The C-Terminal Domain of HIV-1 Integrase: A Swiss Army Knife for the Virus?
Abstract
1. Introduction
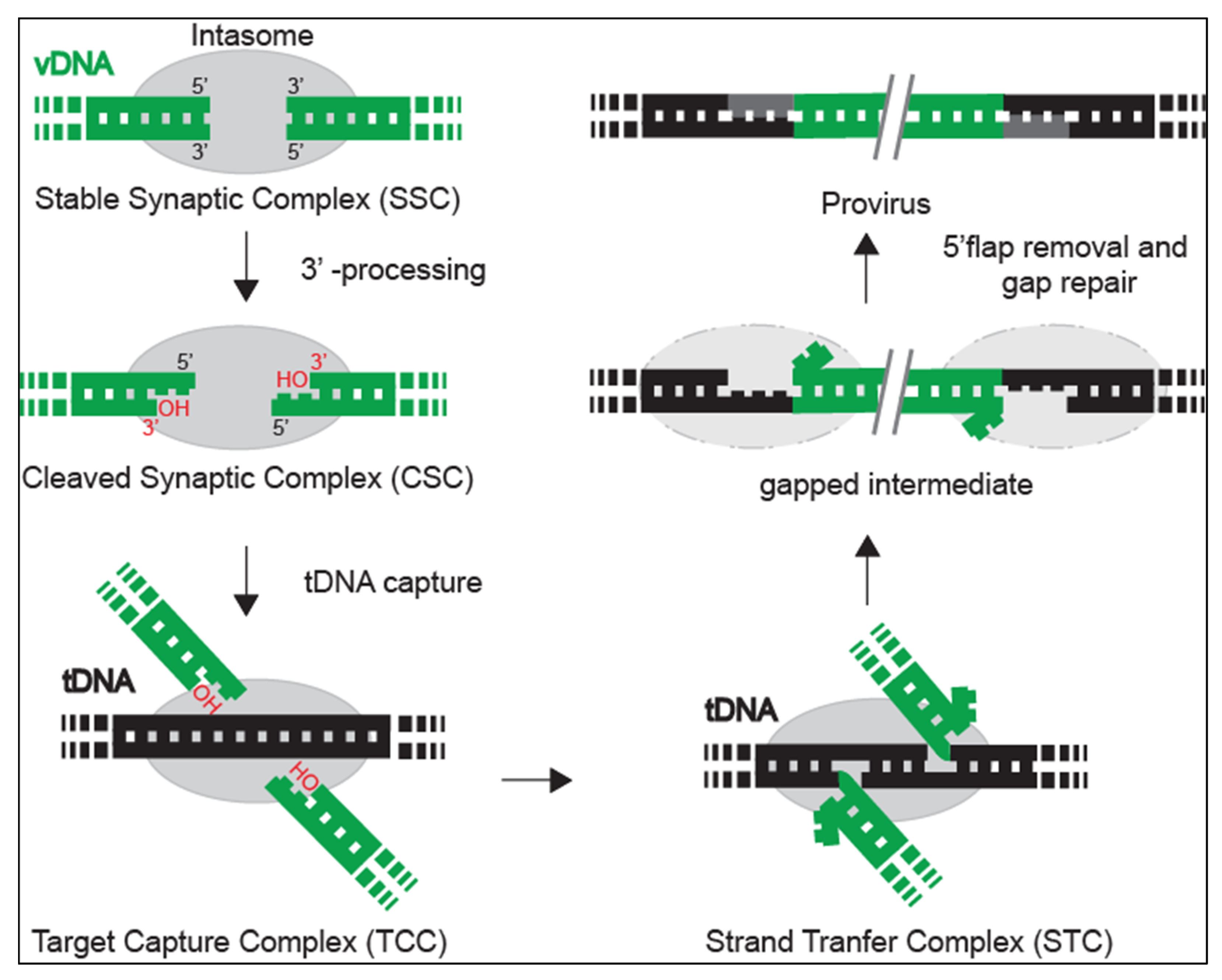
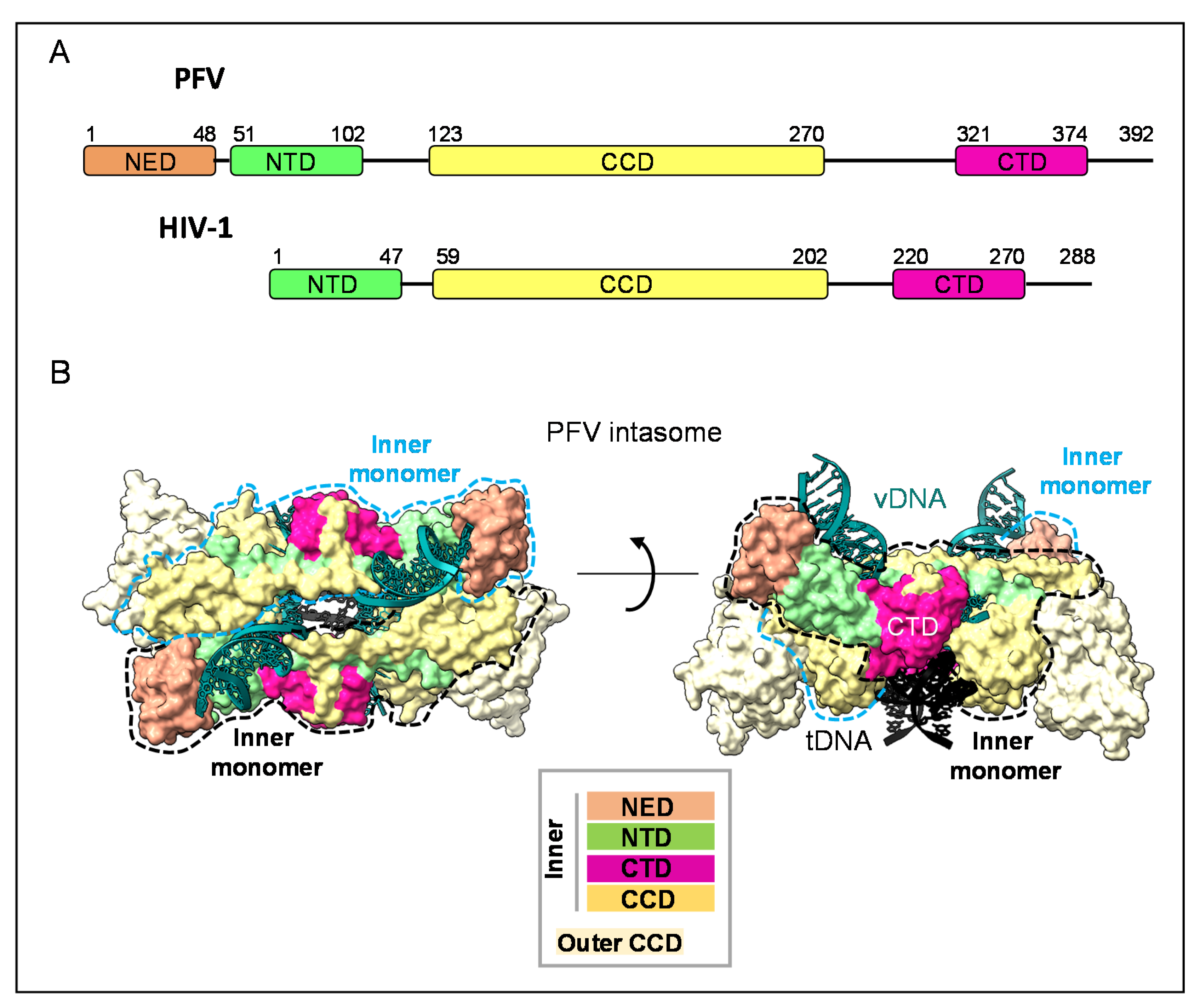
2. Integrase within the Intasome: Conserved Core Organization
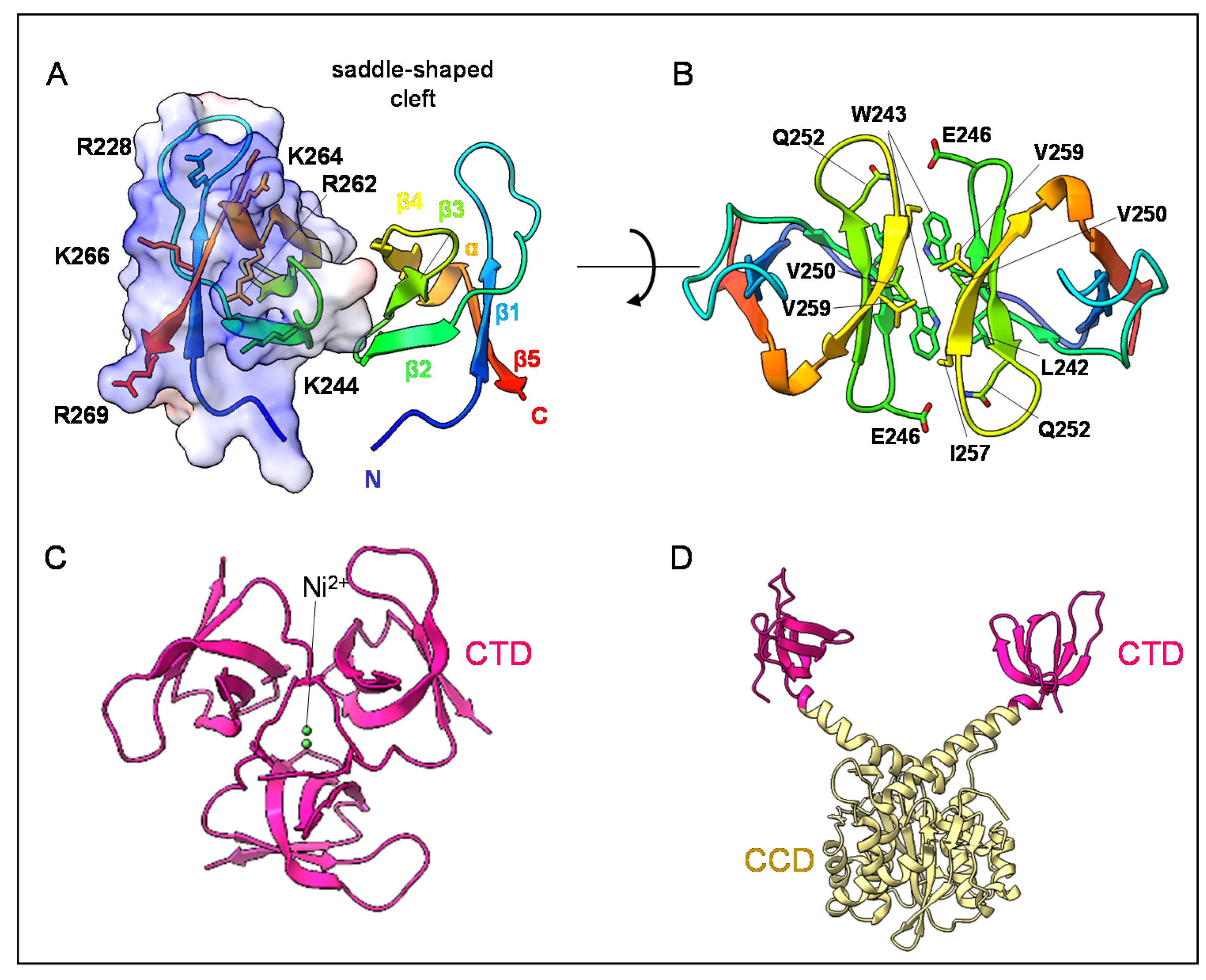
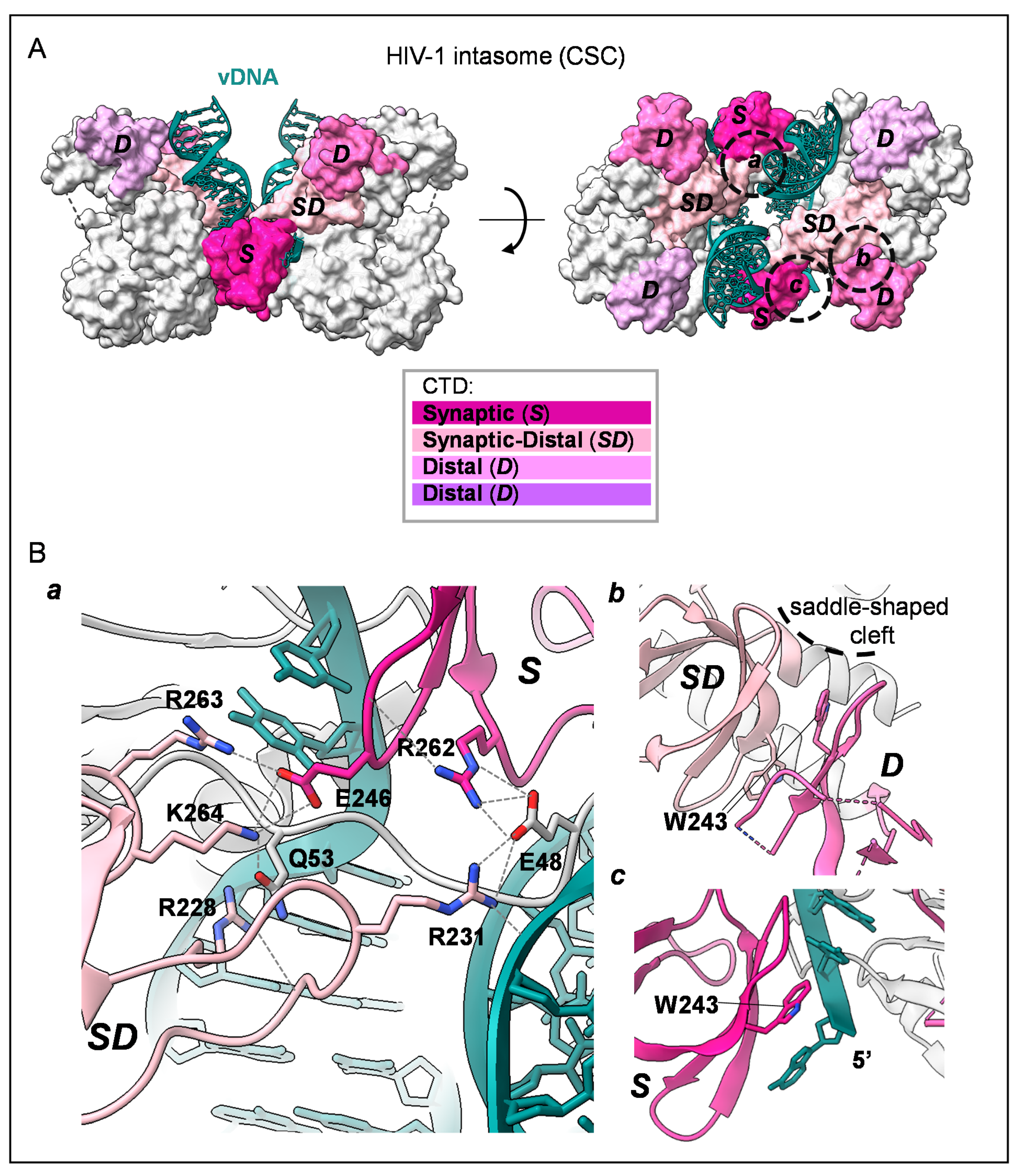
3. The Structure of HIV-1 IN CTD, Interdomains and DNA Interactions
4. Post-Translational Modifications of HIV-1 IN-CTD
5. Interaction of IN-CTD with Reverse Transcriptase
6. Role of the IN-CTD during Nuclear Import
7. Integrase and Chromatin Remodelling System
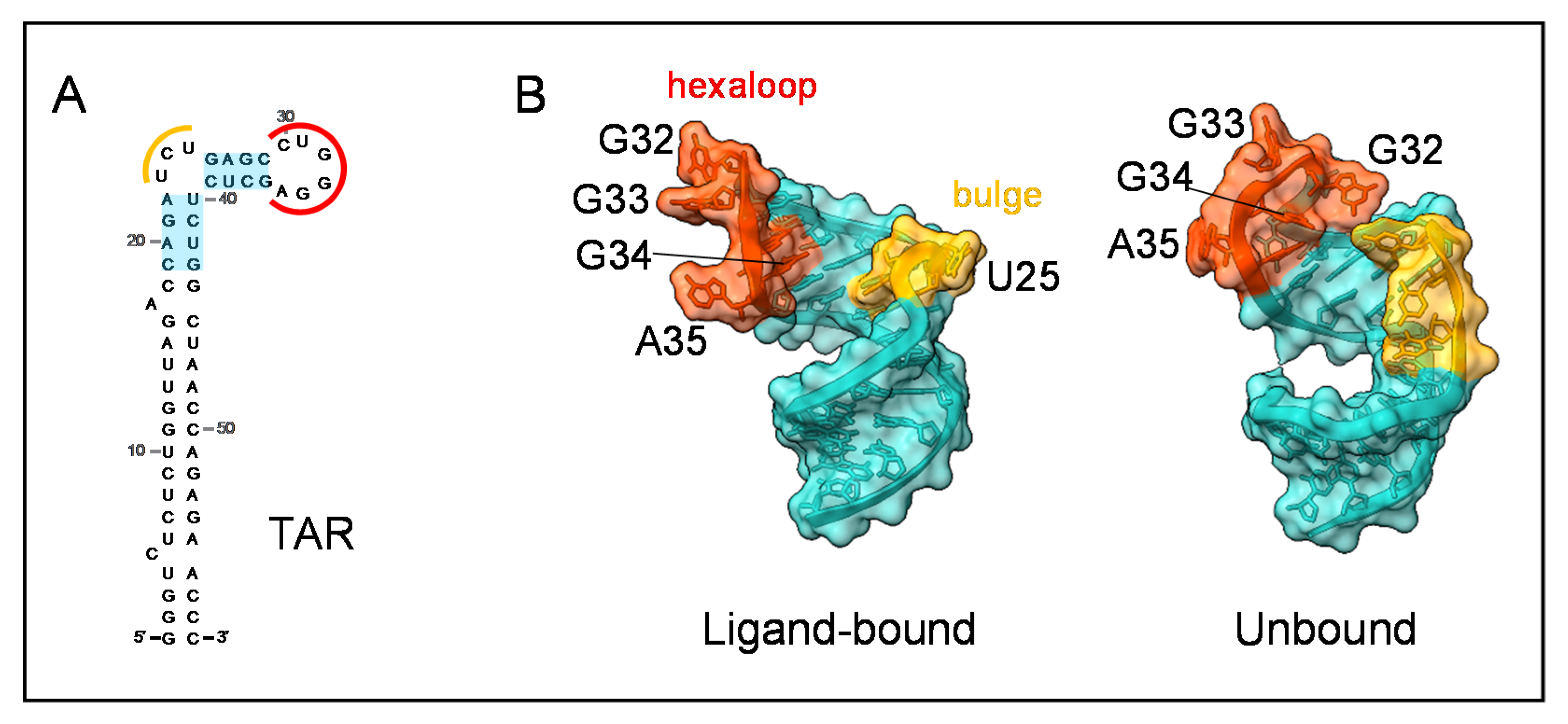
8. RNA-Binding Properties of IN and Impact on Virion Morphogenesis
9. HIV-1 IN in Proviral Transcription
10. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wymant, C.; Bezemer, D.; Blanquart, F.; Ferretti, L.; Gall, A.; Hall, M.; Golubchik, T.; Bakker, M.; Ong, S.H.; Zhao, L.; et al. A highly virulent variant of HIV-1 circulating in the Netherlands. Science 2022, 375, 540–545. [Google Scholar] [CrossRef] [PubMed]
- Maertens, G.N.; Engelman, A.N.; Cherepanov, P. Structure and function of retroviral integrase. Nat. Rev. Microbiol. 2022, 20, 20–34. [Google Scholar] [CrossRef] [PubMed]
- Dobard, C.W.; Briones, M.S.; Chow, S.A. Molecular mechanisms by which human immunodeficiency virus type 1 integrase stimulates the early steps of reverse transcription. J. Virol. 2007, 81, 10037–10046. [Google Scholar] [CrossRef] [PubMed]
- Kobbi, L.; Octobre, G.; Dias, J.; Comisso, M.; Mirande, M. Association of mitochondrial Lysyl-tRNA synthetase with HIV-1 GagPol involves catalytic domain of the synthetase and transframe and integrase domains of Pol. J. Mol. Biol. 2011, 410, 875–886. [Google Scholar] [CrossRef]
- Kovaleski, B.J.; Kennedy, R.; Hong, M.K.; Datta, S.A.; Kleiman, L.; Rein, A.; Musier-Forsyth, K. In vitro characterization of the interaction between HIV-1 Gag and human lysyl-tRNA synthetase. J. Biol. Chem. 2006, 281, 19449–19456. [Google Scholar] [CrossRef]
- Gallay, P.; Hope, T.; Chin, D.; Trono, D. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl. Acad. Sci. USA 1997, 94, 9825–9830. [Google Scholar] [CrossRef]
- Kessl, J.J.; Kutluay, S.B.; Townsend, D.; Rebensburg, S.; Slaughter, A.; Larue, R.C.; Shkriabai, N.; Bakouche, N.; Fuchs, J.R.; Bieniasz, P.D.; et al. HIV-1 Integrase Binds the Viral RNA Genome and Is Essential during Virion Morphogenesis. Cell 2016, 166, 1257–1268.e1212. [Google Scholar] [CrossRef]
- Winans, S.; Goff, S.P. Mutations altering acetylated residues in the CTD of HIV-1 integrase cause defects in proviral transcription at early times after integration of viral DNA. PLoS Pathog. 2020, 16, e1009147. [Google Scholar] [CrossRef]
- Engelman, A. In vivo analysis of retroviral integrase structure and function. Adv. Virus. Res. 1999, 52, 411–426. [Google Scholar] [CrossRef]
- Lu, R.; Ghory, H.Z.; Engelman, A. Genetic analyses of conserved residues in the carboxyl-terminal domain of human immunodeficiency virus type 1 integrase. J. Virol. 2005, 79, 10356–10368. [Google Scholar] [CrossRef]
- Lu, R.; Limon, A.; Devroe, E.; Silver, P.A.; Cherepanov, P.; Engelman, A. Class II integrase mutants with changes in putative nuclear localization signals are primarily blocked at a postnuclear entry step of human immunodeficiency virus type 1 replication. J. Virol. 2004, 78, 12735–12746. [Google Scholar] [CrossRef] [PubMed]
- Craigie, R. Nucleoprotein Intermediates in HIV-1 DNA Integration: Structure and Function of HIV-1 Intasomes. Subcell. Biochem. 2018, 88, 189–210. [Google Scholar] [CrossRef] [PubMed]
- Grandgenett, D.P.; Vora, A.C.; Schiff, R.D. A 32,000-dalton nucleic acid-binding protein from avian retravirus cores possesses DNA endonuclease activity. Virology 1978, 89, 119–132. [Google Scholar] [CrossRef]
- Lesbats, P.; Engelman, A.N.; Cherepanov, P. Retroviral DNA Integration. Chem. Rev. 2016, 116, 12730–12757. [Google Scholar] [CrossRef] [PubMed]
