Human Platelet Lysate Induces Antiviral Responses against Parechovirus A3
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Line, Serum, and Serum Substitute Preparation
2.2. Viruses and Reagents
2.3. Cell Proliferation Assay
2.4. RNA Extraction and RT-qPCR
2.5. Immunofluorescence Assay
2.6. Immunoblotting Analysis
2.7. STAT1 Knockdown
2.8. Statistical Analysis
3. Results
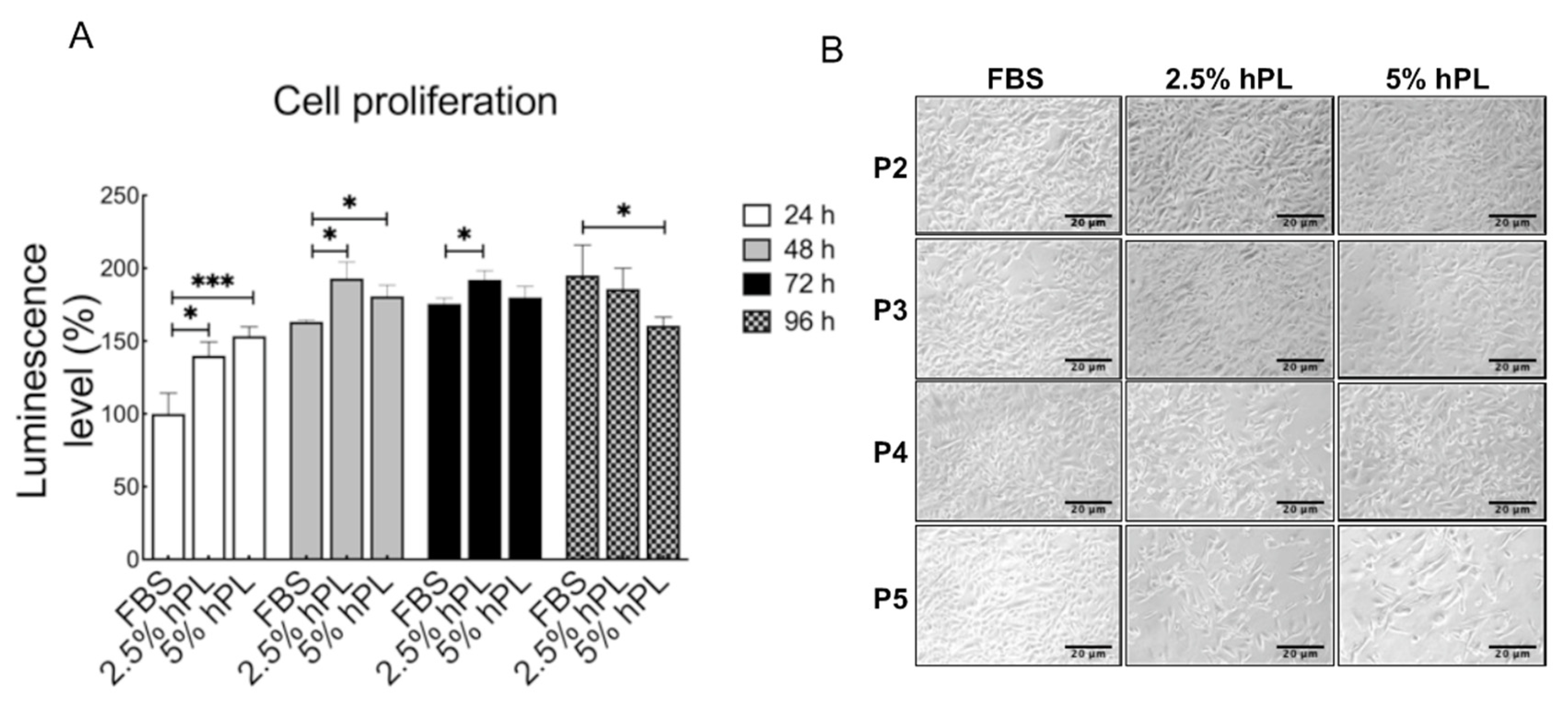
3.1. Comparison of GBM Growth Status between hPL and FBS Culture Mediums
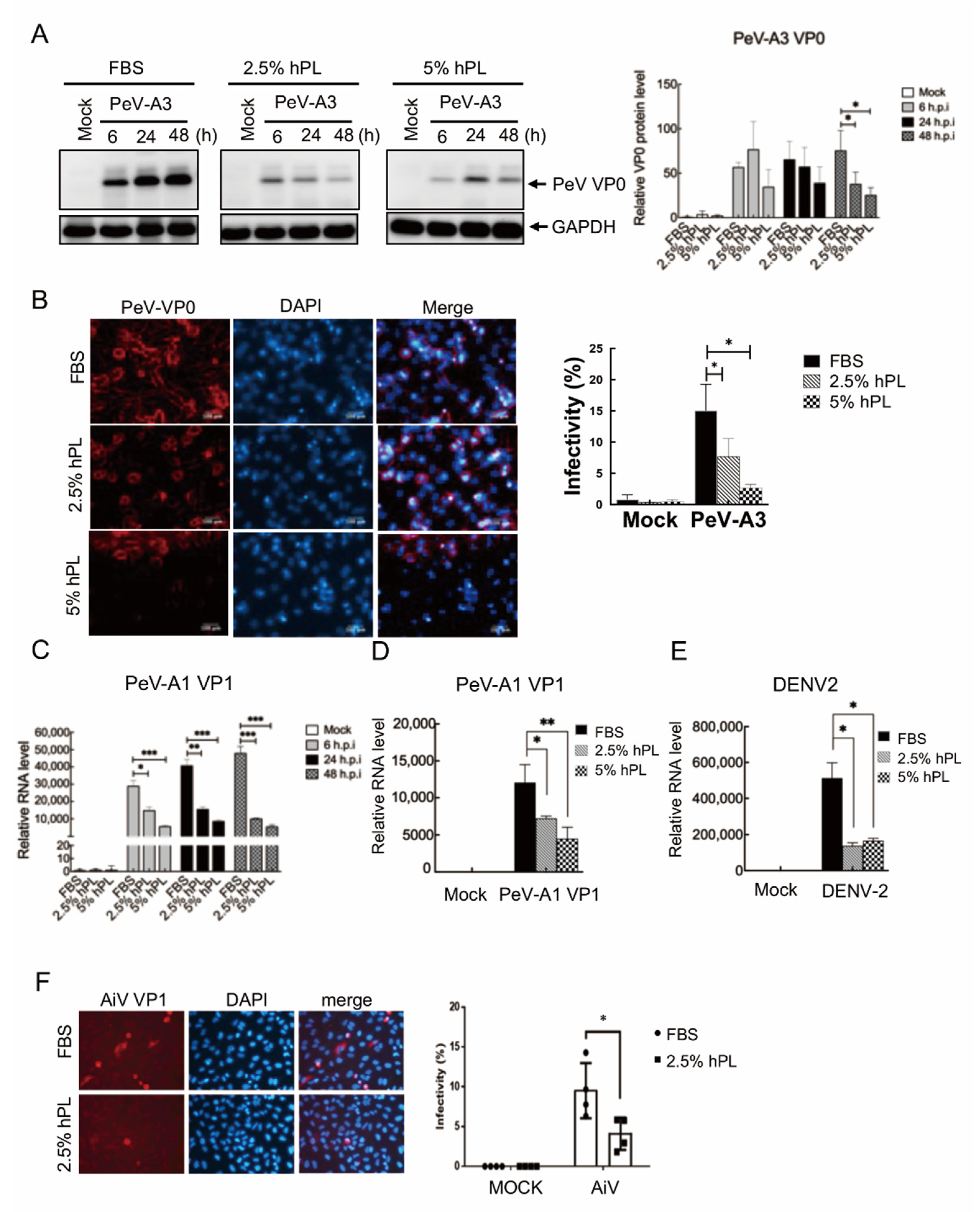
3.2. Downregulation of Viral Replication in hPL Culture Medium-Maintained GBM Cells
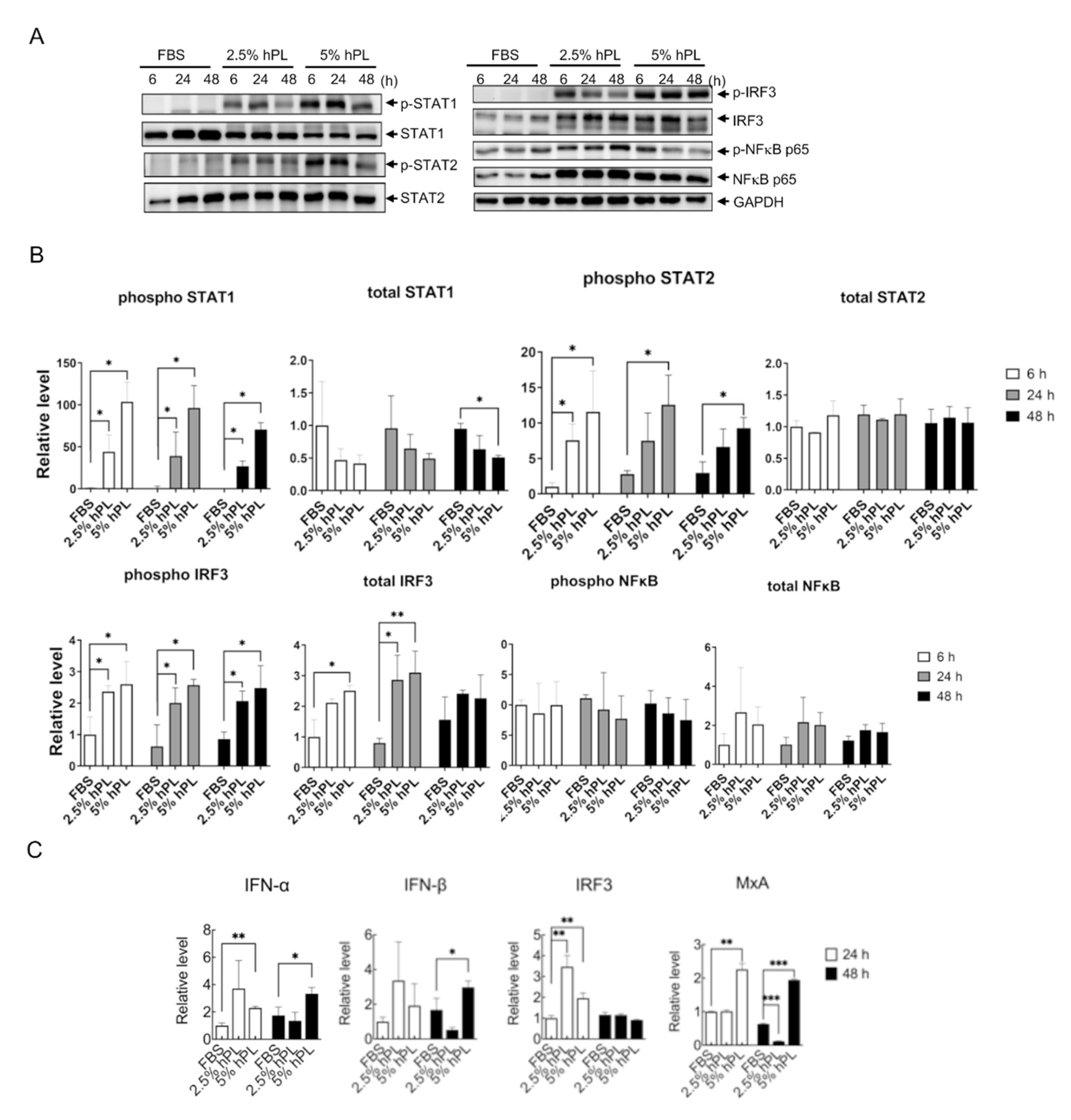
3.3. hPL Induced Type I Interferon Response in GBM Cells
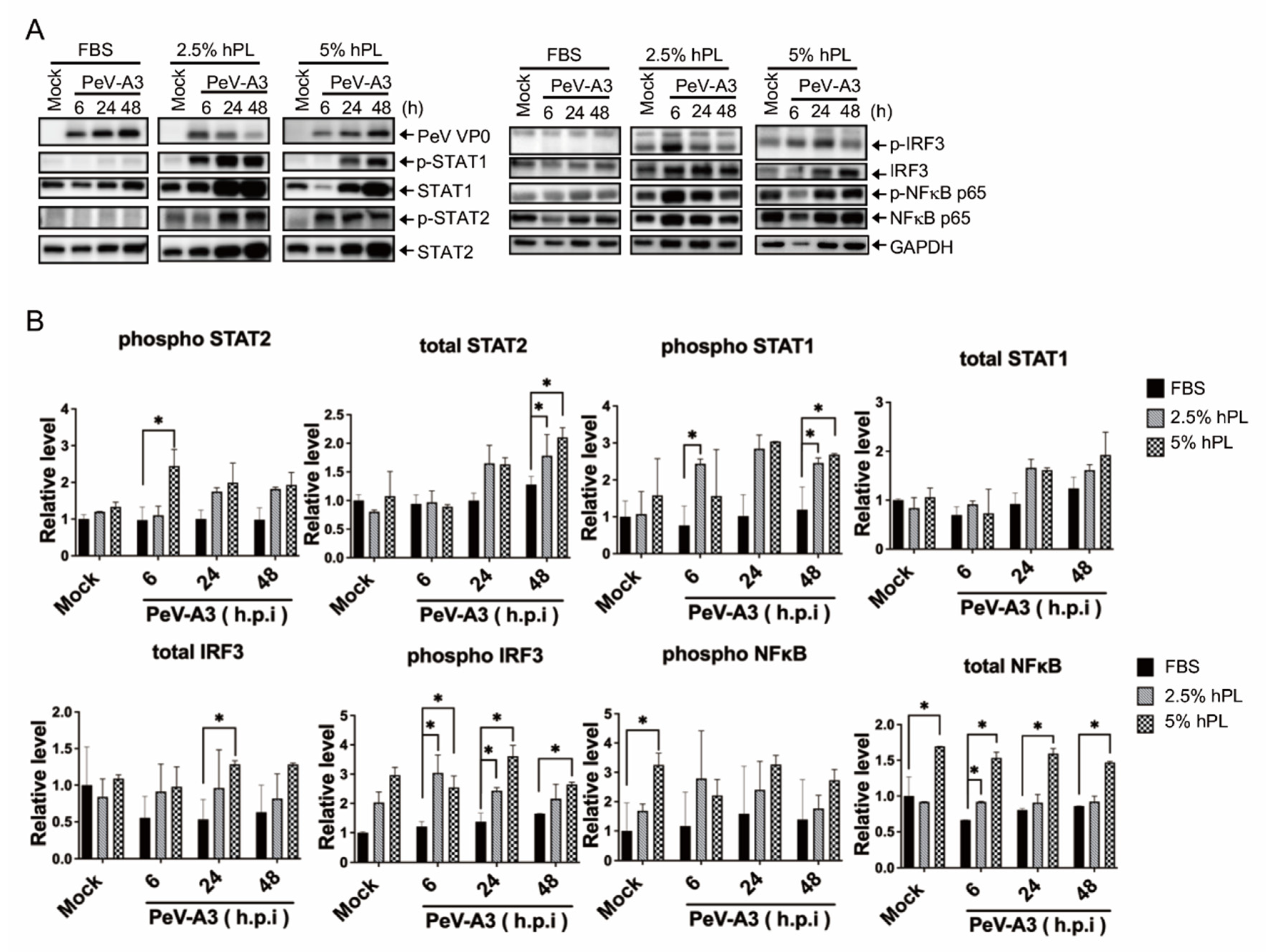
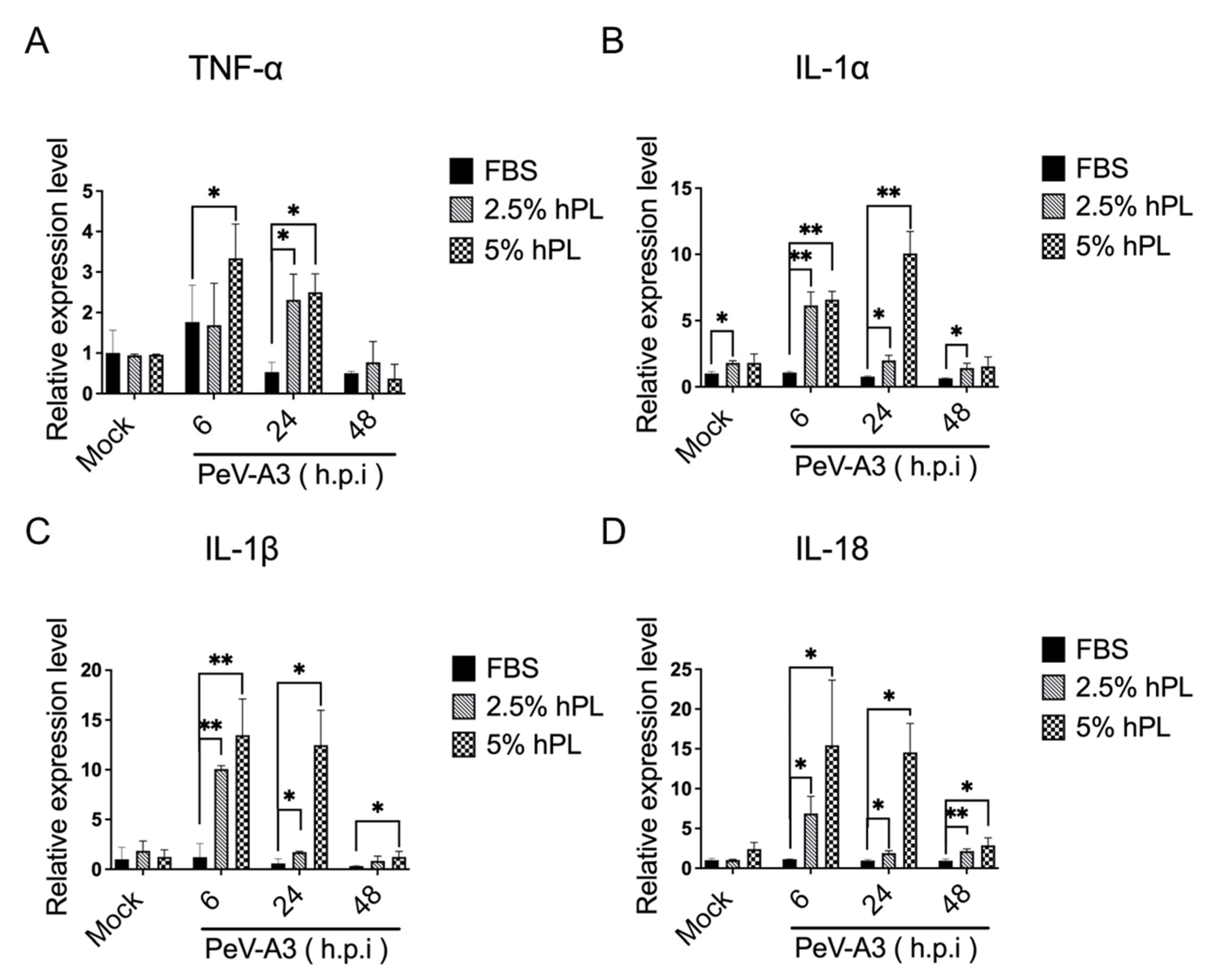
3.4. hPL Enhanced IFN Response in PeV-A3-Infected GBM Cells
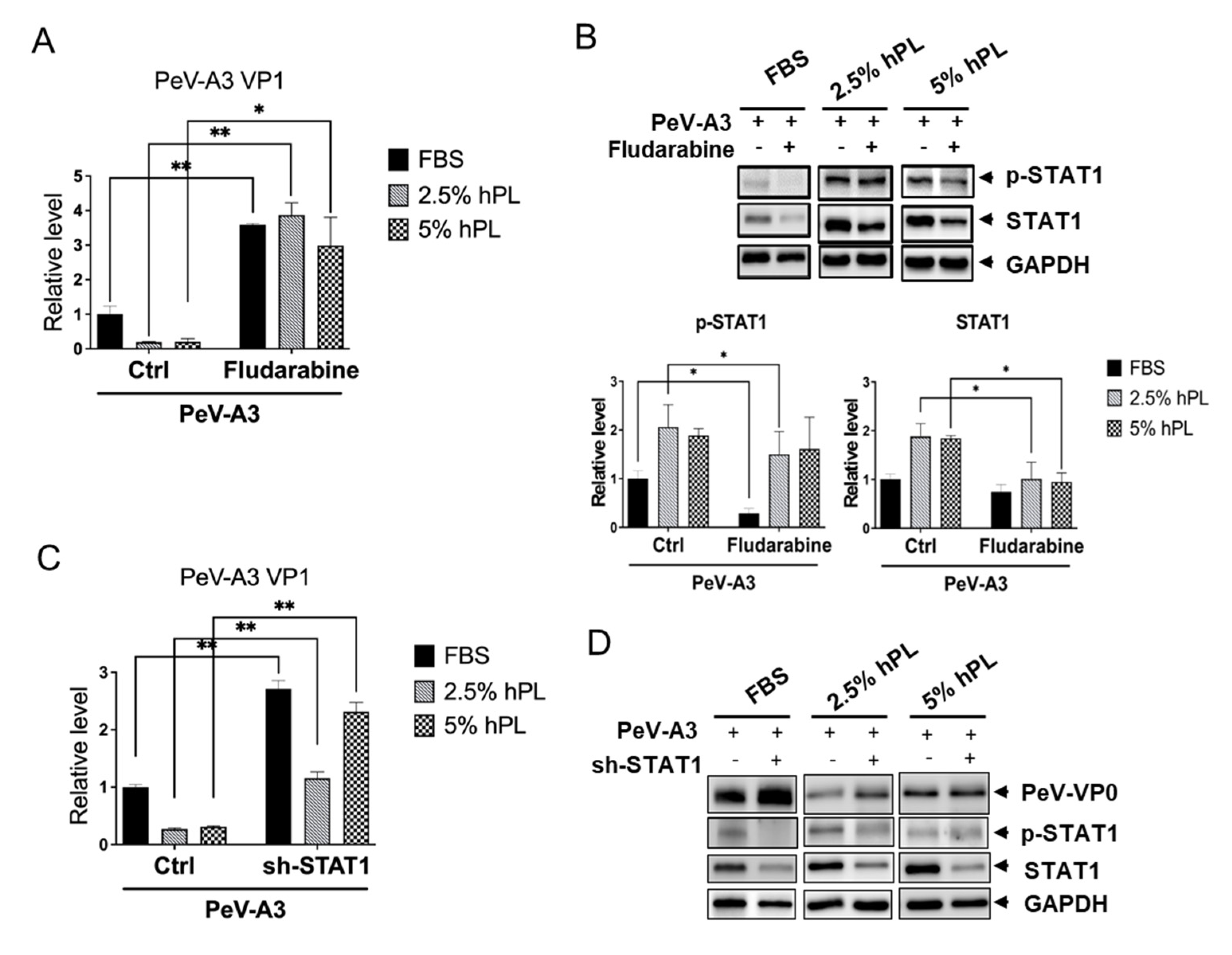
3.5. STAT1 Inhibitor Increased PeV-A3 Infection in hPL Medium-Cultured GBM Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fisher, H.W.; Puck, T.T.; Sato, G. Molecular growth requirements of single mammalian cells: The action of fetuin in promoting cell attachment to glass. Proc. Natl. Acad. Sci. USA 1958, 44, 4–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, B.C.; Chang, J.T.; Huang, T.S.; Chen, J.J.; Chen, Y.S.; Jan, M.W.; Chang, T.H. Parechovirus A Detection by a Comprehensive Approach in a Clinical Laboratory. Viruses 2018, 10, 711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kung, M.H.; Jan, M.W.; Chen, J.J.; Shieh, Y.C.; Chang, T.H. Detection of Parechovirus A1 with Monoclonal Antibody against Capsid Protein VP0. Microorganisms 2020, 8, 1794. [Google Scholar] [CrossRef] [PubMed]
