Design of a Multiepitope Vaccine against Chicken Anemia Virus Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Immunoinformatics of Viral Proteins
2.1.1. Retrieval and Filtering of VP1 and VP2 Protein Sequences
2.1.2. CD8+ T-Cell Epitopes and MHC-I Binding Allele Prediction
2.1.3. CD4+ T-Cell Epitopes and MHC-II Binding Allele Prediction
2.1.4. Immunogenicity, Antigenicity, Conservancy, and Allergenicity of Predicted Epitopes
2.1.5. Prediction of Linear B-Cell Epitopes
2.2. Multiepitope Vaccine Construction
2.2.1. Multiepitope Vaccine (MEV) Design
2.2.2. Physicochemical Properties and Secondary Structure Prediction
2.2.3. Prediction of Tertiary Structure, Refinement, and Validation
2.2.4. Conformational B-Cell Epitopes Prediction
2.2.5. Molecular Docking of MEV with TLR3
2.2.6. Molecular Dynamics of the MEV Construct
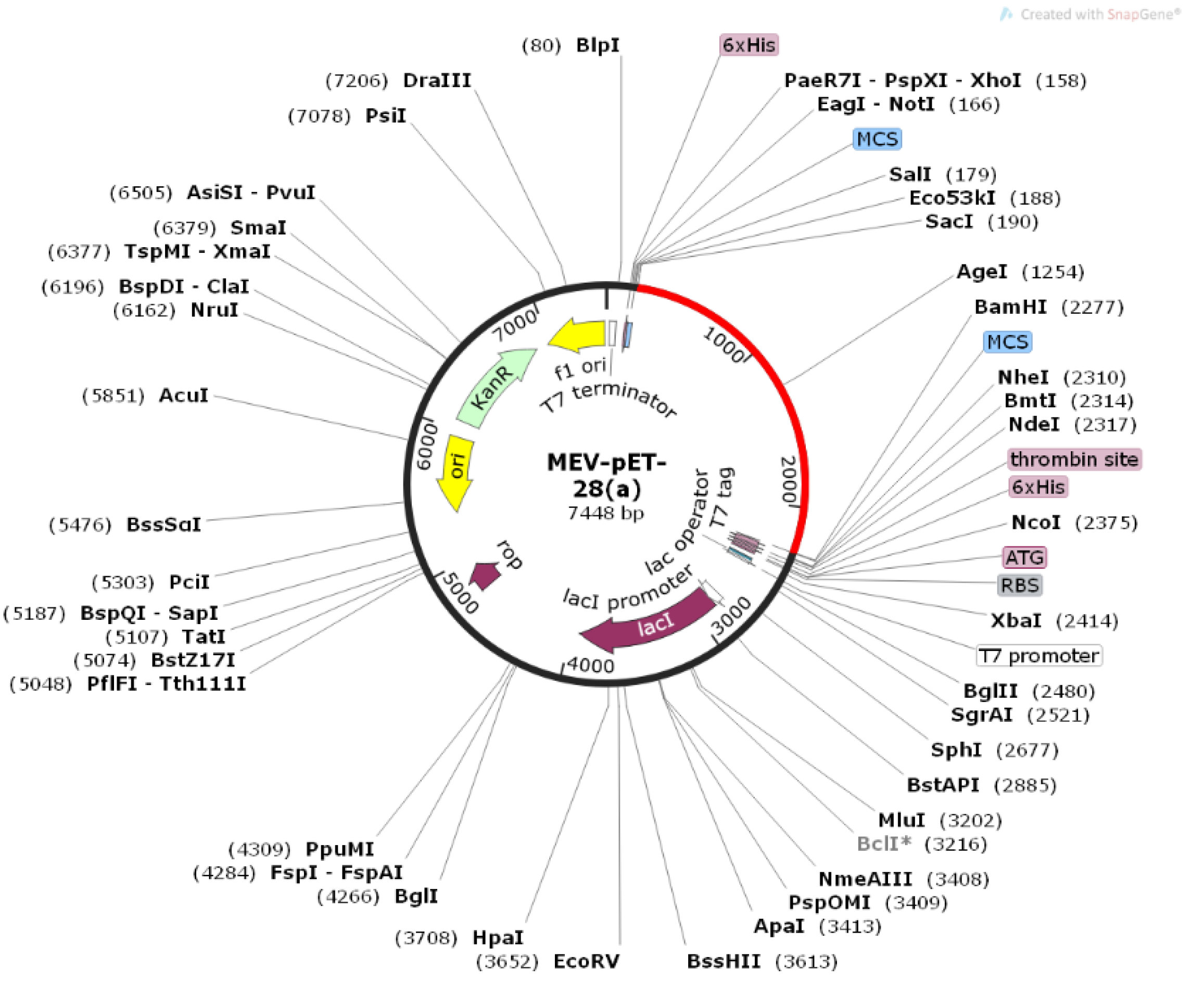
2.2.7. Reverse Translation, Codon Optimization, and In Silico Cloning of the MEV
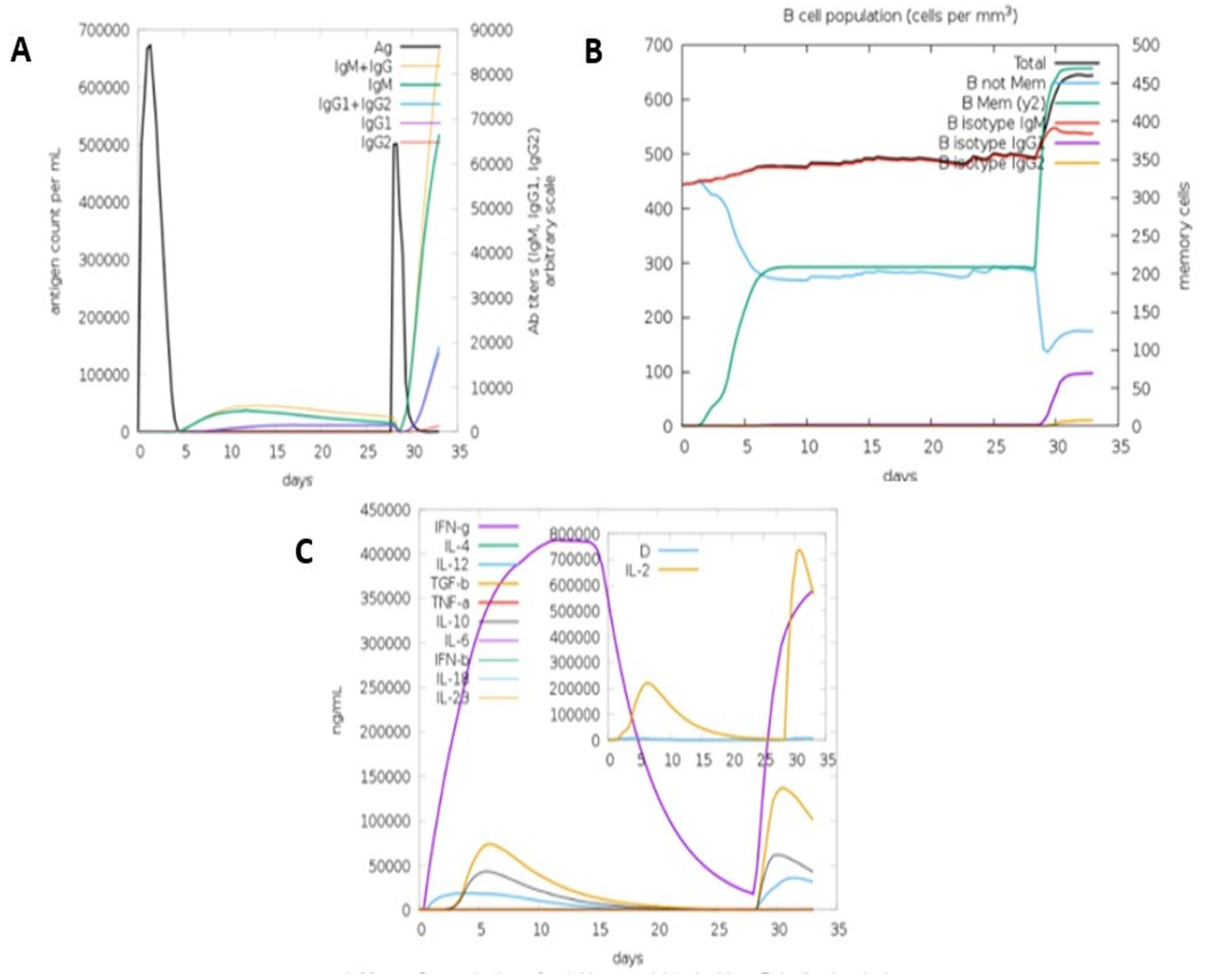
2.2.8. Immune Simulation of MEV Construct
3. Results
3.1. Sequence Alignment, Antigenicity, and Membrane Test
3.2. CD8+ T-Cell Epitopes and MHC-I Binding Allele Prediction
3.3. CD4+ T-Cell Epitopes and MHC-II Binding Allele Prediction, Conservancy, and Allergenicity
3.4. Linear B-Cell Epitopes Prediction
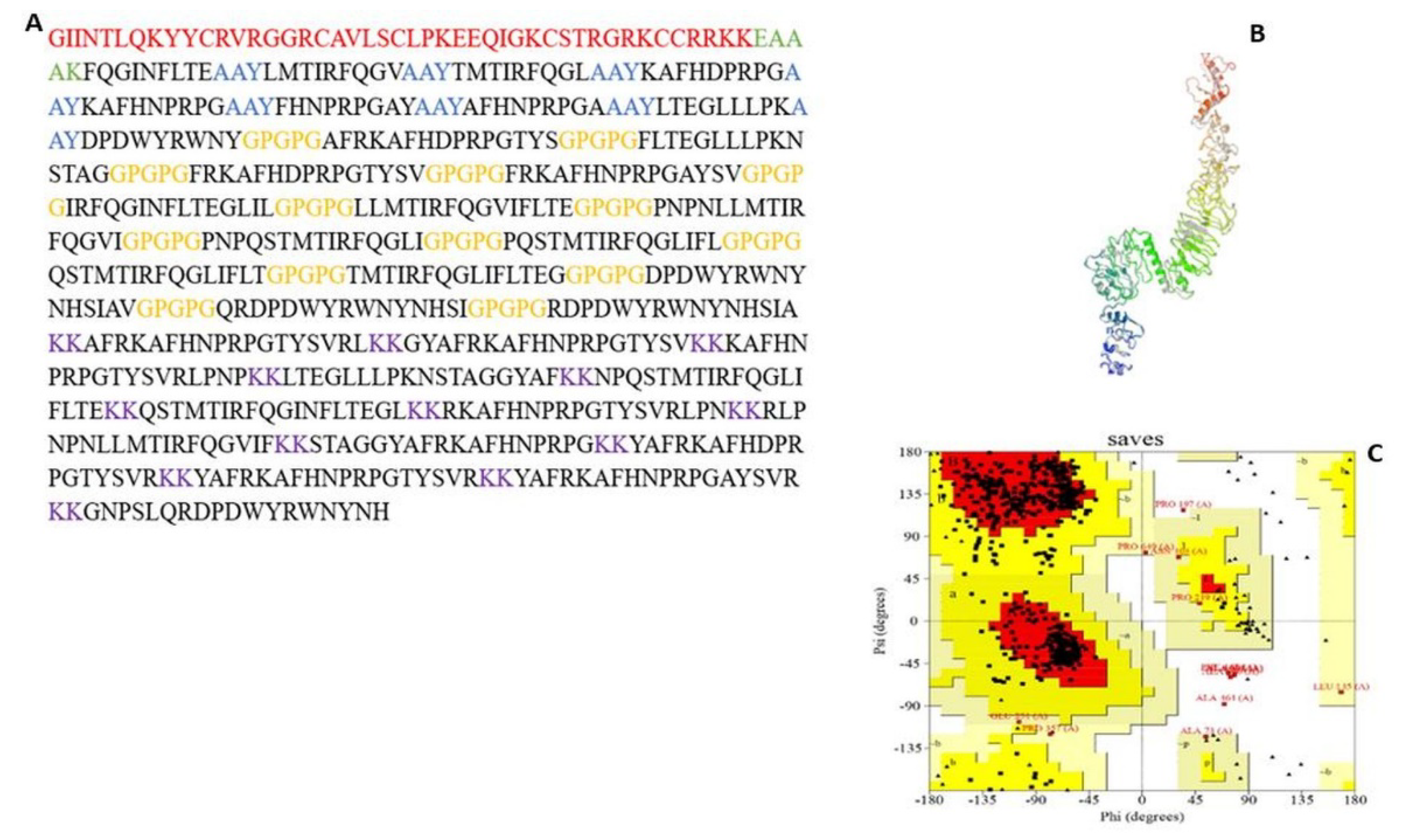
3.5. MEV Construction and Validation
3.5.1. MEV Construction and Screening
3.5.2. Physicochemical Properties and Secondary Structure Prediction
3.5.3. Tertiary Structure Prediction, Refinement, and Validation
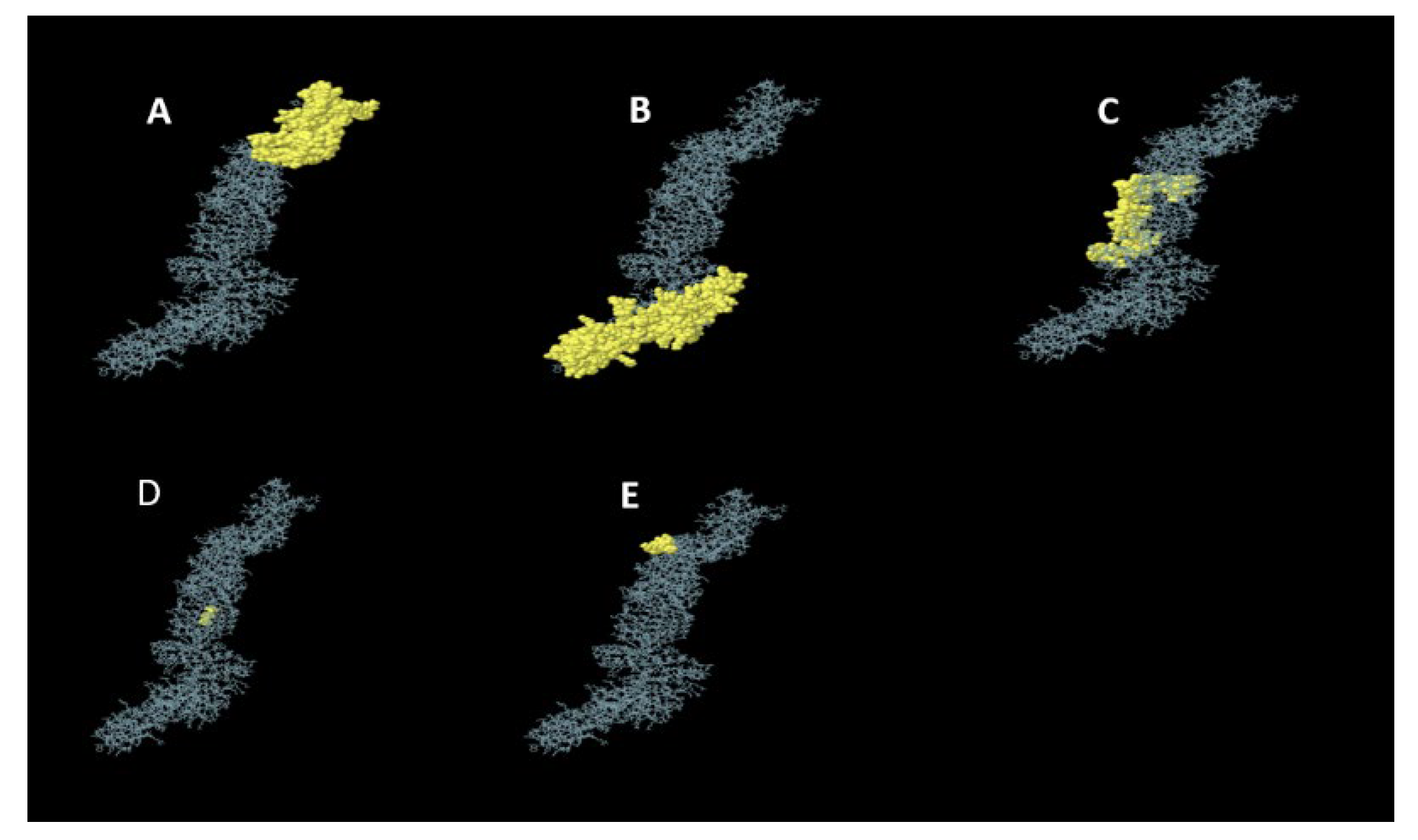
3.5.4. Conformational B-Cell Epitopes Prediction
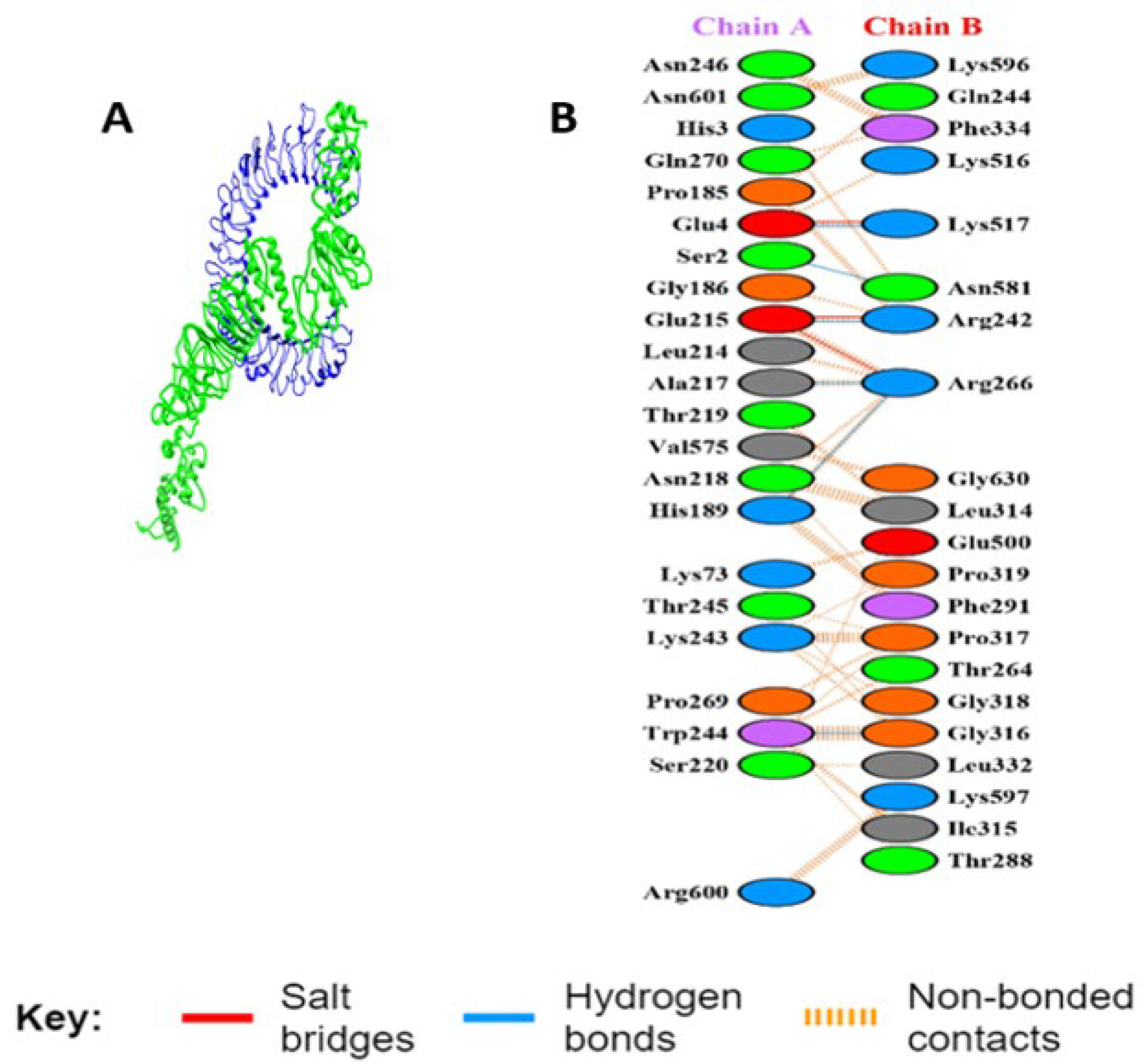
3.5.5. Interaction Analysis of MEV Construct and TLR-3
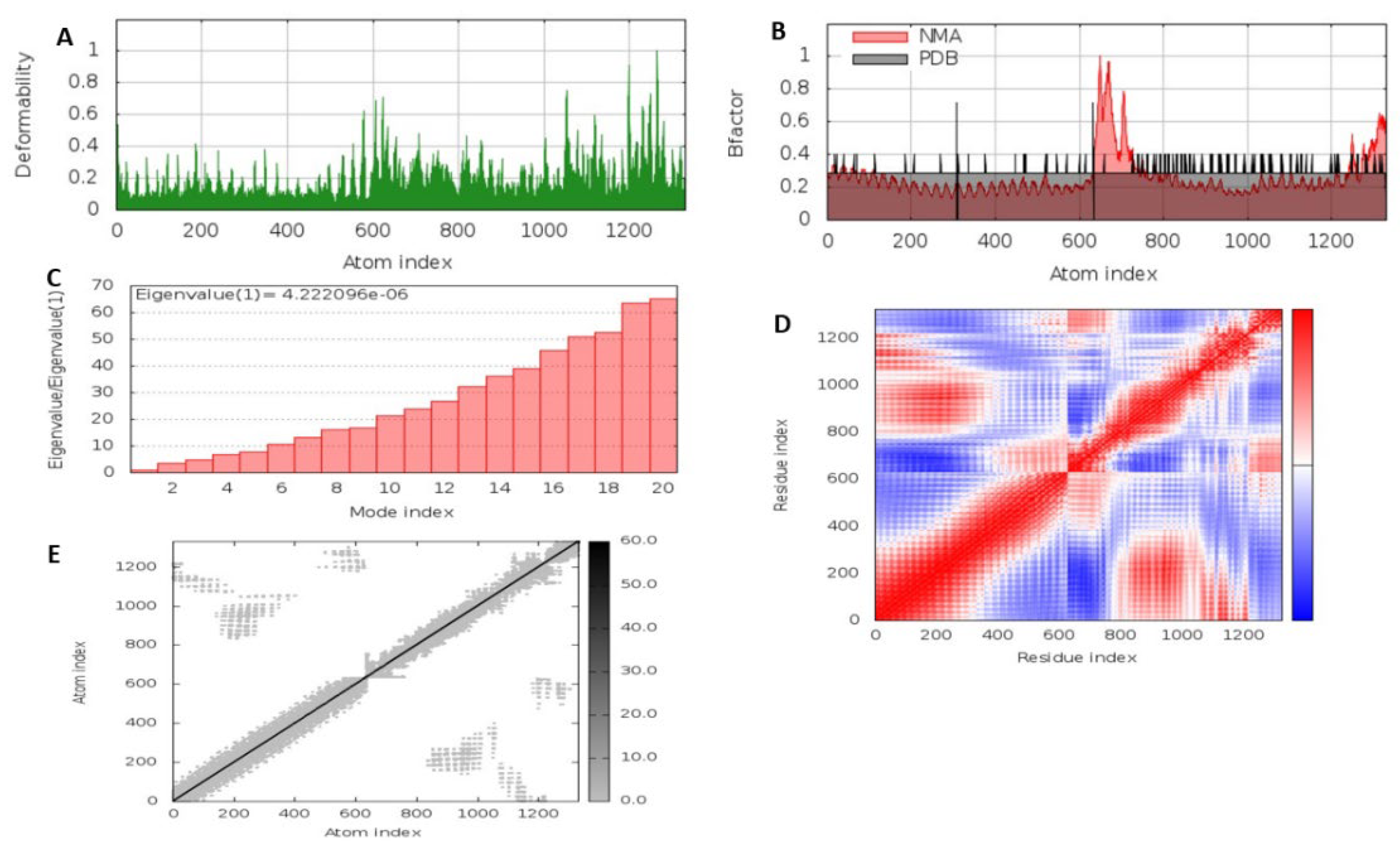
3.5.6. Molecular Dynamics Simulation of the MEV Construct
3.5.7. Reverse Translation, Codon Optimization, and In Silico Cloning
