Comprehensive Linear Epitope Prediction System for Host Specificity in Nodaviridae
Abstract
1. Introduction
2. Materials and Methods
2.1. Multi-Expert Voting Mechanism-Based LE Prediction
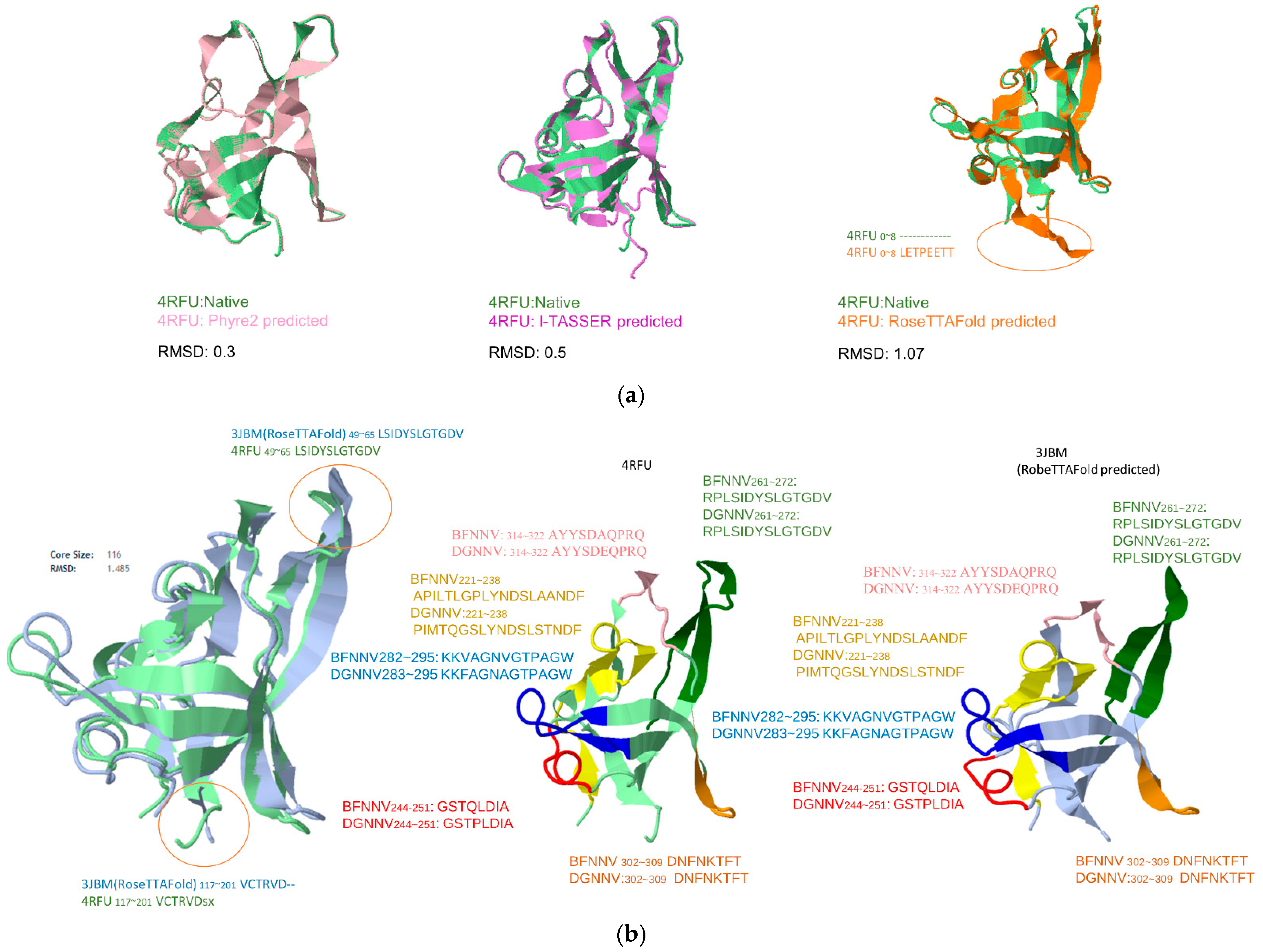
2.2. Validation Method
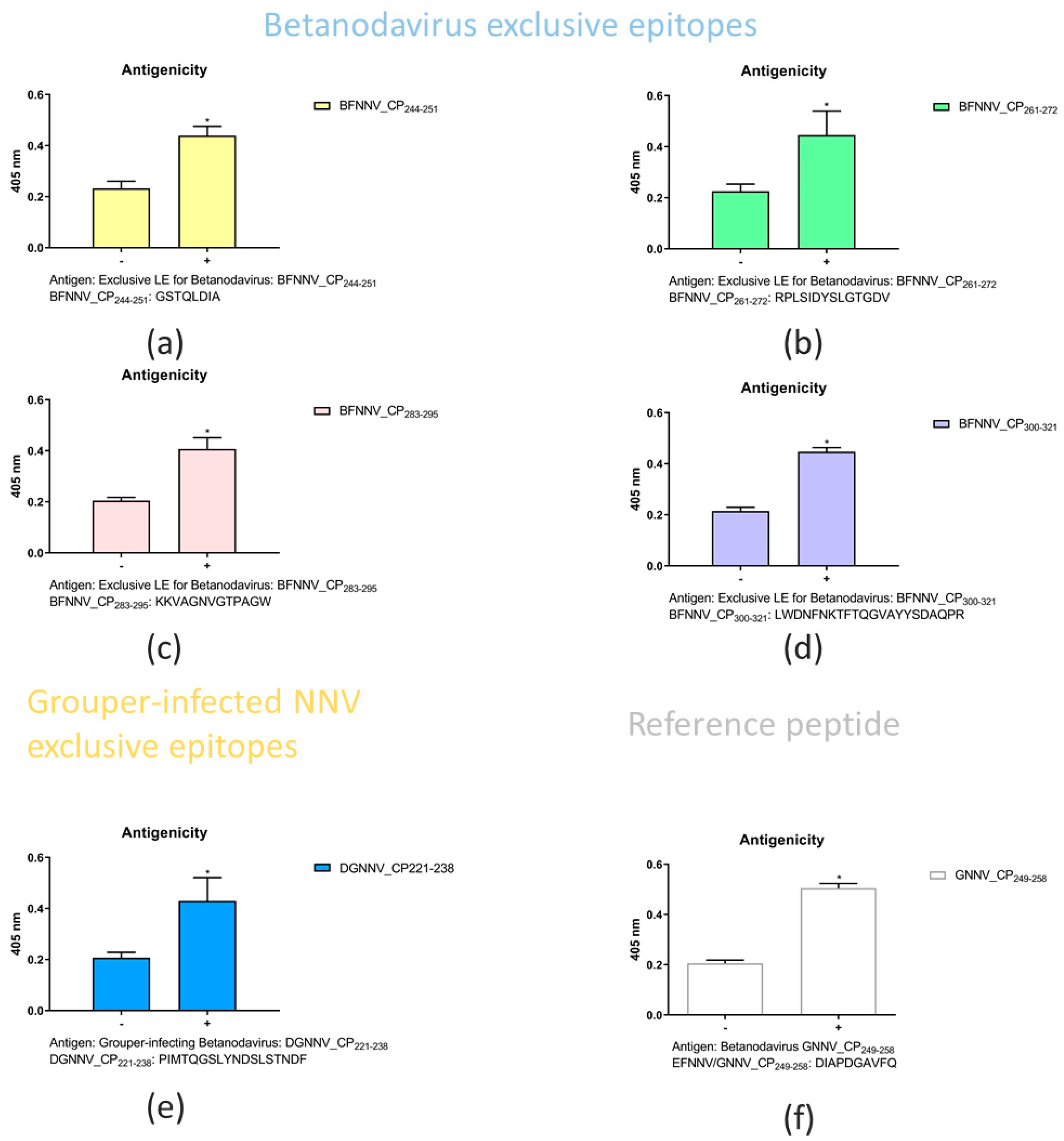
2.3. Biological Assays
3. Results
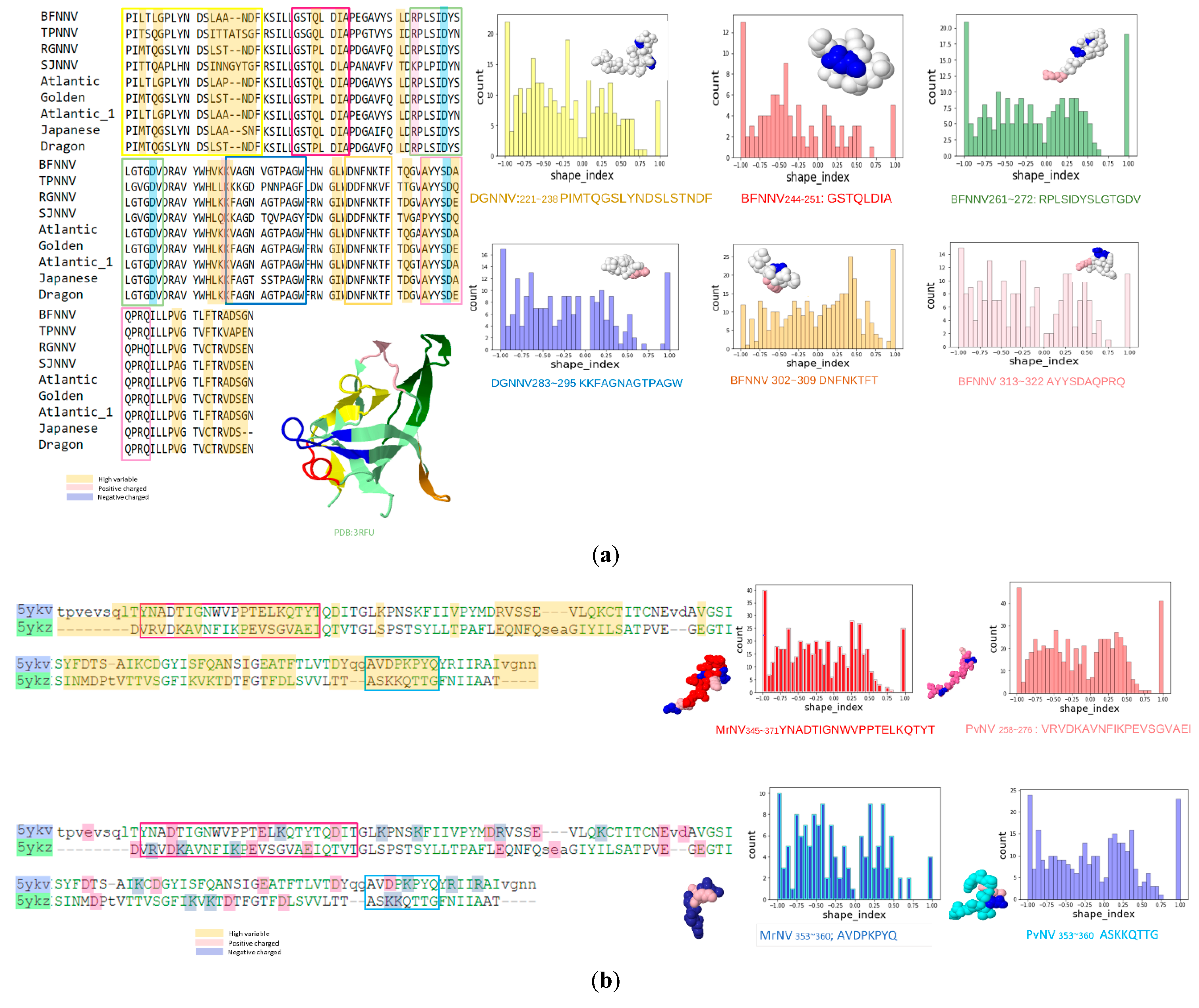
3.1. Prediction Results between Alphanodavirus and Betanodavirus
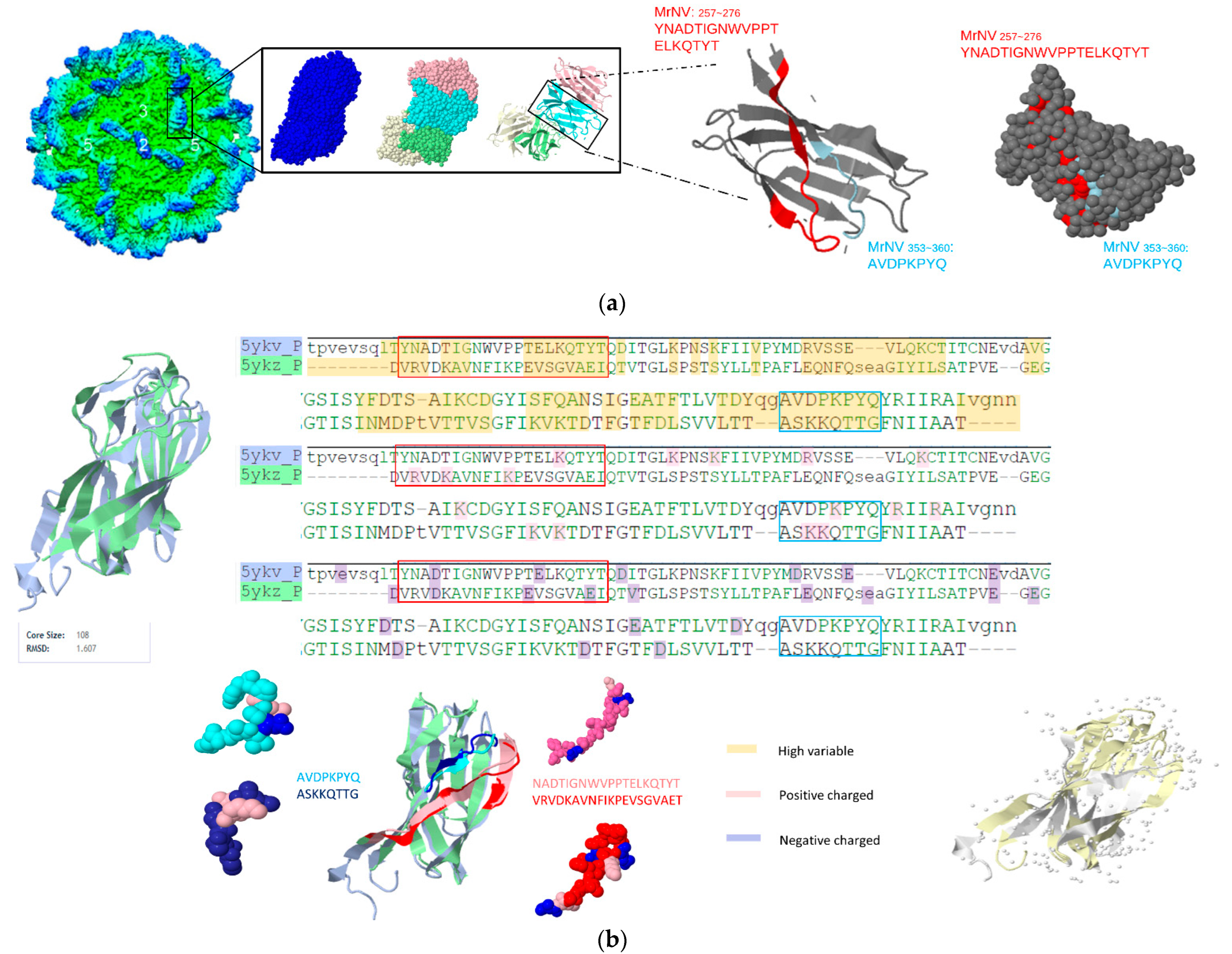
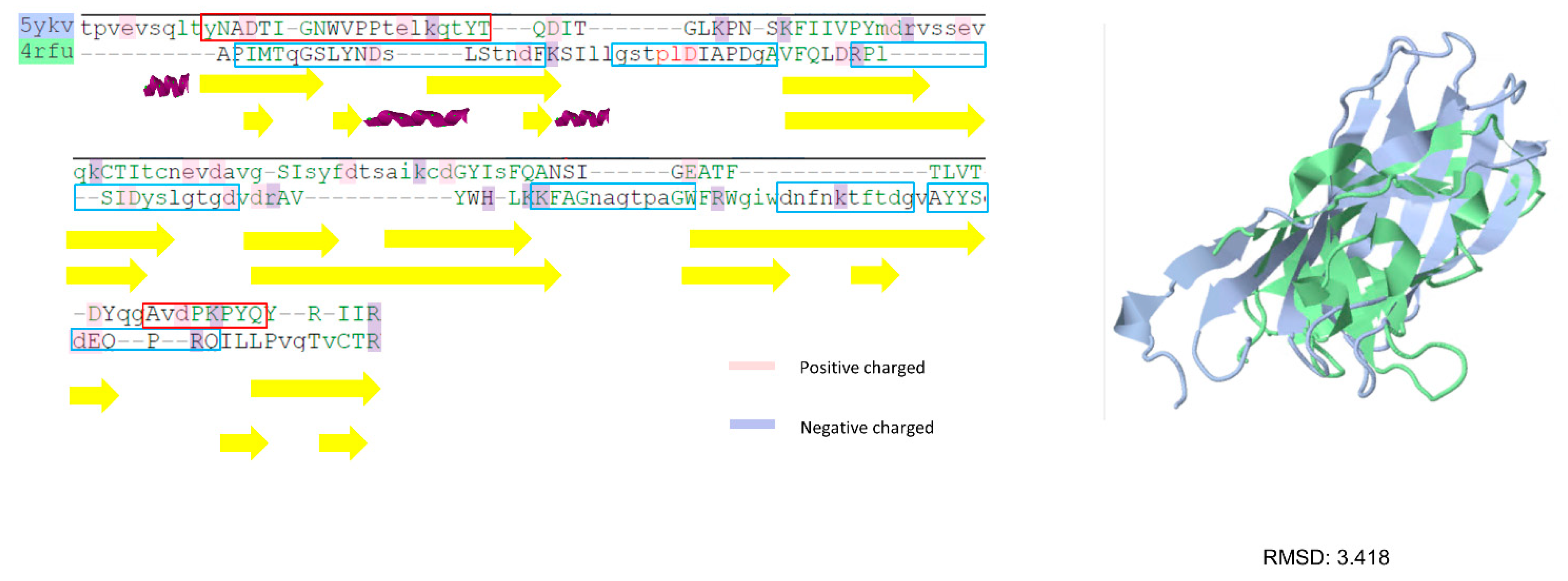
3.2. Prediction Results between Betanodavirus and Gammanodavirus
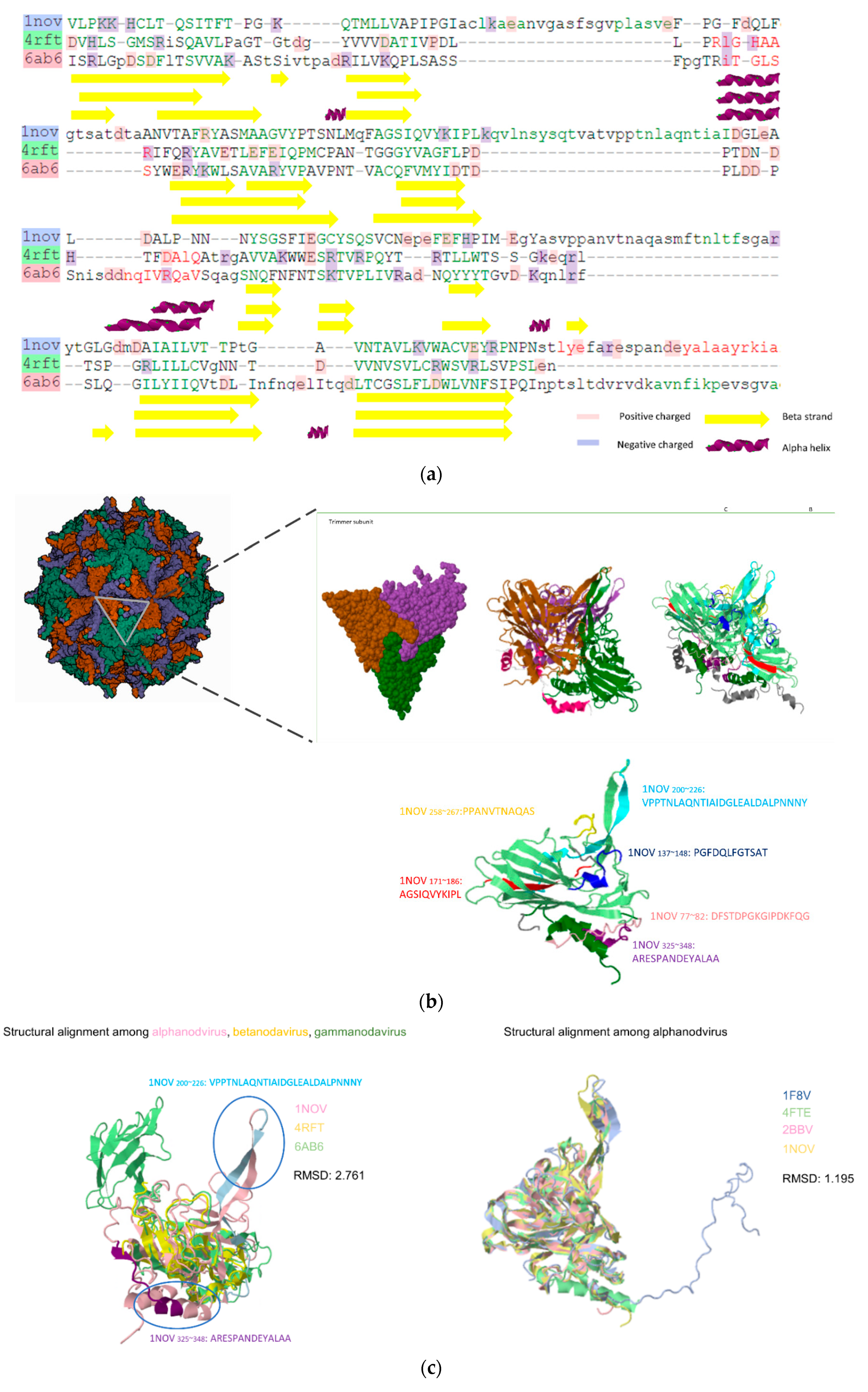
3.3. Prediction Results between Alphanodavirus and Gammanodavirus
3.4. Physicochemical Characteristics of Predicted Residues
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Souto, S.; Olveira, J.; Dopazo, C.; Bandín, I. Reassortant betanodavirus infection in turbot (Scophthalmus maximus). J. Fish Dis. 2016, 39, 1347–1356. [Google Scholar] [CrossRef]
- Costa, J.Z.; Thompson, K.D. Understanding the interaction between Betanodavirus and its host for the development of prophylactic measures for viral encephalopathy and retinopathy. Fish Shellfish Immunol. 2016, 53, 35–49. [Google Scholar] [CrossRef]
- Somrit, M.; Watthammawut, A.; Chotwiwatthanakun, C.; Ounjai, P.; Suntimanawong, W.; Weerachatyanukul, W. C-terminal domain on the outer surface of the Macrobrachium rosenbergii nodavirus capsid is required for Sf9 cell binding and internalization. Virus Res. 2017, 227, 41–48. [Google Scholar] [CrossRef]
- Yong, C.Y.; Yeap, S.K.; Omar, A.R.; Tan, W.S. Advances in the study of nodavirus. PeerJ 2017, 5, e3841. [Google Scholar] [CrossRef]
- Gye, H.J.; Park, M.-J.; Kim, W.-S.; Oh, M.-J.; Nishizawa, T. Heat-denaturation of conformational structures on nervous necrosis virus for generating neutralization antibodies. Aquaculture 2018, 484, 65–70. [Google Scholar] [CrossRef]