- Pecora, A.; Perez Aguirreburualde, M.S.; Ridpath, J.F.; Dus Santos, M.J. Molecular Characterization of Pestiviruses in Fetal Bovine Sera Originating From Argentina: Evidence of Circulation of HoBi-Like Viruses. Front. Vet. Sci. 2019, 6, 359. [Google Scholar] [CrossRef]
- Jochems, C.E.A.; van der Valk, J.B.F.; Stafleu, F.R.; Baumans, V. The Use of Fetal Bovine Serum: Ethical or Scientific Problem? Altern. Lab. Anim. 2002, 30, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Subbiahanadar Chelladurai, K.; Selvan Christyraj, J.D.; Rajagopalan, K.; Yesudhason, B.V.; Venkatachalam, S.; Mohan, M.; Chellathurai Vasantha, N.; Selvan Christyraj, J.R.S. Alternative to FBS in animal cell culture—An overview and future perspective. Heliyon 2021, 7, e07686. [Google Scholar] [CrossRef] [PubMed]
- Semple, J.W.; Italiano, J.E., Jr.; Freedman, J. Platelets and the immune continuum. Nat. Rev. Immunol. 2011, 11, 264–274. [Google Scholar] [CrossRef]
- Burnouf, T.; Strunk, D.; Koh, M.B.; Schallmoser, K. Human platelet lysate: Replacing fetal bovine serum as a gold standard for human cell propagation? Biomaterials 2016, 76, 371–387. [Google Scholar] [CrossRef]
- Golebiewska, E.M.; Poole, A.W. Platelet secretion: From haemostasis to wound healing and beyond. Blood Rev. 2015, 29, 153–162. [Google Scholar] [CrossRef] [Green Version]
- Nebie, O.; Carvalho, K.; Barro, L.; Delila, L.; Faivre, E.; Renn, T.Y.; Chou, M.L.; Wu, Y.W.; Nyam-Erdene, A.; Chou, S.Y.; et al. Human platelet lysate biotherapy for traumatic brain injury: Preclinical assessment. Brain 2021, 144, 3142–3158. [Google Scholar] [CrossRef]
- Copland, I.B.; Garcia, M.A.; Waller, E.K.; Roback, J.D.; Galipeau, J. The effect of platelet lysate fibrinogen on the functionality of MSCs in immunotherapy. Biomaterials 2013, 34, 7840–7850. [Google Scholar] [CrossRef] [PubMed]
