Serological Evidence of Exposure to Onyong-Nyong and Chikungunya Viruses in Febrile Patients of Rural Taita-Taveta County and Urban Kibera Informal Settlement in Nairobi, Kenya
Abstract
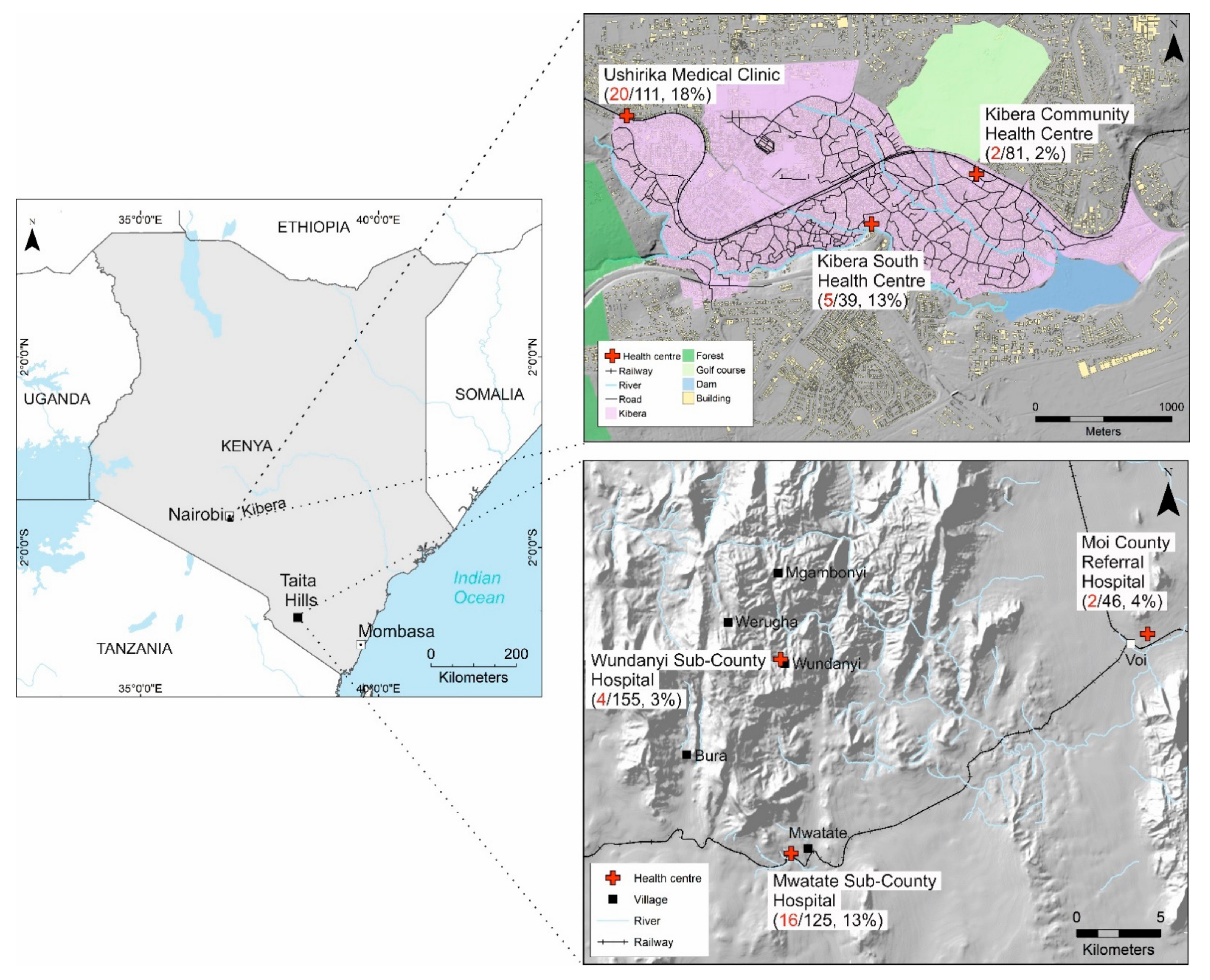
:1. Introduction
2. Materials and Methods
3. Results
3.1. Participant Characteristics
3.2. CHIKV IgG IFA and IgM ELISA Antibody Assay Results
3.3. Plaque Reduction Neutralization Assay Results
3.4. Factors Associated with Alphavirus Seropositivity
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- ICTV. ICTV Master Species List 2021.v1. 2022. Available online: https://talk.ictvonline.org/files/master-species-lists/m/msl/13425 (accessed on 12 April 2022).