- Gye, H.J.; Oh, M.-J.; Nishizawa, T. Lack of nervous necrosis virus (NNV) neutralizing antibodies in convalescent sevenband grouper Hyporthodus septemfasciatus after NNV infection. Vaccine 2018, 36, 1863–1870. [Google Scholar] [CrossRef]
- Ito, Y.; Okinaka, Y.; Mori, K.-I.; Sugaya, T.; Nishioka, T.; Oka, M.; Nakai, T. Variable region of betanodavirus RNA2 is sufficient to determine host specificity. Dis. Aquat. Org. 2008, 79, 199–205. [Google Scholar] [CrossRef]
- Hameed, A.S.; Ninawe, A.; Nakai, T.; Chi, S.; Johnson, K. ICTV virus taxonomy profile: Nodaviridae. J. Gen. Virol. 2019, 100, 3–4. [Google Scholar] [CrossRef]
- Tang, L.; Lin, C.-S.; Krishna, N.K.; Yeager, M.; Schneemann, A.; Johnson, J.E. Virus-like particles of a fish nodavirus display a capsid subunit domain organization different from that of insect nodaviruses. J. Virol. 2002, 76, 6370–6375. [Google Scholar] [CrossRef]
- Vendramin, N.; Toffan, A.; Mancin, M.; Cappellozza, E.; Panzarin, V.; Bovo, G.; Cattoli, G.; Capua, I.; Terregino, C. Comparative pathogenicity study of ten different betanodavirus strains in experimentally infected E uropean sea bass, Dicentrarchus labrax (L.). J. Fish Dis. 2014, 37, 371–383. [Google Scholar] [CrossRef]
- Low, C.F.; Syarul Nataqain, B.; Chee, H.Y.; Rozaini, M.; Najiah, M. Betanodavirus: Dissection of the viral life cycle. J. Fish Dis. 2017, 40, 1489–1496. [Google Scholar] [CrossRef] [PubMed]
- Schneemann, A.; Marshall, D. Specific encapsidation of nodavirus RNAs is mediated through the C terminus of capsid precursor protein alpha. J. Virol. 1998, 72, 8738–8746. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.-C.; Yoshimura, M.; Guan, H.-H.; Wang, T.-Y.; Misumi, Y.; Lin, C.-C.; Chuankhayan, P.; Nakagawa, A.; Chan, S.I.; Tsukihara, T. Crystal structures of a piscine betanodavirus: Mechanisms of capsid assembly and viral infection. PLoS Pathog. 2015, 11, e1005203. [Google Scholar] [CrossRef] [PubMed]
