. ISBN 978-0-12-800014-4. [Google Scholar]
- Markotter, W.; Coertse, J. Bat lyssaviruses. Rev. Sci. Technol. 2018, 37, 385–400. [Google Scholar] [CrossRef] [PubMed]
- Shipley, R.; Wright, E.; Selden, D.; Wu, G.; Aegerter, J.; Fooks, A.R.; Banyard, A.C. Bats and viruses: Emergence of novel lyssaviruses and association of bats with viral zoonoses in the EU. Trop. Med. Infect. Dis. 2019, 4, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, S.-C.; Hsu, C.-L.; Lee, M.-S.; Tu, Y.-C.; Chang, J.-C.; Wu, C.-H.; Lee, S.-H.; Ting, L.-J.; Tsai, K.-R.; Cheng, M.-C.; et al. Lyssavirus in Japanese Pipistrelle, Taiwan. Emerg. Infect. Dis. 2018, 24, 782–785. [Google Scholar] [CrossRef] [Green Version]
- Nokireki, T.; Tammiranta, N.; Kokkonen, U.M.; Kantala, T.; Gadd, T. Tentative novel lyssavirus in a bat in Finland. Transbound. Emerg. Dis. 2018, 65, 593–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coertse, J.; Grobler, C.S.; Sabeta, C.T.; Seamark, E.C.J.; Kearney, T.; Paweska, J.T.; Markotter, W. Lyssaviruses in Insectivorous Bats, South Africa, 2003-2018. Emerg. Infect. Dis. 2020, 26, 3056–3060. [Google Scholar] [CrossRef] [PubMed]
- Gunawardena, P.S.; Marston, D.A.; Ellis, R.J.; Wise, E.L.; Karawita, A.C.; Breed, A.C.; McElhinney, L.M.; Johnson, N.; Banyard, A.C.; Fooks, A.R. Lyssavirus in Indian Flying Foxes, Sri Lanka. Emerg. Infect. Dis. 2016, 22, 1456–1459. [Google Scholar] [CrossRef]
- Genus: Lyssavirus-Rhabdoviridae-Negative-Sense RNA Viruses-ICTV. Available online: https://talk.ictvonline.org/ictv-reports/ictv_online_report/negative-sense-rna-viruses/w/rhabdoviridae/795/genus-lyssavirus (accessed on 26 April 2022).
- Fooks, A.R.; Cliquet, F.; Finke, S.; Freuling, C.; Hemachudha, T.; Mani, R.S.; Müller, T.; Nadin-Davis, S.; Picard-Meyer, E.; Wilde, H.; et al. Rabies. Nat. Rev. Dis. Prim. 2017, 3, 17091. [Google Scholar] [CrossRef]
- Badrane, H.; Bahloul, C.; Perrin, P.; Tordo, N. Evidence of Two Lyssavirus Phylogroups with Distinct Pathogenicity and Immunogenicity. J. Virol. 2001, 75, 3268–3276. [Google Scholar] [CrossRef] [Green Version]
- Nolden, T.; Banyard, A.C.; Finke, S.; Fooks, A.R.; Hanke, D.; Höper, D.; Horton, D.L.; Mettenleiter, T.C.; Müller, T.; Teifke, J.P.; et al. Comparative studies on the genetic, antigenic and pathogenic characteristics of Bokeloh bat lyssavirus. J. Gen. Virol. 2014, 95, 1647–1653. [Google Scholar] [CrossRef] [Green Version]
- Brookes, S.M.; Parsons, G.; Johnson, N.; McElhinney, L.M.; Fooks, A.R. Rabies human diploid cell vaccine elicits cross-neutralising and cross-protecting immune responses against European and Australian bat lyssaviruses. Vaccine 2005, 23, 4101–4109. [Google Scholar] [CrossRef]
- Nokireki, T.; Jakava-Viljanen, M.; Virtala, A.M.; Sihvonen, L. Efficacy of rabies vaccines in dogs and cats and protection in a mouse model against European bat lyssavirus type 2. Acta Vet. Scand. 2017, 59, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Servat, A.; Wasniewski, M.; Cliquet, F. Cross-protection of inactivated rabies vaccines for veterinary use against bat lyssaviruses occurring in Europe. Viruses 2019, 11, 936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marston, D.A.; Horton, D.L.; Ngeleja, C.; Hampson, K.; McElhinney, L.M.; Banyard, A.C.; Haydon, D.; Cleaveland, S.; Rupprecht, C.E.; Bigambo, M.; et al. Ikoma lyssavirus, highly divergent novel lyssavirus in an African civet. Emerg. Infect. Dis. 2012, 18, 664–667. [Google Scholar] [CrossRef] [PubMed]