3.5.8. Immune Simulation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Giotis, E.S.; Rothwell, L.; Scott, A.; Hu, T. Transcriptomic Profiling of Virus-Host Cell Interactions Following Chicken Anaemia Virus (CAV) Infection in an In Vivo Model. PLoS ONE 2015, 1, e0134866. [Google Scholar] [CrossRef]
- Miller, M.M.; Schat, K.A. Chicken Infectious Anemia Virus: An Example of the Ultimate Host–Parasite Relationship. Avian Dis. 2004, 48, 734–745. [Google Scholar] [CrossRef]
- Smyth, J.A.; Moffett, D.A.; Connor, T.J.; McNulty, M.S. Chicken Anaemia Virus Inoculated by the Oral Route Causes Lymphocyte Depletion in the Thymus in 3-Week-Old and 6-Week-Old Chickens. Avian Pathol. 2006, 35, 254–259. [Google Scholar] [CrossRef]
- Hoerr, F.J. Clinical Aspects of Immunosuppression in Poultry. Avian Dis. 2010, 54, 2–15. [Google Scholar] [CrossRef]
- Trinh, D.Q.; Ogawa, H.; Bui, V.N.; Baatartsogt, T.; Kizito, M.K.; Yamaguchi, S.; Imai, K. Characterization of MAbs to Chicken Anemia Virus and Epitope Mapping on Its Viral Protein, VP1. J. Gen. Virol. 2015, 96, 1086–1097. [Google Scholar] [CrossRef]
- Lai, G.; Lin, M.; Lien, Y.; Cheng, J.; Sun, F.; Lee, M.; Chen, H.; Lee, M. Characterization of the DNA Binding Activity of Structural Protein VP1 from Chicken Anaemia Virus. BMC Vet. Res. 2018, 14, 155. [Google Scholar] [CrossRef]
- Rosario, K.; Breitbart, M.; Harrach, B.; Segalés, J.; Delwart, E.; Biagini, P.; Varsani, A. Revisiting the Taxonomy of the Family Circoviridae: Establishment of the Genus Cyclovirus and Removal of the Genus Gyrovirus. Arch. Virol. 2017, 162, 1447–1463. [Google Scholar] [CrossRef]
- Noteborn, M.H.; de Boer, G.F.; van Roozelaar, D.J.; Karreman, C.; Kranenburg, O.; Vos, J.G.; Jeurissen, S.H.; Hoeben, R.C.; Zantema, A.; Koch, G. Characterization of Cloned Chicken Anemia Virus DNA That Contains All Elements for the Infectious Replication Cycle. J. Virol. 1991, 65, 3131–3139. [Google Scholar] [CrossRef]
- Peters, M.A.; Jackson, D.C.; Crabb, B.S.; Browning, G.F. Chicken Anemia Virus VP2 Is a Novel Dual Specificity Protein Phosphatase. J. Biol. Chem. 2002, 277, 39566–39573. [Google Scholar] [CrossRef]