- Capelli, C.; Domenghini, M.; Borleri, G.; Bellavita, P.; Poma, R.; Carobbio, A.; Micò, C.; Rambaldi, A.; Golay, J.; Introna, M. Human platelet lysate allows expansion and clinical grade production of mesenchymal stromal cells from small samples of bone marrow aspirates or marrow filter washouts. Bone Marrow Transplant. 2007, 40, 785–791. [Google Scholar] [CrossRef] [PubMed]
- Xia, W.; Li, H.; Wang, Z.; Xu, R.; Fu, Y.; Zhang, X.; Ye, X.; Huang, Y.; Xiang, A.P.; Yu, W. Human platelet lysate supports ex vivo expansion and enhances osteogenic differentiation of human bone marrow-derived mesenchymal stem cells. Cell Biol. Int. 2011, 35, 639–643. [Google Scholar] [CrossRef] [PubMed]
- Hesler, M.; Kohl, Y.; Wagner, S.; von Briesen, H. Non-pooled Human Platelet Lysate: A Potential Serum Alternative for In Vitro Cell Culture. Altern. Lab. Anim. 2019, 47, 116–127. [Google Scholar] [CrossRef] [Green Version]
- Mohamed, H.E.; Asker, M.E.; Kotb, N.S.; El Habab, A.M. Human platelet lysate efficiency, stability, and optimal heparin concentration required in culture of mammalian cells. Blood Res. 2020, 55, 35–43. [Google Scholar] [CrossRef]
- Shakeel, S.; Dykeman, E.C.; White, S.J.; Ora, A.; Cockburn, J.J.B.; Butcher, S.J.; Stockley, P.G.; Twarock, R. Genomic RNA folding mediates assembly of human parechovirus. Nat. Commun. 2017, 8, 5. [Google Scholar] [CrossRef]
- Britton, P.N.; Dale, R.C.; Nissen, M.D.; Crawford, N.; Elliott, E.; Macartney, K.; Khandaker, G.; Booy, R.; Jones, C.A.; Investigators, P.-A. Parechovirus Encephalitis and Neurodevelopmental Outcomes. Pediatrics 2016, 137, e20152848. [Google Scholar] [CrossRef] [Green Version]