- Banyard, A.C.; Evans, J.S.; Luo, T.R.; Fooks, A.R. Lyssaviruses and bats: Emergence and zoonotic threat. Viruses 2014, 6, 2974–2990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tenzin; Ward, M.P. Review of Rabies Epidemiology and Control in South, South East and East Asia: Past, Present and Prospects for Elimination. Zoonoses Public Health 2012, 59, 451–467. [Google Scholar] [CrossRef] [PubMed]
- Miranda, M.E.G.; Miranda, N.L.J. Rabies Prevention in Asia: Institutionalizing Implementation Capacities. In Rabies and Rabies Vaccines; Springer International Publishing: Berlin/Heidelberg, Germany, 2020; pp. 103–116. [Google Scholar]
- Kuzmin, I.V.; Orciari, L.A.; Arai, Y.T.; Smith, J.S.; Hanlon, C.A.; Kameoka, Y.; Rupprecht, C.E. Bat lyssaviruses (Aravan and Khujand) from Central Asia: Phylogenetic relationships according to N, P and G gene sequences. Virus Res. 2003, 97, 65–79. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, S.; Zhao, J.; Zhang, F.; Hu, R. Isolation of Irkut virus from a Murina leucogaster bat in China. PLoS Negl. Trop. Dis. 2013, 7, e2097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiou, H.Y.; Hsieh, C.H.; Jeng, C.R.; Chan, F.T.; Wang, H.Y.; Pang, V.F. Molecular characterization of cryptically circulating rabies virus from ferret badgers, Taiwan. Emerg. Infect. Dis. 2014, 20, 790–798. [Google Scholar] [CrossRef]
- Chang, J.C.; Tsai, K.J.; Hsu, W.C.; Tu, Y.C.; Chuang, W.C.; Chang, C.Y.; Chang, S.W.; Lin, T.E.; Fang, K.Y.; Chang, Y.F.; et al. Rabies virus infection in ferret badgers (Melogale moschata subaurantiaca) in Taiwan: A retrospective study. J. Wildl. Dis. 2015, 51, 923–928. [Google Scholar] [CrossRef] [PubMed]
- Hsu, W.C.; Hsu, C.L.; Tu, Y.C.; Chang, J.C.; Tsai, K.R.; Lee, F.; Hu, S.C. Standard operating procedure for lyssavirus surveillance of the bat population in taiwan. J. Vis. Exp. 2019, 2019, e59421. [Google Scholar] [CrossRef] [PubMed]
- Hayman, D.T.S.; Banyard, A.C.; Wakeley, P.R.; Harkess, G.; Marston, D.; Wood, J.L.N.; Cunningham, A.A.; Fooks, A.R. A universal real-time assay for the detection of Lyssaviruses. J. Virol. Methods 2011, 177, 87–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franka, R.; Constantine, D.G.; Kuzmin, I.; Velasco-Villa, A.; Reeder, S.A.; Streicker, D.; Orciari, L.A.; Wong, A.J.; Blanton, J.D.; Rupprecht, C.E. A new phylogenetic lineage of Rabies virus associated with western pipistrelle bats (Pipistrellus hesperus). J. Gen. Virol. 2006, 87, 2309–2321. [Google Scholar] [CrossRef] [PubMed]
- Trimarchi, C.V.; Smith, J.S. 9-Diagnostic Evaluation. In Rabies; Jackson, A.C., Wunner, W.H., Eds.; Academic Press: San Diego, CA, USA, 2003; pp. 307–349. ISBN 978-0-12-379077-4. [Google Scholar]