- Noteborn, M.H.M.; Verschueren, C.A.J.; Koch, G.; Eb, A.J. Van Der Simultaneous Expression of Recombinant Baculovirus-Encoded Chicken Anaemia Virus (CAV) Proteins VP1 and VP2 Is Required for Formation of the CAV-Specific Neutralizing Epitope. J. Gen. Virol. 1998, 1, 3073–3077. [Google Scholar] [CrossRef][Green Version]
- Moeini, H.; Omar, A.R.; Rahim, R.A.; Yusoff, K. Improving the Potency of DNA Vaccine against Chicken Anemia Virus (CAV) by Fusing VP1 Protein of CAV to Marek’s Disease Virus (MDV) Type-1 VP22 Protein. Virol. J. 2011, 8, 119. [Google Scholar] [CrossRef]
- Noteborn, M.H.; Todd, D.; Verschueren, C.A.; de Gauw, H.W.; Curran, W.L.; Veldkamp, S.; Douglas, A.J.; McNulty, M.S.; van der EB, A.J.; Koch, G. A Single Chicken Anemia Virus Protein Induces Apoptosis. J. Virol. 1994, 68, 346–351. [Google Scholar] [CrossRef]
- Adair, B.M. Immunopathogenesis of Chicken Anemia Virus Infection. Dev. Comp. Immunol. 2000, 24, 247–255. [Google Scholar] [CrossRef]
- Vaziry, A.; Silim, A.; Bleau, C.; Frenette, D.; Lamontagne, L. Chicken Infectious Anaemia Vaccinal Strain Persists in the Spleen and Thymus of Young Chicks and Induces Thymic Lymphoid Cell Disorders. Avian Pathol. 2011, 40, 377–385. [Google Scholar] [CrossRef]
- Sawant, P.M.; Dhama, K.; Rawool, D.B.; Wani, M.Y.; Tiwari, R.; Singh, S.D.; Singh, R.K. Development of a DNA Vaccine for Chicken Infectious Anemia and Its Immunogenicity Studies Using High Mobility Group Box 1 Protein as a Novel Immunoadjuvant Indicated Induction of Promising Protective Immune Responses. Vaccine 2015, 33, 333–340. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, B.; Liu, Y.; Chen, W.; Dai, Z.; Bi, Y.; Xie, Q. Assessing the Efficacy of an Inactivated Chicken Anemia Virus Vaccine. Vaccine 2015, 33, 1916–1922. [Google Scholar] [CrossRef]