- Britton, P.N.; Jones, C.A.; Macartney, K.; Cheng, A.C. Parechovirus: An important emerging infection in young infants. Med. J. Aust. 2018, 208, 365–369. [Google Scholar] [CrossRef]
- de Crom, S.C.; Rossen, J.W.; van Furth, A.M.; Obihara, C.C. Enterovirus and parechovirus infection in children: A brief overview. Eur. J. Pediatr. 2016, 175, 1023–1029. [Google Scholar] [CrossRef] [Green Version]
- Olijve, L.; Jennings, L.; Walls, T. Human Parechovirus: An Increasingly Recognized Cause of Sepsis-Like Illness in Young Infants. Clin. Microbiol. Rev. 2018, 31, e00047-17. [Google Scholar] [CrossRef] [Green Version]
- Aizawa, Y.; Izumita, R.; Saitoh, A. Human parechovirus type 3 infection: An emerging infection in neonates and young infants. J. Infect. Chemother. 2017, 23, 419–426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renaud, C.; Harrison, C.J. Human Parechovirus 3: The Most Common Viral Cause of Meningoencephalitis in Young Infants. Infect. Dis. Clin. N. Am. 2015, 29, 415–428. [Google Scholar] [CrossRef] [PubMed]
- Sasidharan, A.; Banerjee, D.; Harrison, C.J.; Selvarangan, R. Emergence of Parechovirus A3 as the Leading Cause of Central Nervous System Infection, Surpassing Any Single Enterovirus Type, in Children in Kansas City, Missouri, USA, from 2007 to 2016. J. Clin. Microbiol. 2021, 59, e02935-20. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Paget, M.; Wang, C.; Zhu, Z.; Zheng, H. Innate immune evasion by picornaviruses. Eur. J. Immunol. 2020, 50, 1268–1282. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.T.; Yang, C.S.; Chen, Y.S.; Chen, B.C.; Chiang, A.J.; Chang, Y.H.; Tsai, W.L.; Lin, Y.S.; Chao, D.; Chang, T.H. Genome and infection characteristics of human parechovirus type 1: The interplay between viral infection and type I interferon antiviral system. PLoS ONE 2015, 10, e0116158. [Google Scholar] [CrossRef] [Green Version]