- Corbet, G.B.; Hill, J.E. The Mammals of the Indomalayan Region: A Systematic Review; Oxford University Press: Oxford, UK, 1992; Volume 488. [Google Scholar]
- Huang, J.C.C.; Ho, Y.Y.; Kuo, H.C. Illustrated field keys to the bats (Mammalia: Chiroptera) of Taiwan. J. Threat. Taxa 2020, 12, 15675–15710. [Google Scholar] [CrossRef]
- Ivanova, N.V.; Zemlak, T.S.; Hanner, R.H.; Hebert, P.D.N. Universal primer cocktails for fish DNA barcoding. Mol. Ecol. Notes 2007, 7, 544–548. [Google Scholar] [CrossRef]
- Nadin-Davis, S.A. DNA Barcoding Facilitates Bat Species Identification for Improved Surveillance of Bat-associated Rabies across Canada. Open Zool. J. 2012, 5, 27–37. [Google Scholar] [CrossRef]
- Kress, W.J.; Erickson, D.L. DNA barcodes: Methods and protocols. Methods Mol. Biol. 2012, 858, 3–8. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [Green Version]
- Chernomor, O.; VonHaeseler, A.; Minh, B.Q. Terrace Aware Data Structure for Phylogenomic Inference from Supermatrices. Syst. Biol. 2016, 65, 997–1008. [Google Scholar] [CrossRef] [Green Version]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [Green Version]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT—ScienceOpen. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Marston, D.A.; McElhinney, L.M.; Johnson, N.; Müller, T.; Conzelmann, K.K.; Tordo, N.; Fooks, A.R. Comparative analysis of the full genome sequence of European bat lyssavirus type 1 and type 2 with other lyssaviruses and evidence for a conserved transcription termination and polyadenylation motif in the G-L 3′ non-translated region. J. Gen. Virol. 2007, 88, 1302–1314. [Google Scholar] [CrossRef]
- Gould, A.R.; Kattenbelt, J.A.; Gumley, S.G.; Lunt, R.A. Characterisation of an Australian bat lyssavirus variant isolated from an insectivorous bat. Virus Res. 2002, 89, 1–28. [Google Scholar] [CrossRef]
- McElhinney, L.M.; Marston, D.A.; Wise, E.L.; Freuling, C.M.; Bourhy, H.; Zanoni, R.; Moldal, T.; Kooi, E.A.; Neubauer-Juric, A.; Nokireki, T.; et al. Molecular epidemiology and evolution of european bat lyssavirus 2. Int. J. Mol. Sci. 2018, 19, 156. [Google Scholar] [CrossRef] [Green Version]
- Saito, M.; Oshitani, H.; Orbina, J.R.C.; Tohma, K.; de Guzman, A.S.; Kamigaki, T.; Demetria, C.S.; Manalo, D.L.; Noguchi, A.; Inoue, S.; et al. Genetic Diversity and Geographic Distribution of Genetically Distinct Rabies Viruses in the Philippines. PLoS Negl. Trop. Dis. 2013, 7, e2144. [Google Scholar] [CrossRef]
- Brown, R.P.; Suárez, N.M.; Pestano, J. The Atlas mountains as a biogeographical divide in North–West Africa: Evidence from mtDNA evolution in the Agamid lizard Agama impalearis. Mol. Phylogenet. Evol. 2002, 24, 324–332. [Google Scholar] [CrossRef]
- Wilson, D.E.; Reeder, D.M. Mammal Species of the World: A Taxonomic and Geographic Reference; JHU Press: Baltimore, MD, USA, 2005; Volume 1, ISBN 0801882214. [Google Scholar]
- Shi, H.Y.; Yu, W.; Wu, Y. Nyctalus plancyi (Chinese Noctule) The IUCN Red List of Threatened Species 2020: E.T136828A22044480. Available online: https://www.iucnredlist.org/species/136828/22044480 (accessed on 6 July 2021).