- Tseng, T.-Y.; Liu, Y.-C.; Hsu, Y.-C.; Chang, P.-C.; Hsieh, M.-K.; Shien, J.-H.; Ou, S.-C. Preparation of Chicken Anemia Virus (CAV) Virus-Like Particles and Chicken Interleukin-12 for Vaccine Development Using a Baculovirus Expression System. Pathogens 2019, 8, 262. [Google Scholar] [CrossRef]
- Lee, M.-S.; Hseu, Y.-C.; Lai, G.-H.; Chang, W.-T.; Chen, H.-J.; Huang, C.-H.; Lee, M.-S.; Wang, M.-Y.; Kao, J.-Y.; You, B.-J.; et al. High Yield Expression in a Recombinant E. Coli of a Codon Optimized Chicken Anemia Virus Capsid Protein VP1 Useful for Vaccine Development. Microb. Cell Fact. 2011, 10, 56. [Google Scholar] [CrossRef]
- Dar, H.A.; Waheed, Y.; Najmi, M.H.; Ismail, S.; Hetta, H.F.; Ali, A.; Muhammad, K. Multiepitope Subunit Vaccine Design against COVID-19 Based on the Spike Protein of SARS-CoV-2: An In Silico Analysis. J. Immunol. Res. 2020, 2020, 8893483. [Google Scholar] [CrossRef]
- Abdelmageed, M.I.; Abdelmoneim, A.H.; Mustafa, M.I.; Elfadol, N.M.; Murshed, N.S.; Shantier, S.W.; Makhawi, A.M. Design of a Multiepitope-Based Peptide Vaccine against the e Protein of Human COVID-19: An Immunoinformatics Approach. Biomed Res. Int. 2020, 2020, 2683286. [Google Scholar] [CrossRef]
- Oloomi, M.; Javadi, M.; Asadi Karam, M.R.; Khezerloo, J.K.; Haghri, Z.; Bouzari, S. Protective Multi-Epitope Candidate Vaccine for Urinary Tract Infection. Biotechnol. Rep. 2020, 28, e00564. [Google Scholar] [CrossRef]
- Doytchinova, I.A.; Flower, D.R. VaxiJen: A Server for Prediction of Protective Antigens, Tumour Antigens and Subunit Vaccines. BMC Bioinform. 2007, 8, 4. [Google Scholar] [CrossRef]
- Aziz, F.; Tufail, S.; Shah, M.A.; Salahuddin Shah, M.; Habib, M.; Mirza, O.; Iqbal, M.; Rahman, M. In Silico Epitope Prediction and Immunogenic Analysis for Penton Base Epitope-Focused Vaccine against Hydropericardium Syndrome in Chicken. Virus Res. 2019, 273, 197750. [Google Scholar] [CrossRef]
- Mugunthan, S.P.; Mani Chandra, H. A Computational Reverse Vaccinology Approach for the Design and Development of Multi-Epitopic Vaccine Against Avian Pathogen Mycoplasma Gallisepticum. Front. Vet. Sci. 2021, 8, 721061. [Google Scholar] [CrossRef]