- Dolja, V.V.; Koonin, E.V. Phylogeny of capsid proteins of small icosahedral RNA plant viruses. J. Gen. Virol. 1991, 72, 1481–1486. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.R.; Hryc, C.F.; Jakana, J.; Jiang, H.; Wang, D.; Chiu, W.; Zhong, W.; Tao, Y.J. Crystal structure of a nematode-infecting virus. Proc. Natl. Acad. Sci. USA 2014, 111, 12781–12786. [Google Scholar] [CrossRef]
- Reed, J.; Reed, T.A. A set of constructed type spectra for the practical estimation of peptide secondary structure from circular dichroism. Anal. Biochem. 1997, 254, 36–40. [Google Scholar] [CrossRef]
- Chong, L.C.; Ganesan, H.; Yong, C.Y.; Tan, W.S.; Ho, K.L. Expression, purification and characterization of the dimeric protruding domain of Macrobrachium rosenbergii nodavirus capsid protein expressed in Escherichia coli. PLoS ONE 2019, 14, e0211740. [Google Scholar] [CrossRef]
- Ho, K.L.; Gabrielsen, M.; Beh, P.L.; Kueh, C.L.; Thong, Q.X.; Streetley, J.; Tan, W.S.; Bhella, D. Structure of the Macrobrachium rosenbergii nodavirus: A new genus within the Nodaviridae? PLoS Biol. 2018, 16, e3000038. [Google Scholar] [CrossRef]
- Costa, J.; Adams, A.; Bron, J.; Thompson, K.; Starkey, W.; Richards, R. Identification of B-cell epitopes on the betanodavirus capsid protein. J. Fish Dis. 2007, 30, 419–426. [Google Scholar] [CrossRef]
- Chen, C.-W.; Wu, M.-S.; Huang, Y.-J.; Cheng, C.-A.; Chang, C.-Y. Recognition of linear B-cell epitope of betanodavirus coat protein by RG-M18 neutralizing mAB inhibits giant grouper nervous necrosis virus (GGNNV) infection. PLoS ONE 2015, 10, e0126121. [Google Scholar] [CrossRef]