- Azar, S.R.; Campos, R.K.; Bergren, N.A.; Camargos, V.N.; Rossi, S.L. Epidemic Alphaviruses: Ecology, Emergence and Outbreaks. Microorganisms 2020, 8, 1167. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Mukhopadhyay, S.; Merits, A.; Bolling, B.; Nasar, F.; Coffey, L.L.; Powers, A.; Weaver, S.C.; Ictv Report, C. ICTV Virus Taxonomy Profile: Togaviridae. J. Gen. Virol. 2018, 99, 761–762. [Google Scholar] [CrossRef] [PubMed]
- Tigoi, C.; Lwande, O.; Orindi, B.; Irura, Z.; Ongus, J.; Sang, R. Seroepidemiology of selected arboviruses in febrile patients visiting selected health facilities in the lake/river basin areas of Lake Baringo, Lake Naivasha, and Tana River, Kenya. Vector Borne Zoonotic Dis. 2015, 15, 124–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LaBeaud, A.D.; Banda, T.; Brichard, J.; Muchiri, E.M.; Mungai, P.L.; Mutuku, F.M.; Borland, E.; Gildengorin, G.; Pfeil, S.; Teng, C.Y.; et al. High rates of o’nyong nyong and Chikungunya virus transmission in coastal Kenya. PLoS Negl. Trop. Dis. 2015, 9, e0003436. [Google Scholar] [CrossRef] [PubMed]
- Raulino, R.; Thaurignac, G.; Butel, C.; Villabona-Arenas, C.J.; Foe, T.; Loul, S.; Ndimbo-Kumugo, S.P.; Mbala-Kingebeni, P.; Makiala-Mandanda, S.; Ahuka-Mundeke, S.; et al. Multiplex detection of antibodies to Chikungunya, O’nyong-nyong, Zika, Dengue, West Nile and Usutu viruses in diverse non-human primate species from Cameroon and the Democratic Republic of Congo. PLoS Negl. Trop. Dis. 2021, 15, e0009028. [Google Scholar] [CrossRef] [PubMed]
- Lwande, O.W.; Obanda, V.; Bucht, G.; Mosomtai, G.; Otieno, V.; Ahlm, C.; Evander, M. Global emergence of Alphaviruses that cause arthritis in humans. Infect. Ecol. Epidemiol. 2015, 5, 29853. [Google Scholar] [CrossRef] [Green Version]
- Moizéis, R.N.C.; Fernandes, T.; Guedes, P.; Pereira, H.W.B.; Lanza, D.C.F.; Azevedo, J.W.V.; Galvão, J.M.A.; Fernandes, J.V. Chikungunya fever: A threat to global public health. Pathog. Glob. Health 2018, 112, 182–194. [Google Scholar] [CrossRef]
- Zeller, H.; Van Bortel, W.; Sudre, B. Chikungunya: Its History in Africa and Asia and Its Spread to New Regions in 2013–2014. J. Infect. Dis. 2016, 214, S436–S440. [Google Scholar] [CrossRef] [PubMed]
- Schuffenecker, I.; Iteman, I.; Michault, A.; Murri, S.; Frangeul, L.; Vaney, M.C.; Lavenir, R.; Pardigon, N.; Reynes, J.M.; Pettinelli, F.; et al. Genome microevolution of chikungunya viruses causing the Indian Ocean outbreak. PLoS Med. 2006, 3, e263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maljkovic Berry, I.; Eyase, F.; Pollett, S.; Konongoi, S.L.; Joyce, M.G.; Figueroa, K.; Ofula, V.; Koka, H.; Koskei, E.; Nyunja, A.; et al. Global Outbreaks and Origins of a Chikungunya Virus Variant Carrying Mutations Which May Increase Fitness for Aedes aegypti: Revelations from the 2016 Mandera, Kenya Outbreak. Am. J. Trop. Med. Hyg. 2019, 100, 1249–1257. [Google Scholar] [CrossRef]