- Vainio, O.; Koch, C.; Toivanen, A. B-L Antigens (Class II) of the Chicken Major Histocompatibility Complex Control T-B Cell Interaction. Immunogenetics 1984, 19, 131–140. [Google Scholar] [CrossRef]
- Larsen, M.V.; Lundegaard, C.; Lamberth, K.; Buus, S.; Lund, O.; Nielsen, M. Large-Scale Validation of Methods for Cytotoxic T-Lymphocyte Epitope Prediction. BMC Bioinform. 2007, 8, 424. [Google Scholar] [CrossRef]
- Dhanda, S.K.; Vir, P.; Raghava, G.P.S. Designing of Interferon-Gamma Inducing MHC Class-II Binders. Biol. Direct 2013, 8, 30. [Google Scholar] [CrossRef]
- Nagpal, G.; Usmani, S.S.; Dhanda, S.K.; Kaur, H.; Singh, S.; Sharma, M.; Raghava, G.P.S. Computer-Aided Designing of Immunosuppressive Peptides Based on IL-10 Inducing Potential. Sci. Rep. 2017, 7, 42851. [Google Scholar] [CrossRef]
- Dimitrov, I.; Flower, D.R.; Doytchinova, I. AllerTOP—A Server for in Silico Prediction of Allergens. BMC Bioinform. 2013, 14, S4. [Google Scholar] [CrossRef]
- Gupta, S.; Kapoor, P.; Chaudhary, K.; Gautam, A.; Kumar, R.; Consortium, O.S.D.D.; Raghava, G.P.S. In Silico Approach for Predicting Toxicity of Peptides and Proteins. PLoS ONE 2013, 8, e73957. [Google Scholar] [CrossRef]
- Saha, S.; Raghava, G.P.S. Prediction of Continuous B-Cell Epitopes in an Antigen Using Recurrent Neural Network. Proteins Struct. Funct. Bioinform. 2006, 65, 40–48. [Google Scholar] [CrossRef]
- Yang, J.; Anishchenko, I.; Park, H.; Peng, Z.; Ovchinnikov, S.; Baker, D. Improved Protein Structure Prediction Using Predicted Interresidue Orientations. Proc. Natl. Acad. Sci. USA 2020, 117, 1496–1503. [Google Scholar] [CrossRef]
- Ko, J.; Park, H.; Heo, L.; Seok, C. GalaxyWEB Server for Protein Structure Prediction and Refinement. Nucleic Acids Res. 2012, 40, W294–W297. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A Program to Check the Stereochemical Quality of Protein Structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Ponomarenko, J.; Bui, H.