- Ho, K.L.; Kueh, C.L.; Beh, P.L.; Tan, W.S.; Bhella, D. Cryo-electron microscopy structure of the Macrobrachium rosenbergii nodavirus capsid at 7 angstroms resolution. Sci. Rep. 2017, 7, 2083. [Google Scholar] [CrossRef] [PubMed]
- Zolla-Pazner, S.; Cardozo, T. Structure–function relationships of HIV-1 envelope sequence-variable regions refocus vaccine design. Nat. Rev. Immunol. 2010, 10, 527–535. [Google Scholar] [CrossRef] [PubMed]
- EL-Manzalawy, Y.; Dobbs, D.; Honavar, V. Predicting linear B-cell epitopes using string kernels. J. Mol. Recognit. Interdiscip. J. 2008, 21, 243–255. [Google Scholar] [CrossRef] [PubMed]
- Soria-Guerra, R.E.; Nieto-Gomez, R.; Govea-Alonso, D.O.; Rosales-Mendoza, S. An overview of bioinformatics tools for epitope prediction: Implications on vaccine development. J. Biomed. Inform. 2015, 53, 405–414. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
- Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 2008, 9, 40. [Google Scholar] [CrossRef]
- Baek, M.; DiMaio, F.; Anishchenko, I.; Dauparas, J.; Ovchinnikov, S.; Lee, G.R.; Wang, J.; Cong, Q.; Kinch, L.N.; Schaeffer, R.D. Accurate prediction of protein structures and interactions using a three-track neural network. Science 2021, 373, 871–876. [Google Scholar] [CrossRef]
- Greenbaum, J.A.; Andersen, P.H.; Blythe, M.; Bui, H.H.; Cachau, R.E.; Crowe, J.; Davies, M.; Kolaskar, A.; Lund, O.; Morrison, S. Towards a consensus on datasets and evaluation metrics for developing B-cell epitope prediction tools. J. Mol. Recognit. Interdiscip. J. 2007, 20, 75–82. [Google Scholar] [CrossRef]
- Singh, H.; Ansari, H.R.; Raghava, G.P. Improved method for linear B-cell epitope prediction using antigen’s primary sequence. PLoS ONE 2013, 8, e62216. [Google Scholar] [CrossRef]
- Jespersen, M.C.; Peters, B.; Nielsen, M.; Marcatili, P. BepiPred-2.0: Improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017, 45, W24–W29. [Google Scholar] [CrossRef]