- Eyase, F.; Langat, S.; Berry, I.M.; Mulwa, F.; Nyunja, A.; Mutisya, J.; Owaka, S.; Limbaso, S.; Ofula, V.; Koka, H.; et al. Emergence of a novel chikungunya virus strain bearing the E1:V80A substitution, out of the Mombasa, Kenya 2017–2018 outbreak. PLoS ONE 2020, 15, e0241754. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.M.O.; Tauro, L.B.; Kikuti, M.; Anjos, R.O.; Santos, V.C.; Gonçalves, T.S.F.; Paploski, I.A.D.; Moreira, P.S.S.; Nascimento, L.C.J.; Campos, G.S.; et al. Concomitant Transmission of Dengue, Chikungunya, and Zika Viruses in Brazil: Clinical and Epidemiological Findings From Surveillance for Acute Febrile Illness. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2019, 69, 1353–1359. [Google Scholar] [CrossRef] [PubMed]
- Forrester, N.L.; Palacios, G.; Tesh, R.B.; Savji, N.; Guzman, H.; Sherman, M.; Weaver, S.C.; Lipkin, W.I. Genome-scale phylogeny of the alphavirus genus suggests a marine origin. J. Virol. 2012, 86, 2729–2738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rezza, G.; Chen, R.; Weaver, S.C. O’nyong-nyong fever: A neglected mosquito-borne viral disease. Pathog. Glob. Health 2017, 111, 271–275. [Google Scholar] [CrossRef] [PubMed]
- Lanciotti, R.S.; Ludwig, M.L.; Rwaguma, E.B.; Lutwama, J.J.; Kram, T.M.; Karabatsos, N.; Cropp, B.C.; Miller, B.R. Emergence of epidemic O’nyong-nyong fever in Uganda after a 35-year absence: Genetic characterization of the virus. Virology 1998, 252, 258–268. [Google Scholar] [CrossRef] [Green Version]
- Johnson, B.K.; Gichogo, A.; Gitau, G.; Patel, N.; Ademba, G.; Kirui, R.; Highton, R.B.; Smith, D.H. Recovery of o’nyong-nyong virus from Anopheles funestus in Western Kenya. Trans. R. Soc. Trop. Med. Hyg. 1981, 75, 239–241. [Google Scholar] [CrossRef]
- Gopal, S.; Ma, Y.; Xin, C.; Pitts, J.; Were, L. Characterizing the Spatial Determinants and Prevention of Malaria in Kenya. Int. J. Environ. Res. Public Health 2019, 16, 5078. [Google Scholar] [CrossRef] [Green Version]
- Hortion, J.; Mutuku, F.M.; Eyherabide, A.L.; Vu, D.M.; Boothroyd, D.B.; Grossi-Soyster, E.N.; King, C.H.; Ndenga, B.A.; LaBeaud, A.D. Acute Flavivirus and Alphavirus Infections among Children in Two Different Areas of Kenya, 2015. Am. J. Trop. Med. Hyg. 2019, 100, 170–173. [Google Scholar] [CrossRef] [Green Version]
- Powers, A.M.; Brault, A.C.; Tesh, R.B.; Weaver, S.C. Re-emergence of Chikungunya and O’nyong-nyong viruses: Evidence for distinct geographical lineages and distant evolutionary relationships. J. Gen. Virol. 2000, 81, 471–479. [Google Scholar] [CrossRef]