- Nyctalus plancyi velutinus Taiwan Roadkill Observation Network. Available online: https://v2019.roadkill.tw/bio-taxon/nyctalus-plancyi-velutinus (accessed on 25 March 2022).
- Petit, E.; Mayer, F. Male dispersal in the noctule bat (Nyctalus noctula): Where are the limits? Proc. R. Soc. Lond. Ser. B Biol. Sci. 1999, 266, 1717–1722. [Google Scholar] [CrossRef] [Green Version]
- Wohlgemuth, R.; Devrient, I.; García, A.; Hutterer, R. Long-distance flight of a Lesser noctule (Nyctalus leisleri) after rehabilitation. Myotis 2004, 41, 69–73. [Google Scholar]
- Fukui, D.; Sano, A. Pipistrellus abramus. The IUCN Red List of Threatened Species 2019: E.T17320A22131948. Available online: https://www.iucnredlist.org/species/17320/22131948 (accessed on 6 July 2021).
- Huang, J.C.-C.; Chen, W.-J.; Lin, T.-E. Landscape and Species Traits Co-Drive Roadkills of Bats in a Subtropical Island. Divers 2021, 13, 117. [Google Scholar] [CrossRef]
- Kuzmin, I.V.; Botvinkin, A.D. The behaviour of bats Pipistrellus pipistrellus after experimental inoculation with rabies and rabies-like viruses and some aspects of pathogenesis. Myotis 1996, 34, 93–99. [Google Scholar]
- Johnson, N.; Vos, A.; Neubert, L.; Freuling, C.; Mansfield, K.L.; Kaipf, I.; Denzinger, A.; Hicks, D.; Núñez, A.; Franka, R.; et al. Experimental study of European bat lyssavirus type-2 infection in Daubenton’s bats (Myotis daubentonii). J. Gen. Virol. 2008, 89, 2662–2672. [Google Scholar] [CrossRef] [PubMed]
- Freuling, C.; Vos, A.; Johnson, N.; Kaipf, I.; Denzinger, A.; Neubert, L.; Mansfield, K.; Hicks, D.; Nuñez, A.; Tordo, N.; et al. Experimental infection of serotine bats (Eptesicus serotinus) with European bat lyssavirus type 1a. J. Gen. Virol. 2009, 90, 2493–2502. [Google Scholar] [CrossRef] [PubMed]
- Schatz, J.; Fooks, A.R.; McElhinney, L.; Horton, D.; Echevarria, J.; Vázquez-Moron, S.; Kooi, E.A.; Rasmussen, T.B.; Müller, T.; Freuling, C.M. Bat Rabies Surveillance in Europe. Zoonoses Public Health 2013, 60, 22–34. [Google Scholar] [CrossRef]
- Lumlertdacha, B.; Boongird, K.; Wanghongsa, S.; Wacharapluesadee, S.; Chanhome, L.; Khawplod, P.; Hemachudha, T.; Kuzmin, I.; Rupprecht, C.E. Survey for bat lyssaviruses, Thailand. Emerg. Infect. Dis. 2005, 11, 232–236. [Google Scholar] [CrossRef]
- Arguin, P.M.; Murray-Lillibridge, K.; Miranda, M.E.G.; Smith, J.S.; Calaor, A.B.; Rupprecht, C.E. Serologic evidence of Lyssavirus infections among bats, the Philippines. Emerg. Infect. Dis. 2002, 8, 258–262. [Google Scholar] [CrossRef] [Green Version]
- Kuzmin, I.V.; Niezgoda, M.; Carroll, D.S.; Keeler, N.; Hossain, M.J.; Breiman, R.F.; Ksiazek, T.G.; Rupprecht, C.E. Lyssavirus surveillance in bats, Bangladesh. Emerg. Infect. Dis. 2006, 12, 486–488. [Google Scholar] [CrossRef] [PubMed]
- Klein, A.; Calvelage, S.; Schlottau, K.; Hoffmann, B.; Eggerbauer, E.; Müller, T.; Freuling, C.M. Retrospective Enhanced Bat Lyssavirus Surveillance in Germany between 2018–2020. Viruses 2021, 13, 1538. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).