- Saha, S.; Raghava, G.P.S. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins Struct. Funct. Bioinform. 2006, 65, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-W.; Lin, Y.-C.; Pai, T.-W.; Chang, H.-T. Prediction of B-cell linear epitopes with a combination of support vector machine classification and amino acid propensity identification. J. Biomed. Biotechnol. 2011, 2011, 432830. [Google Scholar] [CrossRef] [PubMed]
- Pruitt, K.D.; Tatusova, T.; Klimke, W.; Maglott, D.R. NCBI Reference Sequences: Current status, policy and new initiatives. Nucleic Acids Res. 2009, 37 (Suppl. 1), D32–D36. [Google Scholar] [CrossRef] [PubMed]
- Consortium, U. UniProt: A hub for protein information. Nucleic Acids Res. 2015, 43, D204–D212. [Google Scholar] [CrossRef]
- Batzoglou, S.; Chan, C.X.; Zhou, Y. T-coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000, 1, 1. [Google Scholar]
- Yao, B.; Zhang, L.; Liang, S.; Zhang, C. SVMTriP: A method to predict antigenic epitopes using support vector machine to integrate tri-peptide similarity and propensity. PLoS ONE 2012, 7, e45152. [Google Scholar] [CrossRef]
- Chen, J.; Liu, H.; Yang, J.; Chou, K.-C. Prediction of linear B-cell epitopes using amino acid pair antigenicity scale. Amino Acids 2007, 33, 423–428. [Google Scholar] [CrossRef]
- Stormo, G.D.; Schneider, T.D.; Gold, L.; Ehrenfeucht, A. Use of the ‘Perceptron’ algorithm to distinguish translational initiation sites in E. coli. Nucleic Acids Res. 1982, 10, 2997–3011. [Google Scholar] [CrossRef]
- Saha, S.; Raghava, G.P.S. BcePred: Prediction of continuous B-cell epitopes in antigenic sequences using physico-chemical properties. In Proceedings of the International Conference on Artificial Immune Systems, Catania, Italy, 13–16 September 2004; Springer: Berlin/Heidelberg, Germany, 2004; pp. 197–204. [Google Scholar]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef]