- Solomon Tsegaye, T.; Gnirss, K.; Rahe-Meyer, N.; Kiene, M.; Kramer-Kuhl, A.; Behrens, G.; Munch, J.; Pohlmann, S. Platelet activation suppresses HIV-1 infection of T cells. Retrovirology 2013, 10, 48. [Google Scholar] [CrossRef] [Green Version]
- Jeyaraman, M.; Muthu, S.; Khanna, M.; Jain, R.; Anudeep, T.C.; Muthukanagaraj, P.; Siddesh, S.E.; Gulati, A.; Satish, A.S.; Jeyaraman, N.; et al. Platelet lysate for COVID-19 pneumonia-a newer adjunctive therapeutic avenue. Stem Cell Investig. 2021, 8, 11. [Google Scholar] [CrossRef]
- Chen, B.C.; Cheng, M.F.; Huang, T.S.; Liu, Y.C.; Tang, C.W.; Chen, C.S.; Chen, Y.S. Detection and identification of human parechoviruses from clinical specimens. Diagn. Microbiol. Infect. Dis. 2009, 65, 254–260. [Google Scholar] [CrossRef]
- Chang, J.T.; Yang, C.S.; Chen, B.C.; Chen, Y.S.; Chang, T.H. Complete genome sequence of the first human parechovirus type 3 isolated in Taiwan. J. Chin. Med. Assoc. 2017, 80, 737–739. [Google Scholar] [CrossRef]
- Lin, Y.L.; Liao, C.L.; Chen, L.K.; Yeh, C.T.; Liu, C.I.; Ma, S.H.; Huang, Y.Y.; Huang, Y.L.; Kao, C.L.; King, C.C. Study of Dengue virus infection in SCID mice engrafted with human K562 cells. J. Virol. 1998, 72, 9729–9737. [Google Scholar] [CrossRef] [Green Version]
- Chang, J.T.; Chen, Y.S.; Chen, B.C.; Chao, D.; Chang, T.H. Complete genome sequence of the first aichi virus isolated in taiwan. Genome Announc. 2013, 1, e00107-12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.S.; Chen, B.C.; Lin, Y.S.; Chang, J.T.; Huang, T.S.; Chen, J.J.; Chang, T.H. Detection of Aichi virus with antibody targeting of conserved viral protein 1 epitope. Appl. Microbiol. Biotechnol. 2013, 97, 8529–8536. [Google Scholar] [CrossRef] [PubMed]