- Jozwik, I.K.; Passos, D.O.; Lyumkis, D. Structural Biology of HIV Integrase Strand Transfer Inhibitors. Trends Pharmacol. Sci. 2020, 41, 611–626. [Google Scholar] [CrossRef] [PubMed]
- Engelman, A.; Hickman, A.B.; Craigie, R. The core and carboxyl-terminal domains of the integrase protein of human immunodeficiency virus type 1 each contribute to nonspecific DNA binding. J. Virol. 1994, 68, 5911–5917. [Google Scholar] [CrossRef]
- Cai, M.; Zheng, R.; Caffrey, M.; Craigie, R.; Clore, G.M.; Gronenborn, A.M. Solution structure of the N-terminal zinc binding domain of HIV-1 integrase. Nat. Struct. Biol. 1997, 4, 567–577. [Google Scholar] [CrossRef]
- Lee, S.P.; Xiao, J.; Knutson, J.R.; Lewis, M.S.; Han, M.K. Zn2+ promotes the self-association of human immunodeficiency virus type-1 integrase in vitro. Biochemistry 1997, 36, 173–180. [Google Scholar] [CrossRef]
- Zheng, R.; Jenkins, T.M.; Craigie, R. Zinc folds the N-terminal domain of HIV-1 integrase, promotes multimerization, and enhances catalytic activity. Proc. Natl. Acad. Sci. USA 1996, 93, 13659–13664. [Google Scholar] [CrossRef]
- Dyda, F.; Hickman, A.B.; Jenkins, T.M.; Engelman, A.; Craigie, R.; Davies, D.R. Crystal structure of the catalytic domain of HIV-1 integrase: Similarity to other polynucleotidyl transferases. Science 1994, 266, 1981–1986. [Google Scholar] [CrossRef]
- Cheng, Q.; Swalla, B.M.; Beck, M.; Alcaraz, R., Jr.; Gumport, R.I.; Gardner, J.F. Specificity determinants for bacteriophage Hong Kong 022 integrase: Analysis of mutants with relaxed core-binding specificities. Mol. Microbiol. 2000, 36, 424–436. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Y.; Ling, H.; Yang, W.; Craigie, R. Structure of a two-domain fragment of HIV-1 integrase: Implications for domain organization in the intact protein. EMBO J. 2001, 20, 7333–7343. [Google Scholar] [CrossRef] [PubMed]
- Passos, D.O.; Li, M.; Jozwik, I.K.; Zhao, X.Z.; Santos-Martins, D.; Yang, R.; Smith, S.J.; Jeon, Y.; Forli, S.; Hughes, S.H.; et al. Structural basis for strand-transfer inhibitor binding to HIV intasomes. Science 2020, 367, 810–814. [Google Scholar] [CrossRef] [PubMed]
- Passos, D.O.; Li, M.; Yang, R.; Rebensburg, S.V.; Ghirlando, R.; Jeon, Y.; Shkriabai, N.; Kvaratskhelia, M.; Craigie, R.; Lyumkis, D. Cryo-EM structures and atomic model of the HIV-1 strand transfer complex intasome. Science 2017, 355, 89–92. [Google Scholar] [CrossRef] [PubMed]
- Hare, S.; Gupta, S.S.; Valkov, E.; Engelman, A.; Cherepanov, P. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature 2010, 464, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Hare, S.; Maertens, G.N.; Cherepanov, P. 3′-processing and strand transfer catalysed by retroviral integrase in crystallo. EMBO J. 2012, 31, 3020–3028. [Google Scholar] [CrossRef]
- Maertens, G.N.; Hare, S.; Cherepanov, P. The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature 2010, 468, 326–329. [Google Scholar] [CrossRef]
- Ballandras-Colas, A.; Maskell, D.P.; Serrao, E.; Locke, J.; Swuec, P.; Jonsson, S.R.; Kotecha, A.; Cook, N.J.; Pye, V.E.; Taylor, I.A.; et al. A supramolecular assembly mediates lentiviral DNA integration. Science 2017, 355, 93–95. [Google Scholar] [CrossRef]
- Cook, N.J.; Li, W.; Berta, D.; Badaoui, M.; Ballandras-Colas, A.; Nans, A.; Kotecha, A.; Rosta, E.; Engelman, A.N.; Cherepanov, P. Structural basis of second-generation HIV integrase inhibitor action and viral resistance. Science 2020, 367, 806–810. [Google Scholar] [CrossRef]
- Lee, G.E.; Kim, J.; Shin, C.G. Single residue mutation in integrase catalytic core domain affects feline foamy viral DNA integration. Biosci. Biotechnol. Biochem. 2019, 83, 270–280. [Google Scholar] [CrossRef]
- Lutzke, R.A.; Vink, C.; Plasterk, R.H. Characterization of the minimal DNA-binding domain of the HIV integrase protein. Nucleic Acids Res. 1994, 22, 4125–4131. [Google Scholar] [CrossRef] [PubMed]