- Vihinen, M.; Torkkila, E.; Riikonen, P. Accuracy of protein flexibility predictions. Proteins Struct. Funct. Bioinform. 1994, 19, 141–149. [Google Scholar] [CrossRef]
- Emini, E.A.; Hughes, J.V.; Perlow, D.; Boger, J. Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J. Virol. 1985, 55, 836–839. [Google Scholar] [CrossRef] [PubMed]
- Grantham, R. Amino acid difference formula to help explain protein evolution. Science 1974, 185, 862–864. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Xiong, Y.; Zhao, M.; Zou, H.; Ye, X.; Liu, J. Prediction of conformational B-cell epitopes from 3D structures by random forests with a distance-based feature. BMC Bioinform. 2011, 12, 341. [Google Scholar] [CrossRef] [PubMed]
- Frishman, D.; Argos, P. Knowledge-based protein secondary structure assignment. Proteins Struct. Funct. Bioinform. 1995, 23, 566–579. [Google Scholar] [CrossRef]
- Jones, S.; Thornton, J.M. Analysis of protein-protein interaction sites using surface patches. J. Mol. Biol. 1997, 272, 121–132. [Google Scholar] [CrossRef]
- Sweredoski, M.J.; Baldi, P. PEPITO: Improved discontinuous B-cell epitope prediction using multiple distance thresholds and half sphere exposure. Bioinformatics 2008, 24, 1459–1460. [Google Scholar] [CrossRef]
- Sanner, M.F.; Olson, A.J.; Spehner, J.C. Reduced surface: An efficient way to compute molecular surfaces. Biopolymers 1996, 38, 305–320. [Google Scholar] [CrossRef]
- Rubinstein, N.D.; Mayrose, I.; Martz, E.; Pupko, T. Epitopia: A web-server for predicting B-cell epitopes. BMC Bioinform. 2009, 10, 287. [Google Scholar] [CrossRef]
- Zhou, Q.; Grinspun, E.; Zorin, D.; Jacobson, A. Mesh arrangements for solid geometry. ACM Trans. Graph. (TOG) 2016, 35, 39. [Google Scholar] [CrossRef]