-H.; Li, W.; Fusseder, N.; Bourne, P.E.; Sette, A.; Peters, B. ElliPro: A New Structure-Based Tool for the Prediction of Antibody Epitopes. BMC Bioinform. 2008, 9, 514. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Jabłońska, J.; Pravda, L.; Vařeková, R.S.; Thornton, J.M. PDBsum: Structural Summaries of PDB Entries. Protein Sci. 2018, 27, 129–134. [Google Scholar] [CrossRef]
- López-Blanco, J.R.; Aliaga, J.I.; Quintana-Ortí, E.S.; Chacón, P. IMODS: Internal Coordinates Normal Mode Analysis Server. Nucleic Acids Res. 2014, 42, W271–W276. [Google Scholar] [CrossRef]
- Noteborn, M.H.; Koch, G. Chicken Anaemia Virus Infection: Molecular Basis of Pathogenicity. Avian Pathol. 1995, 24, 11–31. [Google Scholar] [CrossRef]
- Natesan, S.; Kataria, J.M.; Dhama, K.; Rahul, S.; Baradhwaj, N. Biological and Molecular Characterization of Chicken Anaemia Virus Isolates of Indian Origin. Virus Res. 2006, 118, 78–86. [Google Scholar] [CrossRef]
- Pestka, S.; Krause, C.D.; Walter, M.R. Interferons, Interferon-like Cytokines, and Their Receptors. Immunol. Rev. 2004, 202, 8–32. [Google Scholar] [CrossRef]
- Rojas, J.M.; Avia, M.; Martín, V.; Sevilla, N. IL-10: A Multifunctional Cytokine in Viral Infections. J. Immunol. Res. 2017, 2017, 6104054. [Google Scholar] [CrossRef]
- Shen, S.Y.; Chang, W.C.; Yi, H.H.; Tsai, S.-S.; Liu, H.J.; Liao, P.-C.; Chuang, K.P. Development of a Subunit Vaccine Containing Recombinant Chicken Anemia Virus VP1 and Pigeon IFN-γ. Vet. Immunol. Immunopathol. 2015, 167, 200–204. [Google Scholar] [CrossRef]
- Meza, B.; Ascencio, F.; Sierra-Beltrán, A.P.; Torres, J.; Angulo, C. A Novel Design of a Multi-Antigenic, Multistage and Multi-Epitope Vaccine against Helicobacter Pylori: An in Silico Approach. Infect. Genet. Evol. 2017, 49, 309–317. [Google Scholar] [CrossRef]
- Onile, O.S.; Fadahunsi, A.I.; Adekunle, A.A.; Oyeyemi, B.F.; Anumudu, C.I. An Immunoinformatics Approach for the Design of a Multi-Epitope Subunit Vaccine for Urogenital Schistosomiasis. PeerJ 2020, 8, 1–21. [Google Scholar] [CrossRef]