- Kung, M.H.; Lin, Y.S.; Chang, T.H. Aichi virus 3C protease modulates LC3- and SQSTM1/p62-involved antiviral response. Theranostics 2020, 10, 9200–9213. [Google Scholar] [CrossRef]
- Chang, Y.T.; Kung, M.H.; Hsu, T.H.; Hung, W.T.; Chen, Y.S.; Yen, L.C.; Chang, T.H. Aichi Virus Induces Antiviral Host Defense in Primary Murine Intestinal Epithelial Cells. Viruses 2019, 11, 763. [Google Scholar] [CrossRef] [Green Version]
- Mesev, E.V.; LeDesma, R.A.; Ploss, A. Decoding type I and III interferon signalling during viral infection. Nat. Microbiol. 2019, 4, 914–924. [Google Scholar] [CrossRef] [PubMed]
- Frank, D.A.; Mahajan, S.; Ritz, J. Fludarabine-induced immunosuppression is associated with inhibition of STAT1 signaling. Nat. Med. 1999, 5, 444–447. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Song, Q.; Huang, W.; Lin, Y.; Wang, X.; Wang, C.; Willard, B.; Zhao, C.; Nan, J.; Holvey-Bates, E.; et al. A virus-induced conformational switch of STAT1-STAT2 dimers boosts antiviral defenses. Cell Res. 2021, 31, 206–218. [Google Scholar] [CrossRef]
- Tylek, T.; Schilling, T.; Schlegelmilch, K.; Ries, M.; Rudert, M.; Jakob, F.; Groll, J. Platelet lysate outperforms FCS and human serum for co-culture of primary human macrophages and hMSCs. Sci. Rep. 2019, 9, 3533. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.-J.; Sun, Y.-C.; Christopher, K.; Pai, A.S.-I.; Lu, C.-J.; Hu, F.-R.; Lin, S.-Y.; Chen, W.-L. Comparison of corneal epitheliotrophic capacities among human platelet lysates and other blood derivatives. PLoS ONE 2017, 12, e0171008. [Google Scholar] [CrossRef]