- Woerner, A.M.; Marcus-Sekura, C.J. Characterization of a DNA binding domain in the C-terminus of HIV-1 integrase by deletion mutagenesis. Nucleic Acids Res. 1993, 21, 3507–3511. [Google Scholar] [CrossRef] [PubMed]
- Lodi, P.J.; Ernst, J.A.; Kuszewski, J.; Hickman, A.B.; Engelman, A.; Craigie, R.; Clore, G.M.; Gronenborn, A.M. Solution structure of the DNA binding domain of HIV-1 integrase. Biochemistry 1995, 34, 9826–9833. [Google Scholar] [CrossRef] [PubMed]
- Teyra, J.; Huang, H.; Jain, S.; Guan, X.; Dong, A.; Liu, Y.; Tempel, W.; Min, J.; Tong, Y.; Kim, P.M.; et al. Comprehensive Analysis of the Human SH3 Domain Family Reveals a Wide Variety of Non-canonical Specificities. Structure 2017, 25, 1598–1610.e1593. [Google Scholar] [CrossRef]
- Pek, J.W.; Anand, A.; Kai, T. Tudor domain proteins in development. Development 2012, 139, 2255–2266. [Google Scholar] [CrossRef]
- Eissenberg, J.C. Structural biology of the chromodomain: Form and function. Gene 2012, 496, 69–78. [Google Scholar] [CrossRef]
- Kanja, M.; Cappy, P.; Levy, N.; Oladosu, O.; Schmidt, S.; Rossolillo, P.; Winter, F.; Gasser, R.; Moog, C.; Ruff, M.; et al. NKNK: A New Essential Motif in the C-Terminal Domain of HIV-1 Group M Integrases. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- Chen, J.C.; Krucinski, J.; Miercke, L.J.; Finer-Moore, J.S.; Tang, A.H.; Leavitt, A.D.; Stroud, R.M. Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: A model for viral DNA binding. Proc. Natl. Acad. Sci. USA 2000, 97, 8233–8238. [Google Scholar] [CrossRef]
- Engelman, A. Most of the avian genome appears available for retroviral DNA integration. Bioessays 1994, 16, 797–799. [Google Scholar] [CrossRef]
- Khan, E.; Mack, J.P.; Katz, R.A.; Kulkosky, J.; Skalka, A.M. Retroviral integrase domains: DNA binding and the recognition of LTR sequences. Nucleic Acids Res. 1991, 19, 851–860. [Google Scholar] [CrossRef]
- Vink, C.; Lutzke, R.A.; Plasterk, R.H. Formation of a stable complex between the human immunodeficiency virus integrase protein and viral DNA. Nucleic Acids Res. 1994, 22, 4103–4110. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Eijkelenboom, A.P.; Sprangers, R.; Hard, K.; Puras Lutzke, R.A.; Plasterk, R.H.; Boelens, R.; Kaptein, R. Refined solution structure of the C-terminal DNA-binding domain of human immunovirus-1 integrase. Proteins 1999, 36, 556–564. [Google Scholar] [CrossRef]
- Tsirkone, V.G.; Blokken, J.; De Wit, F.; Breemans, J.; De Houwer, S.; Debyser, Z.; Christ, F.; Strelkov, S.V. N-terminal half of transportin SR2 interacts with HIV integrase. J. Biol. Chem. 2017, 292, 9699–9710. [Google Scholar] [CrossRef] [PubMed]
- Serrao, E.; Ballandras-Colas, A.; Cherepanov, P.; Maertens, G.N.; Engelman, A.N. Key determinants of target DNA recognition by retroviral intasomes. Retrovirology 2015, 12, 39. [Google Scholar] [CrossRef] [PubMed]
- Dar, M.J.; Monel, B.; Krishnan, L.; Shun, M.C.; Di Nunzio, F.; Helland, D.E.; Engelman, A. Biochemical and virological analysis of the 18-residue C-terminal tail of HIV-1 integrase. Retrovirology 2009, 6, 94. [Google Scholar] [CrossRef]
- Elliott, J.L.; Eschbach, J.E.; Koneru, P.C.; Li, W.; Puray-Chavez, M.; Townsend, D.; Lawson, D.Q.; Engelman, A.N.; Kvaratskhelia, M.; Kutluay, S.B. Integrase-RNA interactions underscore the critical role of integrase in HIV-1 virion morphogenesis. Elife 2020, 9, e54311. [Google Scholar] [CrossRef]
- Cereseto, A.; Manganaro, L.; Gutierrez, M.I.; Terreni, M.; Fittipaldi, A.; Lusic, M.; Marcello, A.; Giacca, M. Acetylation of HIV-1 integrase by p300 regulates viral integration. EMBO J. 2005, 24, 3070–3081. [Google Scholar] [CrossRef]
- Manganaro, L.; Lusic, M.; Gutierrez, M.I.; Cereseto, A.; Del Sal, G.; Giacca, M. Concerted action of cellular JNK and Pin1 restricts HIV-1 genome integration to activated CD4+ T lymphocytes. Nat. Med. 2010, 16, 329–333. [Google Scholar] [CrossRef]