- Henss, L.; Yue, C.; Kandler, J.; Faddy, H.M.; Simmons, G.; Panning, M.; Lewis-Ximenez, L.L.; Baylis, S.A.; Schnierle, B.S. Establishment of an Alphavirus-Specific Neutralization Assay to Distinguish Infections with Different Members of the Semliki Forest complex. Viruses 2019, 11, 82. [Google Scholar] [CrossRef] [Green Version]
- Ling, J.; Smura, T.; Lundström, J.O.; Pettersson, J.H.; Sironen, T.; Vapalahti, O.; Lundkvist, Å.; Hesson, J.C. Introduction and Dispersal of Sindbis Virus from Central Africa to Europe. J. Virol. 2019, 93, e00620-19. [Google Scholar] [CrossRef] [Green Version]
- Adouchief, S.; Smura, T.; Sane, J.; Vapalahti, O.; Kurkela, S. Sindbis virus as a human pathogen-epidemiology, clinical picture and pathogenesis. Rev. Med. Virol. 2016, 26, 221–241. [Google Scholar] [CrossRef] [PubMed]
- Nyaruaba, R.; Mwaliko, C.; Mwau, M.; Mousa, S.; Wei, H. Arboviruses in the East African Community partner states: A review of medically important mosquito-borne Arboviruses. Pathog. Glob. Health 2019, 113, 209–228. [Google Scholar] [CrossRef]
- Ochieng, C.; Lutomiah, J.; Makio, A.; Koka, H.; Chepkorir, E.; Yalwala, S.; Mutisya, J.; Musila, L.; Khamadi, S.; Richardson, J.; et al. Mosquito-borne arbovirus surveillance at selected sites in diverse ecological zones of Kenya; 2007–2012. Virol. J. 2013, 10, 140. [Google Scholar] [CrossRef] [Green Version]
- Nyamwaya, D.K.; Otiende, M.; Omuoyo, D.O.; Githinji, G.; Karanja, H.K.; Gitonga, J.N.; de Laurent, Z.R.; Otieno, J.R.; Sang, R.; Kamau, E.; et al. Endemic chikungunya fever in Kenyan children: A prospective cohort study. BMC Infect. Dis. 2021, 21, 186. [Google Scholar] [CrossRef] [PubMed]
- Geser, A.; Henderson, B.E.; Christensen, S. A multipurpose serological survey in Kenya. 2. Results of arbovirus serological tests. Bull. World Health Organ. 1970, 43, 539–552. Available online: https://pubmed.ncbi.nlm.nih.gov/5313066/ (accessed on 3 January 2022). [PubMed]
- Grossi-Soyster, E.N.; Cook, E.A.J.; de Glanville, W.A.; Thomas, L.F.; Krystosik, A.R.; Lee, J.; Wamae, C.N.; Kariuki, S.; Fèvre, E.M.; LaBeaud, A.D. Serological and spatial analysis of alphavirus and flavivirus prevalence and risk factors in a rural community in western Kenya. PLoS Negl. Trop. Dis. 2017, 11, e0005998. [Google Scholar] [CrossRef] [Green Version]
- Fritzell, C.; Rousset, D.; Adde, A.; Kazanji, M.; Van Kerkhove, M.D.; Flamand, C. Current challenges and implications for dengue, chikungunya and Zika seroprevalence studies worldwide: A scoping review. PLoS Negl. Trop. Dis. 2018, 12, e0006533. [Google Scholar] [CrossRef] [Green Version]
- Kimata, Y.; Borus, P.; Nzunza, R.; Ofula, V.; Chepkorir, E.; Waihenya, R.; Sang, R. Serological Evidence of Chikungunya Virus Infection Among Suspected Measles Cases in Selected Regions of Kenya: 2008–2014. Vector Borne Zoonotic Dis. 2020, 20, 903–909. [Google Scholar] [CrossRef]