- Li, C.Y.; Wu, X.Y.; Tong, J.B.; Yang, X.X.; Zhao, J.L.; Zheng, Q.F.; Zhao, G.B.; Ma, Z.J. Comparative analysis of human mesenchymal stem cells from bone marrow and adipose tissue under xeno-free conditions for cell therapy. Stem Cell Res. Ther. 2015, 6, 55. [Google Scholar] [CrossRef] [Green Version]
- Bartek, J.; Hodny, Z.; Lukas, J. Cytokine loops driving senescence. Nat. Cell Biol. 2008, 10, 887–889. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, M.; Ikeda, H.; Sato, Y.; Nakanuma, Y. Proinflammatory cytokine-induced cellular senescence of biliary epithelial cells is mediated via oxidative stress and activation of ATM pathway: A culture study. Free Radic. Res. 2008, 42, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Sipe, J.B.; Zhang, J.; Waits, C.; Skikne, B.; Garimella, R.; Anderson, H.C. Localization of bone morphogenetic proteins (BMPs)-2, -4, and -6 within megakaryocytes and platelets. Bone 2004, 35, 1316–1322. [Google Scholar] [CrossRef]
- Li, Q.; Wijesekera, O.; Salas, S.J.; Wang, J.Y.; Zhu, M.; Aprhys, C.; Chaichana, K.L.; Chesler, D.A.; Zhang, H.; Smith, C.L.; et al. Mesenchymal Stem Cells from Human Fat Engineered to Secrete BMP4 Are Nononcogenic, Suppress Brain Cancer, and Prolong Survival. Clin. Cancer Res. 2014, 20, 2375–2387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piccirillo, S.G.; Reynolds, B.A.; Zanetti, N.; Lamorte, G.; Binda, E.; Broggi, G.; Brem, H.; Olivi, A.; Dimeco, F.; Vescovi, A.L. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature 2006, 444, 761–765. [Google Scholar] [CrossRef] [PubMed]
- Hemeda, H.; Kalz, J.; Walenda, G.; Lohmann, M.; Wagner, W. Heparin concentration is critical for cell culture with human platelet lysate. Cytotherapy 2013, 15, 1174–1181. [Google Scholar] [CrossRef]
- Lin, Y.L.; Lei, H.Y.; Lin, Y.S.; Yeh, T.M.; Chen, S.H.; Liu, H.S. Heparin inhibits dengue-2 virus infection of five human liver cell lines. Antivir. Res. 2002, 56, 93–96. [Google Scholar] [CrossRef]