- Terreni, M.; Valentini, P.; Liverani, V.; Gutierrez, M.I.; Di Primio, C.; Di Fenza, A.; Tozzini, V.; Allouch, A.; Albanese, A.; Giacca, M.; et al. GCN5-dependent acetylation of HIV-1 integrase enhances viral integration. Retrovirology 2010, 7, 18. [Google Scholar] [CrossRef]
- Zamborlini, A.; Coiffic, A.; Beauclair, G.; Delelis, O.; Paris, J.; Koh, Y.; Magne, F.; Giron, M.L.; Tobaly-Tapiero, J.; Deprez, E.; et al. Impairment of human immunodeficiency virus type-1 integrase SUMOylation correlates with an early replication defect. J. Biol. Chem. 2011, 286, 21013–21022. [Google Scholar] [CrossRef]
- Zheng, Y.; Jayappa, K.D.; Ao, Z.; Qiu, X.; Su, R.C.; Yao, X. Noncovalent SUMO-interaction motifs in HIV integrase play important roles in SUMOylation, cofactor binding, and virus replication. Virol. J. 2019, 16, 42. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Yao, X. Posttranslational modifications of HIV-1 integrase by various cellular proteins during viral replication. Viruses 2013, 5, 1787–1801. [Google Scholar] [CrossRef] [PubMed]
- Roth, S.Y.; Denu, J.M.; Allis, C.D. Histone acetyltransferases. Annu. Rev. Biochem. 2001, 70, 81–120. [Google Scholar] [CrossRef]
- Carrozza, M.J.; Utley, R.T.; Workman, J.L.; Cote, J. The diverse functions of histone acetyltransferase complexes. Trends Genet. 2003, 19, 321–329. [Google Scholar] [CrossRef]
- Topper, M.; Luo, Y.; Zhadina, M.; Mohammed, K.; Smith, L.; Muesing, M.A. Posttranslational acetylation of the human immunodeficiency virus type 1 integrase carboxyl-terminal domain is dispensable for viral replication. J. Virol. 2007, 81, 3012–3017. [Google Scholar] [CrossRef] [PubMed]
- Allouch, A.; Cereseto, A. Identification of cellular factors binding to acetylated HIV-1 integrase. Amino Acids 2011, 41, 1137–1145. [Google Scholar] [CrossRef] [PubMed]
- Cherepanov, P.; Maertens, G.; Proost, P.; Devreese, B.; Van Beeumen, J.; Engelborghs, Y.; De Clercq, E.; Debyser, Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 2003, 278, 372–381. [Google Scholar] [CrossRef] [PubMed]
- Emiliani, S.; Mousnier, A.; Busschots, K.; Maroun, M.; Van Maele, B.; Tempe, D.; Vandekerckhove, L.; Moisant, F.; Ben-Slama, L.; Witvrouw, M.; et al. Integrase mutants defective for interaction with LEDGF/p75 are impaired in chromosome tethering and HIV-1 replication. J. Biol. Chem. 2005, 280, 25517–25523. [Google Scholar] [CrossRef]
- Marshall, H.M.; Ronen, K.; Berry, C.; Llano, M.; Sutherland, H.; Saenz, D.; Bickmore, W.; Poeschla, E.; Bushman, F.D. Role of PSIP1/LEDGF/p75 in lentiviral infectivity and integration targeting. PLoS ONE 2007, 2, e1340. [Google Scholar] [CrossRef]
- Busschots, K.; Voet, A.; De Maeyer, M.; Rain, J.C.; Emiliani, S.; Benarous, R.; Desender, L.; Debyser, Z.; Christ, F. Identification of the LEDGF/p75 binding site in HIV-1 integrase. J. Mol. Biol. 2007, 365, 1480–1492. [Google Scholar] [CrossRef]
- Li, Z.; Wu, S.; Wang, J.; Li, W.; Lin, Y.; Ji, C.; Xue, J.; Chen, J. Evaluation of the interactions of HIV-1 integrase with small ubiquitin-like modifiers and their conjugation enzyme Ubc9. Int. J. Mol. Med. 2012, 30, 1053–1060. [Google Scholar] [CrossRef] [PubMed]
- Kerscher, O. SUMO junction-what’s your function? New insights through SUMO-interacting motifs. EMBO Rep. 2007, 8, 550–555. [Google Scholar] [CrossRef] [PubMed]
- Wei, P.; Garber, M.E.; Fang, S.M.; Fischer, W.H.; Jones, K.A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 1998, 92, 451–462. [Google Scholar] [CrossRef]
- Mousnier, A.; Kubat, N.; Massias-Simon, A.; Segeral, E.; Rain, J.C.; Benarous, R.; Emiliani, S.; Dargemont, C. von Hippel Lindau binding protein 1-mediated degradation of integrase affects HIV-1 gene expression at a postintegration step. Proc. Natl. Acad. Sci. USA 2007, 104, 13615–13620. [Google Scholar] [CrossRef]
- Jaspart, A.; Calmels, C.; Cosnefroy, O.; Bellecave, P.; Pinson, P.; Claverol, S.; Guyonnet-Duperat, V.; Dartigues, B.; Benleulmi, M.S.; Mauro, E.; et al. GCN2 phosphorylates HIV-1 integrase and decreases HIV-1 replication by limiting viral integration. Sci. Rep. 2017, 7, 2283. [Google Scholar] [CrossRef]