- Bradford, J.R.; Westhead, D.R. Improved prediction of protein–protein binding sites using a support vector machines approach. Bioinformatics 2005, 21, 1487–1494. [Google Scholar] [CrossRef]
- Tittle, J.S.; Perotti, V.J. The perception of shape and curvedness from binocular stereopsis and structure from motion. Percept. Psychophys. 1997, 59, 1167–1179. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lin, C.-F.; Jiang, H.-K.; Chen, N.-C.; Wang, T.-Y.; Chen, T.-Y. Novel subunit vaccine with linear array epitope protect giant grouper against nervous necrosis virus infection. Fish Shellfish Immunol. 2018, 74, 551–558. [Google Scholar] [CrossRef] [PubMed]
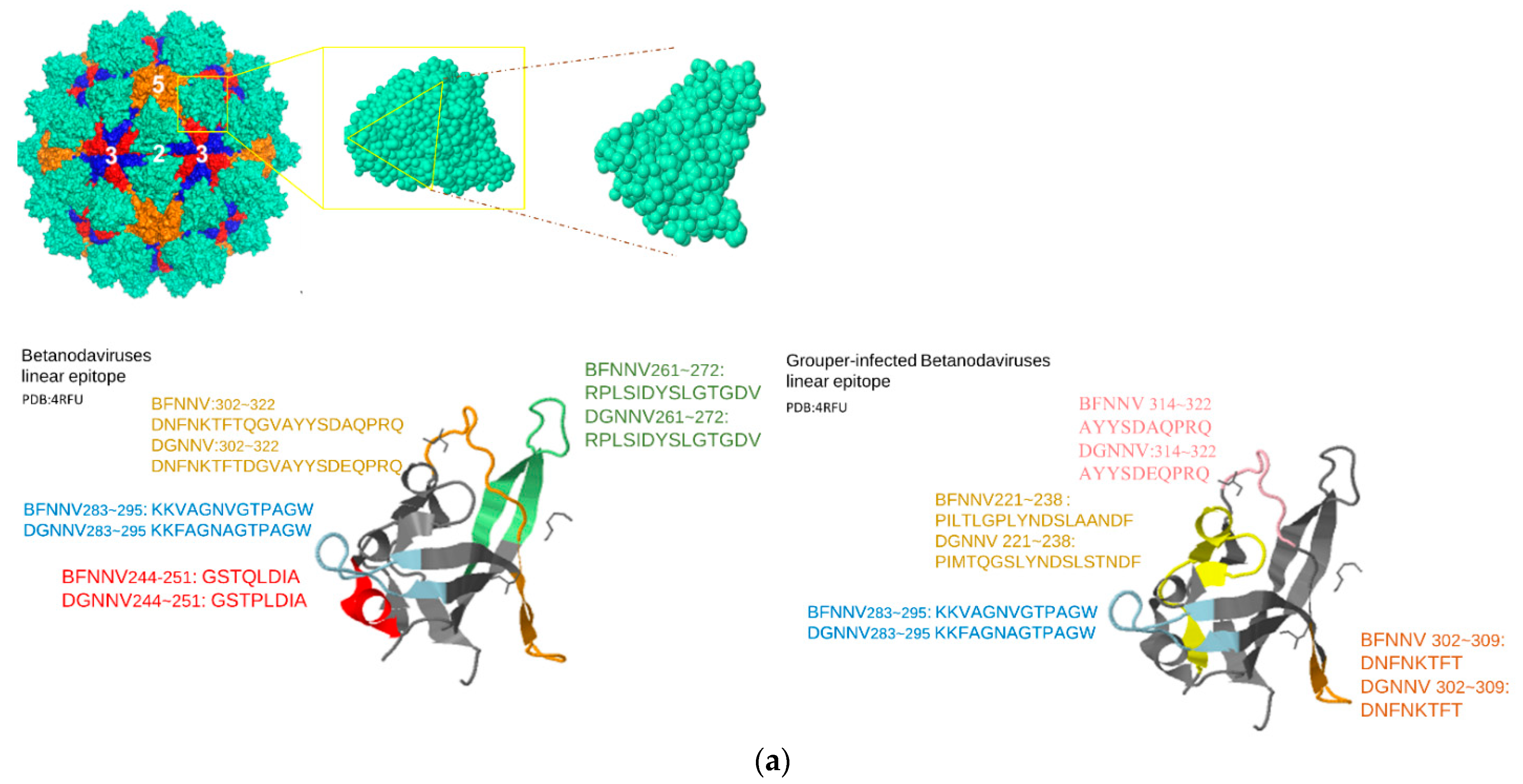
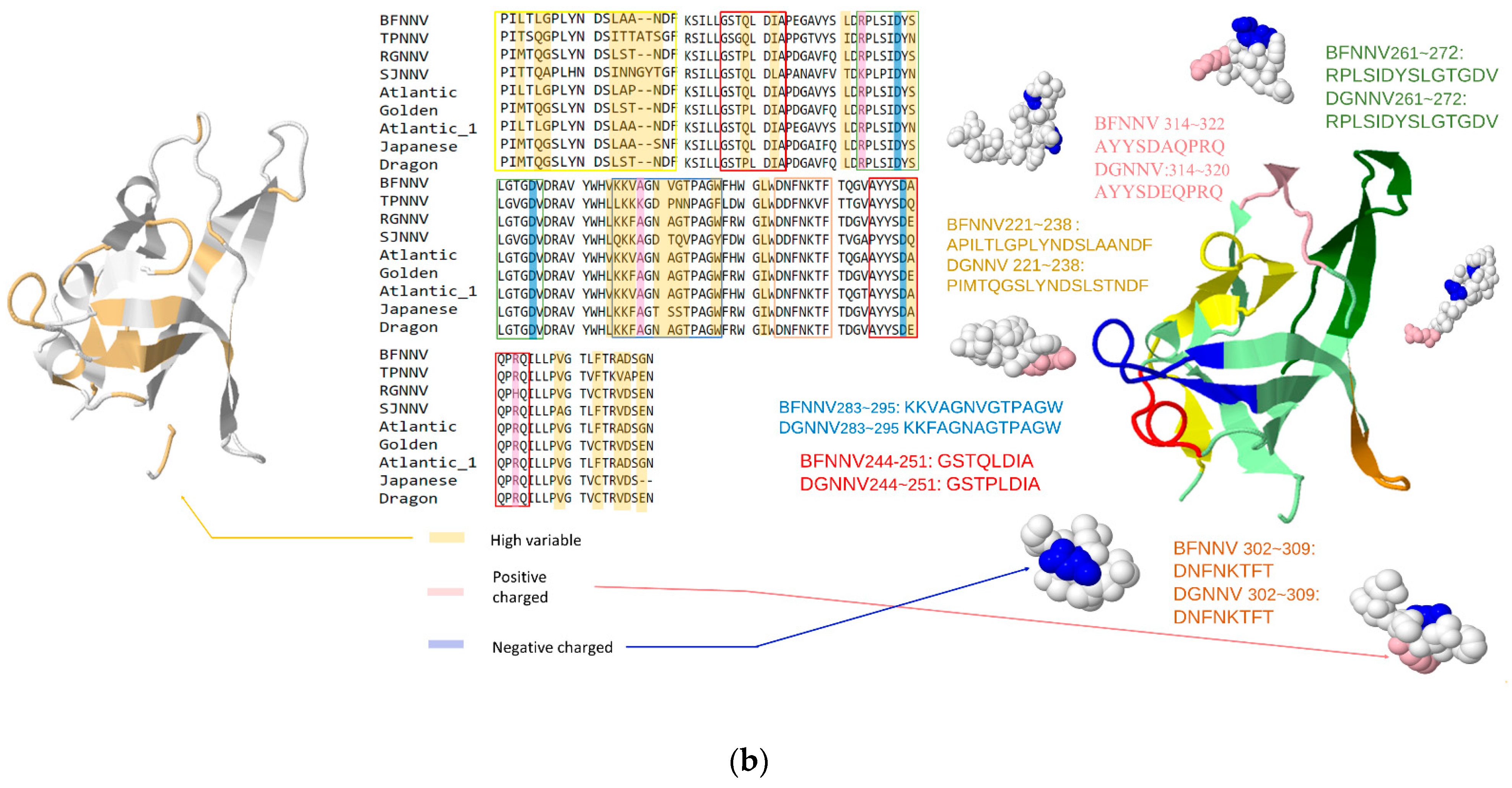
| Genus | Selected Species | Hosts | RCSB ID | NCBI GenBank |
|---|---|---|---|---|
| Alphanodviruses | Flock House virus (FHV) | Barley/Saccharomyces Cerevisiae/Moth/Beetle | 4FSJ 6ITB 6ITF 4RFT | EF690538.1 |
| Black Beetle virus (BBV) | Beetle | 2BBV | X00956.1 | |
| Drosophila melanogaster American nodavirus (DmANV) | Bee | GQ342966.1 | ||
| Nodamura virus (Nov) | Moth/Mosquito/Bee Wild swine | 1NOV | NC_002691.1 | |
| Boolarra virus (Bov) | Moth | NC_004145.1 | ||
| Pariacoto virus (Pav) | Moth | 1F8V | NC_003692.1 | |
| Betanodaviruses | Striped jack nervous necrosis virus (SJNNV) | Bass | NC_003449.1 | |
| Tiger puffer nervous necrosis virus (TPNNV) | Puffer | NC_013461.1 | ||
| Atlantic halibut nodavirus (AHNV) | Halibut | AY962682.1 | ||
| Golden pompano nervous necrosis virus (GPNNV) | Pompano | HQ859934.1 | ||
| Atlantic cod nodavirus (ACNV) | Cod | ABU95413.1 | ||
| Japanese flounder nervous necrosis virus (JFNNV) | Flounder | BAB00609.2 | ||
| Dragon grouper nervous necrosis virus (DGNNV) | Grouper | 3JBM | AAG22496.1 | |
| Barfin flounder nervous necrosis virus (BFNNV) | Flounder | NC_013459.1 | ||