- Masika, M.M.; Korhonen, E.; Smura, T.; Uusitalo, R.; Vapalahti, K.; Mwaengo, D.; Jääskeläinen, A.J.; Anzala, O.; Vapalahti, O.; Huhtamo, E. Detection of dengue virus type 2 of Indian origin in acute febrile patients in rural Kenya. PLoS Negl. Trop. Dis. 2020, 14, e0008099. [Google Scholar] [CrossRef] [Green Version]
- Abera, T.A.; Vuorinne, I.; Munyao, M.; Pellikka, P.; Heiskanen, J. Taita Taveta County, Kenya—2020 Land cover map and reference database. 2021. Available online: https://data.mendeley.com/datasets/xv24ngy2dz (accessed on 6 January 2022). [CrossRef]
- UN-HABITAT. Informal Settlements’ Vulnerability Mapping in Kenya: The Case of Kibera. 2020. Available online: https://data.unhabitat.org/documents/GUO-UN-Habitat::the-case-of-kibera-slum-mapping-report/about (accessed on 12 April 2022).
- Kounkuey Design Initiative. Kounkuey Design Initiative—Open Data. 2015. Available online: https://www.kounkuey.org/about/open_data (accessed on 28 February 2022).
- Clark, B.J.F.; Pellikka, P.K.E. The development of a land use change detection methodology for mapping the Taita Hills, South-East Kenya. In Proceedings of the 31st International Symposium of Remote Sensing of the Environment, St. Petersburg, Russia, 20–24 June 2005. [Google Scholar]
- Kallio-Kokko, H.; Laakkonen, J.; Rizzoli, A.; Tagliapietra, V.; Cattadori, I.; Perkins, S.E.; Hudson, P.J.; Cristofolini, A.; Versini, W.; Vapalahti, O.; et al. Hantavirus and arenavirus antibody prevalence in rodents and humans in Trentino, Northern Italy. Epidemiol. Infect. 2006, 134, 830–836. [Google Scholar] [CrossRef]
- Hozé, N.; Diarra, I.; Sangaré, A.K.; Pastorino, B.; Pezzi, L.; Kouriba, B.; Sagara, I.; Dabo, A.; Djimdé, A.; Thera, M.A.; et al. Model-based assessment of Chikungunya and O’nyong-nyong virus circulation in Mali in a serological cross-reactivity context. Nat. Commun. 2021, 12, 6735. [Google Scholar] [CrossRef]
- Chua, C.-L.; Sam, I.C.; Chiam, C.-W.; Chan, Y.-F. The neutralizing role of IgM during early Chikungunya virus infection. PLoS ONE 2017, 12, e0171989. [Google Scholar] [CrossRef] [Green Version]
- Suhrbier, A.; Jaffar-Bandjee, M.-C.; Gasque, P. Arthritogenic alphaviruses—An overview. Nat. Rev. Rheumatol. 2012, 8, 420–429. [Google Scholar] [CrossRef]
- Inziani, M.; Adungo, F.; Awando, J.; Kihoro, R.; Inoue, S.; Morita, K.; Obimbo, E.; Onyango, F.; Mwau, M. Seroprevalence of yellow fever, dengue, West Nile and chikungunya viruses in children in Teso South Sub-County, Western Kenya. Int. J. Infect. Dis. IJID Off. Publ. Int. Soc. Infect. Dis. 2020, 91, 104–110. [Google Scholar] [CrossRef] [Green Version]
- Kasili, S.; Odemba, N.; Ngere, F.G.; Kamanza, J.B.; Muema, A.M.; Kutima, H.L. Entomological assessment of the potential for malaria transmission in Kibera slum of Nairobi, Kenya. J. Vector Borne Dis. 2009, 46, 273–279. [Google Scholar]