- Cosnefroy, O.; Jaspart, A.; Calmels, C.; Parissi, V.; Fleury, H.; Ventura, M.; Reigadas, S.; Andreola, M.L. Activation of GCN2 upon HIV-1 infection and inhibition of translation. Cell Mol. Life Sci. 2013, 70, 2411–2421. [Google Scholar] [CrossRef]
- Del Pino, J.; Jimenez, J.L.; Ventoso, I.; Castello, A.; Munoz-Fernandez, M.A.; de Haro, C.; Berlanga, J.J. GCN2 has inhibitory effect on human immunodeficiency virus-1 protein synthesis and is cleaved upon viral infection. PLoS ONE 2012, 7, e47272. [Google Scholar] [CrossRef] [PubMed]
- Zila, V.; Margiotta, E.; Turonova, B.; Muller, T.G.; Zimmerli, C.E.; Mattei, S.; Allegretti, M.; Borner, K.; Rada, J.; Muller, B.; et al. Cone-shaped HIV-1 capsids are transported through intact nuclear pores. Cell 2021, 184, 1032–1046.e1018. [Google Scholar] [CrossRef]
- Hehl, E.A.; Joshi, P.; Kalpana, G.V.; Prasad, V.R. Interaction between human immunodeficiency virus type 1 reverse transcriptase and integrase proteins. J. Virol. 2004, 78, 5056–5067. [Google Scholar] [CrossRef]
- Tekeste, S.S.; Wilkinson, T.A.; Weiner, E.M.; Xu, X.; Miller, J.T.; Le Grice, S.F.; Clubb, R.T.; Chow, S.A. Interaction between Reverse Transcriptase and Integrase Is Required for Reverse Transcription during HIV-1 Replication. J. Virol. 2015, 89, 12058–12069. [Google Scholar] [CrossRef]
- Wilkinson, T.A.; Januszyk, K.; Phillips, M.L.; Tekeste, S.S.; Zhang, M.; Miller, J.T.; Le Grice, S.F.; Clubb, R.T.; Chow, S.A. Identifying and characterizing a functional HIV-1 reverse transcriptase-binding site on integrase. J. Biol. Chem. 2009, 284, 7931–7939. [Google Scholar] [CrossRef] [PubMed]
- Hehl, A.B.; Marti, M. Secretory protein trafficking in Giardia intestinalis. Mol. Microbiol. 2004, 53, 19–28. [Google Scholar] [CrossRef]
- Wu, X.; Liu, H.; Xiao, H.; Conway, J.A.; Hehl, E.; Kalpana, G.V.; Prasad, V.; Kappes, J.C. Human immunodeficiency virus type 1 integrase protein promotes reverse transcription through specific interactions with the nucleoprotein reverse transcription complex. J. Virol. 1999, 73, 2126–2135. [Google Scholar] [CrossRef] [PubMed]
- Cen, S.; Khorchid, A.; Javanbakht, H.; Gabor, J.; Stello, T.; Shiba, K.; Musier-Forsyth, K.; Kleiman, L. Incorporation of lysyl-tRNA synthetase into human immunodeficiency virus type 1. J. Virol. 2001, 75, 5043–5048. [Google Scholar] [CrossRef] [PubMed]
- Phongsavanh, X.; Al-Qatabi, N.; Shaban, M.S.; Khoder-Agha, F.; El Asri, M.; Comisso, M.; Guerois, R.; Mirande, M. How HIV-1 Integrase Associates with Human Mitochondrial Lysyl-tRNA Synthetase. Viruses 2020, 12, 1202. [Google Scholar] [CrossRef] [PubMed]
- Benleulmi, M.S.; Matysiak, J.; Robert, X.; Miskey, C.; Mauro, E.; Lapaillerie, D.; Lesbats, P.; Chaignepain, S.; Henriquez, D.R.; Calmels, C.; et al. Modulation of the functional association between the HIV-1 intasome and the nucleosome by histone amino-terminal tails. Retrovirology 2017, 14, 54. [Google Scholar] [CrossRef] [PubMed]
- Ao, Z.; Danappa Jayappa, K.; Wang, B.; Zheng, Y.; Kung, S.; Rassart, E.; Depping, R.; Kohler, M.; Cohen, E.A.; Yao, X. Importin alpha3 interacts with HIV-1 integrase and contributes to HIV-1 nuclear import and replication. J. Virol. 2010, 84, 8650–8663. [Google Scholar] [CrossRef]
- Fassati, A.; Gorlich, D.; Harrison, I.; Zaytseva, L.; Mingot, J.M. Nuclear import of HIV-1 intracellular reverse transcription complexes is mediated by importin 7. EMBO J. 2003, 22, 3675–3685. [Google Scholar] [CrossRef]
- Hearps, A.C.; Jans, D.A. HIV-1 integrase is capable of targeting DNA to the nucleus via an importin alpha/beta-dependent mechanism. Biochem. J. 2006, 398, 475–484. [Google Scholar] [CrossRef]
- De Houwer, S.; Demeulemeester, J.; Thys, W.; Taltynov, O.; Zmajkovicova, K.; Christ, F.; Debyser, Z. Identification of residues in the C-terminal domain of HIV-1 integrase that mediate binding to the transportin-SR2 protein. J. Biol. Chem. 2012, 287, 34059–34068. [Google Scholar] [CrossRef]