- Tahir ul Qamar, M.; Rehman, A.; Tusleem, K.; Ashfaq, U.A.; Qasim, M.; Zhu, X.; Fatima, I.; Shahid, F.; Chen, L.-L. Designing of a next Generation Multiepitope Based Vaccine (MEV) against SARS-COV-2: Immunoinformatics and in Silico Approaches. PLoS ONE 2020, 15, e0244176. [Google Scholar] [CrossRef]
- Vercammen, E.; Staal, J.; Beyaert, R. Sensing of Viral Infection and Activation of Innate Immunity by Toll-like Receptor 3. Clin. Microbiol. Rev. 2008, 21, 13–25. [Google Scholar] [CrossRef]

| Viral Protein | CD8+ T-Cell Epitopes | Antigenicity Score | Immunogenicity | MHCI-Alleles |
|---|---|---|---|---|
| VP1 | FQGINFLTE | 0.5863 | 0.2621 | HLA-A*02:06 |
| LMTIRFQGV | 1.0143 | 0.2292 | HLA-A*02:06, HLA-A*02:03, HLA-B*08:01, HLA-A*02:01 | |
| TMTIRFQGL | 1.1407 | 0.2292 | HLA-B*08:01 | |
| KAFHDPRPG | 0.7871 | 0.1189 | HLA-A*30:01 | |
| KAFHNPRPG | 0.5687 | 0.0910 | HLA-A*30:01 | |
| FHNPRPGAY | 0.6146 | 0.0782 | HLA-A*30:02, HLA-B*35:01 | |
| AFHNPRPGA | 0.5667 | 0.0524 | HLA-A*30:01 | |
| LTEGLLLPK | 0.5185 | 0.0295 | HLA-A*11:01 | |
| VP2 | DPDWYRWNY | 0.9093 | 0.4584 | HLA-B*35:01 |
| Viral Protein | CD4+ T-Cell Epitopes | Antigenicity Score | MHCII-Alleles |
|---|---|---|---|
| VP1 | AFRKAFHDPRPGTYS | 0.5598 | HLA-DRB5*01:01 |
| FLTEGLLLPKNSTAG | 0.5757 | HLA-DRB1*01:01, HLA-DPA1*03:01 | |
| FRKAFHDPRPGTYSV | 0.6011 | HLA-DRB5*01:01 | |
| FRKAFHNPRPGAYSV | 0.5000 | HLA-DRB5*01:01 | |
| IRFQGINFLTEGLIL | 0.7592 | HLA-DPA1*01:03, HLA-DPA1*03:01, HLA-DPA1*02:01 | |
| LLMTIRFQGVIFLTE | 0.8222 | HLA-DRB1*04:05, HLA-DRB1*01:01, HLA-DRB1*15:01, HLA-DPA1*02:01, HLA-DPA1*03:01, HLA-DPA1*01:03 | |
| PNPNLLMTIRFQGVI | 0.8698 | HLA-DRB4*01:01, HLA-DPA1*03:01, HLA-DPA1*01:03, HLA-DRB1*11: 01, HLA-DRB1*01:01 | |
| PNPQSTMTIRFQGLI | 0.8712 | HLA-DRB4*01:01 | |
| PQSTMTIRFQGLIFL | 0.7590 | HLA-DRB1*15:01, HLA-DRB4*01:01, HLA-DPA1*02:01, HLA-DPA1*03:01 | |