- Karelehto, E.; Cristella, C.; Yu, X.; Sridhar, A.; Hulsdouw, R.; de Haan, K.; van Eijk, H.; Koekkoek, S.; Pajkrt, D.; de Jong, M.D.; et al. Polarized Entry of Human Parechoviruses in the Airway Epithelium. Front. Cell. Infect. Microbiol. 2018, 8, 294. [Google Scholar] [CrossRef]
- Sellberg, F.; Berglund, E.; Ronaghi, M.; Strandberg, G.; Lof, H.; Sommar, P.; Lubenow, N.; Knutson, F.; Berglund, D. Composition of growth factors and cytokines in lysates obtained from fresh versus stored pathogen-inactivated platelet units. Transfus. Apher. Sci. 2016, 55, 333–337. [Google Scholar] [CrossRef]
- Gao, C.; Wen, C.; Li, Z.; Lin, S.; Gao, S.; Ding, H.; Zou, P.; Xing, Z.; Yu, Y. Fludarabine Inhibits Infection of Zika Virus, SFTS Phlebovirus, and Enterovirus A71. Viruses 2021, 13, 774. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jan, M.-W.; Chiu, C.-Y.; Chen, J.-J.; Chang, T.-H.; Tsai, K.-J. Human Platelet Lysate Induces Antiviral Responses against Parechovirus A3. Viruses 2022, 14, 1499. https://doi.org/10.3390/v14071499
Jan M-W, Chiu C-Y, Chen J-J, Chang T-H, Tsai K-J. Human Platelet Lysate Induces Antiviral Responses against Parechovirus A3. Viruses. 2022; 14(7):1499. https://doi.org/10.3390/v14071499
Chicago/Turabian StyleJan, Ming-Wei, Chih-Yun Chiu, Jih-Jung Chen, Tsung-Hsien Chang, and Kuen-Jer Tsai. 2022. "Human Platelet Lysate Induces Antiviral Responses against Parechovirus A3" Viruses 14, no. 7: 1499. https://doi.org/10.3390/v14071499
APA StyleJan, M.-W., Chiu, C.-Y., Chen, J.-J., Chang, T.-H., & Tsai, K.-J. (2022). Human Platelet Lysate Induces Antiviral Responses against Parechovirus A3. Viruses, 14(7), 1499. https://doi.org/10.3390/v14071499