- Larue, R.; Gupta, K.; Wuensch, C.; Shkriabai, N.; Kessl, J.J.; Danhart, E.; Feng, L.; Taltynov, O.; Christ, F.; Van Duyne, G.D.; et al. Interaction of the HIV-1 intasome with transportin 3 protein (TNPO3 or TRN-SR2). J. Biol. Chem. 2012, 287, 34044–34058. [Google Scholar] [CrossRef] [PubMed]
- Dixit, U.; Bhutoria, S.; Wu, X.; Qiu, L.; Spira, M.; Mathew, S.; Harris, R.; Adams, L.J.; Cahill, S.; Pathak, R.; et al. INI1/SMARCB1 Rpt1 domain mimics TAR RNA in binding to integrase to facilitate HIV-1 replication. Nat. Commun. 2021, 12, 2743. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Rodriguez, G.; Gazi, A.; Monel, B.; Frabetti, S.; Scoca, V.; Mueller, F.; Schwartz, O.; Krijnse-Locker, J.; Charneau, P.; Di Nunzio, F. Remodeling of the Core Leads HIV-1 Preintegration Complex into the Nucleus of Human Lymphocytes. J. Virol. 2020, 94. [Google Scholar] [CrossRef] [PubMed]
- Tabasi, M.; Nombela, I.; Janssens, J.; Lahousse, A.P.; Christ, F.; Debyser, Z. Role of Transportin-SR2 in HIV-1 Nuclear Import. Viruses 2021, 13, 829. [Google Scholar] [CrossRef]
- Jayappa, K.D.; Ao, Z.; Yang, M.; Wang, J.; Yao, X. Identification of critical motifs within HIV-1 integrase required for importin alpha3 interaction and viral cDNA nuclear import. J. Mol. Biol. 2011, 410, 847–862. [Google Scholar] [CrossRef]
- Kvaratskhelia, M.; Sharma, A.; Larue, R.C.; Serrao, E.; Engelman, A. Molecular mechanisms of retroviral integration site selection. Nucleic Acids Res. 2014, 42, 10209–10225. [Google Scholar] [CrossRef]
- Sowd, G.A.; Serrao, E.; Wang, H.; Wang, W.; Fadel, H.J.; Poeschla, E.M.; Engelman, A.N. A critical role for alternative polyadenylation factor CPSF6 in targeting HIV-1 integration to transcriptionally active chromatin. Proc. Natl. Acad. Sci. USA 2016, 113, E1054–E1063. [Google Scholar] [CrossRef]
- Wang, G.P.; Ciuffi, A.; Leipzig, J.; Berry, C.C.; Bushman, F.D. HIV integration site selection: Analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007, 17, 1186–1194. [Google Scholar] [CrossRef]
- Cherepanov, P.; Devroe, E.; Silver, P.A.; Engelman, A. Identification of an evolutionarily conserved domain in human lens epithelium-derived growth factor/transcriptional co-activator p75 (LEDGF/p75) that binds HIV-1 integrase. J. Biol. Chem. 2004, 279, 48883–48892. [Google Scholar] [CrossRef]
- Eidahl, J.O.; Crowe, B.L.; North, J.A.; McKee, C.J.; Shkriabai, N.; Feng, L.; Plumb, M.; Graham, R.L.; Gorelick, R.J.; Hess, S.; et al. Structural basis for high-affinity binding of LEDGF PWWP to mononucleosomes. Nucleic Acids Res. 2013, 41, 3924–3936. [Google Scholar] [CrossRef]
- Llano, M.; Vanegas, M.; Hutchins, N.; Thompson, D.; Delgado, S.; Poeschla, E.M. Identification and characterization of the chromatin-binding domains of the HIV-1 integrase interactor LEDGF/p75. J. Mol. Biol. 2006, 360, 760–773. [Google Scholar] [CrossRef] [PubMed]
- Cherepanov, P. LEDGF/p75 interacts with divergent lentiviral integrases and modulates their enzymatic activity in vitro. Nucleic Acids Res. 2007, 35, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Mauro, E.; Lesbats, P.; Lapaillerie, D.; Chaignepain, S.; Maillot, B.; Oladosu, O.; Robert, X.; Fiorini, F.; Kieffer, B.; Bouaziz, S.; et al. Human H4 tail stimulates HIV-1 integration through binding to the carboxy-terminal domain of integrase. Nucleic Acids Res 2019, 47, 3607–3618. [Google Scholar] [CrossRef]
- Lapaillerie, D.; Lelandais, B.; Mauro, E.; Lagadec, F.; Tumiotto, C.; Miskey, C.; Ferran, G.; Kuschner, N.; Calmels, C.; Metifiot, M.; et al. Modulation of the intrinsic chromatin binding property of HIV-1 integrase by LEDGF/p75. Nucleic Acids Res. 2021, 49, 11241–11256. [Google Scholar] [CrossRef]
- Kalpana, G.V.; Marmon, S.; Wang, W.; Crabtree, G.R.; Goff, S.P. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science 1994, 266, 2002–2006. [Google Scholar] [CrossRef] [PubMed]