| Redspotted grouper nervous necrosis virus (RGNNV) | Grouper | 3JBM | NC_008041.1 | |
| Epinephelus coioides nervous necrosis virus (GNNV/EFNNV) | Grouper | 4RFU (P-domain only) 4RFT (S-domain only) 4WIZ | MG874758.1 | |
| Gammanodaviruses | Macrobrachium rosenbergii nodavirus (MrNV) | Macrobrachium rosenbergii | 6H2B 6JJC 5ykv | NC_005095.1 |
| Penaeus vannamei nodavirus (PvNV) | Whiteleg Shrimp | 5YKZ (P-domain only) 5YL0 | NC_014977.1 |
| Nodaviridae | Predictive LEs of Representative Peptide | Residue Location | IEDB(CSS) | SVM_Classifier |
|---|---|---|---|---|
| Grouper-infecting betanodavirus | PILTLGPLYNDSLAANDF PIMTQGSLYNDSLSTNDF | BFNNV: 221~238 DGNNV: 221~238 | 136550 | Y |
| KKVAGNVGTPAGW KKFAGNAGTPAGW | BFNNV: 283~295 DGNNV: 283~295 | N/A | Y | |
| DNFNKTFT DNFNKTFT | BFNNV: 302~309 DGNNV: 302~309 | N/A | Y | |
| AYYSDAQPRQ AYYSDEQPRQ | BFNNV: 313~322 DGNNV: 313~322 | N/A | Y |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shih, T.-C.; Ho, L.-P.; Chou, H.-Y.; Wu, J.-L.; Pai, T.-W. Comprehensive Linear Epitope Prediction System for Host Specificity in Nodaviridae. Viruses 2022, 14, 1357. https://doi.org/10.3390/v14071357
Shih T-C, Ho L-P, Chou H-Y, Wu J-L, Pai T-W. Comprehensive Linear Epitope Prediction System for Host Specificity in Nodaviridae. Viruses. 2022; 14(7):1357. https://doi.org/10.3390/v14071357
Chicago/Turabian StyleShih, Tao-Chuan, Li-Ping Ho, Hsin-Yiu Chou, Jen-Leih Wu, and Tun-Wen Pai. 2022. "Comprehensive Linear Epitope Prediction System for Host Specificity in Nodaviridae" Viruses 14, no. 7: 1357. https://doi.org/10.3390/v14071357
APA StyleShih, T.-C., Ho, L.-P., Chou, H.-Y., Wu, J.-L., & Pai, T.-W. (2022). Comprehensive Linear Epitope Prediction System for Host Specificity in Nodaviridae. Viruses, 14(7), 1357. https://doi.org/10.3390/v14071357