| QSTMTIRFQGLIFLT | 0.8821 | HLA-DRB1*15:01, HLA-DRB4*01:01, HLA-DPA1*02:01, HLA-DPA1*03:01 | |
| TMTIRFQGLIFLTEG | 0.8422 | HLA-DRB1*04:05, HLA-DRB1*15:01, HLA-DRB4*01:01, HLA-DPA1*02:01, HLA-DPA1*03:01 | |
| VP2 | DPDWYRWNYNHSIAV | 0.7586 | HLA-DRB1*13:02, HLA-DRB3*01:01 |
| QRDPDWYRWNYNHSI | 0.8368 | HLA-DRB1*07:01, HLA-DRB3*01:01 | |
| RDPDWYRWNYNHSIA | 0.8539 | HLA-DRB1*07:01, HLA-DRB3*01:01 |
| Viral Protein | B-Cell Epitopes | Antigenicity Score | ABCpred Score |
|---|---|---|---|
| VP1 | AFRKAFHNPRPGTYSVRL | 0.6040 | 0.84 |
| GYAFRKAFHNPRPGTYSV | 0.5284 | 0.71 | |
| KAFHNPRPGTYSVRLPNP | 0.6401 | 0.89 | |
| LTEGLLLPKNSTAGGYAF | 0.6674 | 0.77 | |
| NPQSTMTIRFQGLIFLTE | 0.8296 | 0.65 | |
| QSTMTIRFQGINFLTEGL | 0.8772 | 0.77 | |
| RKAFHNPRPGTYSVRLPN | 0.7101 | 0.86 | |
| RLPNPNLLMTIRFQGVIF | 0.5641 | 0.74 | |
| STAGGYAFRKAFHNPRPG | 0.6224 | 0.65 | |
| YAFRKAFHDPRPGTYSVR | 0.6751 | 0.74 | |
| YAFRKAFHNPRPGAYSVR | 0.8513 | 0.78 | |
| YAFRKAFHNPRPGTYSVR | 0.7547 | 0.88 | |
| VP2 | GNPSLQRDPDWYRWNYNH | 0.5966 | 0.80 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fatoba, A.J.; Adeleke, V.T.; Maharaj, L.; Okpeku, M.; Adeniyi, A.A.; Adeleke, M.A. Design of a Multiepitope Vaccine against Chicken Anemia Virus Disease. Viruses 2022, 14, 1456. https://doi.org/10.3390/v14071456
Fatoba AJ, Adeleke VT, Maharaj L, Okpeku M, Adeniyi AA, Adeleke MA. Design of a Multiepitope Vaccine against Chicken Anemia Virus Disease. Viruses. 2022; 14(7):1456. https://doi.org/10.3390/v14071456
Chicago/Turabian StyleFatoba, Abiodun Joseph, Victoria T. Adeleke, Leah Maharaj, Moses Okpeku, Adebayo A. Adeniyi, and Matthew A. Adeleke. 2022. "Design of a Multiepitope Vaccine against Chicken Anemia Virus Disease" Viruses 14, no. 7: 1456. https://doi.org/10.3390/v14071456
APA StyleFatoba, A. J., Adeleke, V. T., Maharaj, L., Okpeku, M., Adeniyi, A. A., & Adeleke, M. A. (2022). Design of a Multiepitope Vaccine against Chicken Anemia Virus Disease. Viruses, 14(7), 1456. https://doi.org/10.3390/v14071456