- Maroun, M.; Delelis, O.; Coadou, G.; Bader, T.; Segeral, E.; Mbemba, G.; Petit, C.; Sonigo, P.; Rain, J.C.; Mouscadet, J.F.; et al. Inhibition of early steps of HIV-1 replication by SNF5/Ini1. J. Biol. Chem. 2006, 281, 22736–22743. [Google Scholar] [CrossRef] [PubMed]
- Sorin, M.; Yung, E.; Wu, X.; Kalpana, G.V. HIV-1 replication in cell lines harboring INI1/hSNF5 mutations. Retrovirology 2006, 3, 56. [Google Scholar] [CrossRef] [PubMed]
- Yung, E.; Sorin, M.; Pal, A.; Craig, E.; Morozov, A.; Delattre, O.; Kappes, J.; Ott, D.; Kalpana, G.V. Inhibition of HIV-1 virion production by a transdominant mutant of integrase interactor 1. Nat. Med. 2001, 7, 920–926. [Google Scholar] [CrossRef]
- Yung, E.; Sorin, M.; Wang, E.J.; Perumal, S.; Ott, D.; Kalpana, G.V. Specificity of interaction of INI1/hSNF5 with retroviral integrases and its functional significance. J. Virol. 2004, 78, 2222–2231. [Google Scholar] [CrossRef]
- Das, S.; Cano, J.; Kalpana, G.V. Multimerization and DNA binding properties of INI1/hSNF5 and its functional significance. J. Biol. Chem. 2009, 284, 19903–19914. [Google Scholar] [CrossRef]
- Lesbats, P.; Botbol, Y.; Chevereau, G.; Vaillant, C.; Calmels, C.; Arneodo, A.; Andreola, M.L.; Lavigne, M.; Parissi, V. Functional coupling between HIV-1 integrase and the SWI/SNF chromatin remodeling complex for efficient in vitro integration into stable nucleosomes. PLoS Pathog. 2011, 7, e1001280. [Google Scholar] [CrossRef] [PubMed]
- Maillot, B.; Levy, N.; Eiler, S.; Crucifix, C.; Granger, F.; Richert, L.; Didier, P.; Godet, J.; Pradeau-Aubreton, K.; Emiliani, S.; et al. Structural and functional role of INI1 and LEDGF in the HIV-1 preintegration complex. PLoS ONE 2013, 8, e60734. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, M.; Yant, S.R.; Tsai, L.; O’Sullivan, C.; Bam, R.A.; Tsai, A.; Niedziela-Majka, A.; Stray, K.M.; Sakowicz, R.; Cihlar, T. Non-catalytic site HIV-1 integrase inhibitors disrupt core maturation and induce a reverse transcription block in target cells. PLoS ONE 2013, 8, e74163. [Google Scholar] [CrossRef] [PubMed]
- Desimmie, B.A.; Schrijvers, R.; Demeulemeester, J.; Borrenberghs, D.; Weydert, C.; Thys, W.; Vets, S.; Van Remoortel, B.; Hofkens, J.; De Rijck, J.; et al. LEDGINs inhibit late stage HIV-1 replication by modulating integrase multimerization in the virions. Retrovirology 2013, 10, 57. [Google Scholar] [CrossRef] [PubMed]
- Fontana, J.; Jurado, K.A.; Cheng, N.; Ly, N.L.; Fuchs, J.R.; Gorelick, R.J.; Engelman, A.N.; Steven, A.C. Distribution and Redistribution of HIV-1 Nucleocapsid Protein in Immature, Mature, and Integrase-Inhibited Virions: A Role for Integrase in Maturation. J. Virol. 2015, 89, 9765–9780. [Google Scholar] [CrossRef]
- Jurado, K.A.; Wang, H.; Slaughter, A.; Feng, L.; Kessl, J.J.; Koh, Y.; Wang, W.; Ballandras-Colas, A.; Patel, P.A.; Fuchs, J.R.; et al. Allosteric integrase inhibitor potency is determined through the inhibition of HIV-1 particle maturation. Proc. Natl. Acad. Sci. USA 2013, 110, 8690–8695. [Google Scholar] [CrossRef]
- Madison, M.K.; Lawson, D.Q.; Elliott, J.; Ozanturk, A.N.; Koneru, P.C.; Townsend, D.; Errando, M.; Kvaratskhelia, M.; Kutluay, S.B. Allosteric HIV-1 Integrase Inhibitors Lead to Premature Degradation of the Viral RNA Genome and Integrase in Target Cells. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Sharma, A.; Slaughter, A.; Jena, N.; Feng, L.; Kessl, J.J.; Fadel, H.J.; Malani, N.; Male, F.; Wu, L.; Poeschla, E.; et al. A new class of multimerization selective inhibitors of HIV-1 integrase. PLoS Pathog. 2014, 10, e1004171. [Google Scholar] [CrossRef]
- Quillent, C.; Borman, A.M.; Paulous, S.; Dauguet, C.; Clavel, F. Extensive regions of pol are required for efficient human immunodeficiency virus polyprotein processing and particle maturation. Virology 1996, 219, 29–36. [Google Scholar] [CrossRef]
- Elliott, J.L.; Kutluay, S.B. Going beyond Integration: The Emerging Role of HIV-1 Integrase in Virion Morphogenesis. Viruses 2020, 12, 1005. [Google Scholar] [CrossRef]
- Fitzon, T.; Leschonsky, B.; Bieler, K.; Paulus, C.; Schroder, J.; Wolf, H.; Wagner, R. Proline residues in the HIV-1 NH2-terminal capsid domain: Structure determinants for proper core assembly and subsequent steps of early replication. Virology 2000, 268, 294–307. [Google Scholar] [CrossRef] [PubMed]
- Forshey, B.M.; von Schwedler, U.; Sundquist, W.I.; Aiken, C. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J. Virol. 2002, 76, 5667–5677. [Google Scholar] [CrossRef] [PubMed]
- Reicin, A.S.; Ohagen, A.; Yin, L.; Hoglund, S.; Goff, S.P. The role of Gag in human immunodeficiency virus type 1 virion morphogenesis and early steps of the viral life cycle. J. Virol. 1996, 70, 8645–8652. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Murakami, T.; Agresta, B.E.; Campbell, S.; Freed, E.O.; Levin, J.G. Human immunodeficiency virus type 1 N-terminal capsid mutants that exhibit aberrant core morphology and are blocked in initiation of reverse transcription in infected cells. J. Virol. 2001, 75, 9357–9366. [Google Scholar] [CrossRef] [PubMed]
- Chavali, S.S.; Bonn-Breach, R.; Wedekind, J.E. Face-time with TAR: Portraits of an HIV-1 RNA with diverse modes of effector recognition relevant for drug discovery. J. Biol. Chem. 2019, 294, 9326–9341. [Google Scholar] [CrossRef] [PubMed]
- Jonkers, I.; Lis, J.T. Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2015, 16, 167–177. [Google Scholar] [CrossRef]
- Mousseau, G.; Valente, S.T. Role of Host Factors on the Regulation of Tat-Mediated HIV-1 Transcription. Curr. Pharm. Des. 2017, 23, 4079–4090. [Google Scholar] [CrossRef]
- He, N.; Liu, M.; Hsu, J.; Xue, Y.; Chou, S.; Burlingame, A.; Krogan, N.J.; Alber, T.; Zhou, Q. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol. Cell 2010, 38, 428–438. [Google Scholar] [CrossRef]
- Pham, V.V.; Salguero, C.; Khan, S.N.; Meagher, J.L.; Brown, W.C.; Humbert, N.; de Rocquigny, H.; Smith, J.L.; D’Souza, V.M. HIV-1 Tat interactions with cellular 7SK and viral TAR RNAs identifies dual structural mimicry. Nat. Commun. 2018, 9, 4266. [Google Scholar] [CrossRef]
- Schulze-Gahmen, U.; Hurley, J.H. Structural mechanism for HIV-1 TAR loop recognition by Tat and the super elongation complex. Proc. Natl. Acad. Sci. USA 2018, 115, 12973–12978. [Google Scholar] [CrossRef]
- Sobhian, B.; Laguette, N.; Yatim, A.; Nakamura, M.; Levy, Y.; Kiernan, R.; Benkirane, M. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol. Cell 2010, 38, 439–451. [Google Scholar] [CrossRef] [PubMed]
- Ne, E.; Palstra, R.J.; Mahmoudi, T. Transcription: Insights From the HIV-1 Promoter. Int. Rev. Cell Mol. Biol. 2018, 335, 191–243. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Koneru, P.C.; Li, W.; Pathirage, C.; Engelman, A.N.; Kvaratskhelia, M.; Musier-Forsyth, K. HIV-1 integrase binding to genomic RNA 5′-UTR induces local structural changes in vitro and in virio. Retrovirology 2021, 18, 37. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rocchi, C.; Gouet, P.; Parissi, V.; Fiorini, F. The C-Terminal Domain of HIV-1 Integrase: A Swiss Army Knife for the Virus? Viruses 2022, 14, 1397. https://doi.org/10.3390/v14071397
Rocchi C, Gouet P, Parissi V, Fiorini F. The C-Terminal Domain of HIV-1 Integrase: A Swiss Army Knife for the Virus? Viruses. 2022; 14(7):1397. https://doi.org/10.3390/v14071397
Chicago/Turabian StyleRocchi, Cecilia, Patrice Gouet, Vincent Parissi, and Francesca Fiorini. 2022. "The C-Terminal Domain of HIV-1 Integrase: A Swiss Army Knife for the Virus?" Viruses 14, no. 7: 1397. https://doi.org/10.3390/v14071397
APA StyleRocchi, C., Gouet, P., Parissi, V., & Fiorini, F. (2022). The C-Terminal Domain of HIV-1 Integrase: A Swiss Army Knife for the Virus? Viruses, 14(7), 1397. https://doi.org/10.3390/v14071397