- Eastwood, G.; Sang, R.C.; Lutomiah, J.; Tunge, P.; Weaver, S.C. Sylvatic Mosquito Diversity in Kenya-Considering Enzootic Ecology of Arboviruses in an Era of Deforestation. Insects 2020, 11, 342. [Google Scholar] [CrossRef]
- Karungu, S.; Atoni, E.; Ogalo, J.; Mwaliko, C.; Agwanda, B.; Yuan, Z.; Hu, X. Mosquitoes of Etiological Concern in Kenya and Possible Control Strategies. Insects 2019, 10, 173. [Google Scholar] [CrossRef] [Green Version]
- Munyao, V.; Karisa, J.; Munyao, C.M.; Ngari, M.; Menza, N.; Peshu, N.; Rono, M.; Mbogo, C.; Mwangangi, J. Surveillance of Culicine Mosquitoes in Six Villages of Taita-Taveta County, Kenya, With Host Determinations From Blood-Fed Females. J. Med. Entomol. 2020, 57, 1972–1982. [Google Scholar] [CrossRef]
- Clements, T.L.; Rossi, C.A.; Irish, A.K.; Kibuuka, H.; Eller, L.A.; Robb, M.L.; Kataaha, P.; Michael, N.L.; Hensley, L.E.; Schoepp, R.J. Chikungunya and O’nyong-nyong Viruses in Uganda: Implications for Diagnostics. Open Forum Infect. Dis. 2019, 6, ofz001. [Google Scholar] [CrossRef]
- Jääskeläinen, A.J.; Kareinen, L.; Smura, T.; Kallio-Kokko, H.; Vapalahti, O. Chikungunya virus infections in Finnish travellers 2009–2019. Infect. Ecol. Epidemiol. 2020, 10, 1798096. [Google Scholar] [CrossRef]
- Silva, J.V.J., Jr.; Ludwig-Begall, L.F.; Oliveira-Filho, E.F.D.; Oliveira, R.A.S.; Durães-Carvalho, R.; Lopes, T.R.R.; Silva, D.E.A.; Gil, L.H.V.G. A scoping review of Chikungunya virus infection: Epidemiology, clinical characteristics, viral co-circulation complications, and control. Acta Trop. 2018, 188, 213–224. [Google Scholar] [CrossRef]
- Lemant, J.; Boisson, V.; Winer, A.; Thibault, L.; Andre, H.; Tixier, F.; Lemercier, M.; Antok, E.; Cresta, M.P.; Grivard, P.; et al. Serious acute chikungunya virus infection requiring intensive care during the Reunion Island outbreak in 2005–2006. Crit. Care Med. 2008, 36, 2536–2541. [Google Scholar] [CrossRef]
- Harring, J.; Granger, D.; Jespersen, D.; Theel, E. Comparison of the euroimmun Chikungunya Immunoglobulin (Ig)M and IgG Enzyme Immunoassays (Eias) to a Commercially Available Immunofluorescence Assay. Open Forum Infect. Dis. 2015, 2, 235. [Google Scholar] [CrossRef] [Green Version]
- De Salazar, P.M.; Valadere, A.M.; Goodman, C.H.; Johnson, B.W. Evaluation of three commercially-available chikungunya virus immunoglobulin G immunoassays. Rev. Panam. Salud Publica 2017, 41, e62. [Google Scholar] [CrossRef]
- Valentine, M.J.; Murdock, C.C.; Kelly, P.J. Sylvatic cycles of arboviruses in non-human primates. Parasites Vectors 2019, 12, 463. [Google Scholar] [CrossRef]
| Parameter | Taita-Taveta | Kibera | Total (%) |
|---|---|---|---|
| n | 326 (59%) | 231 (41%) | 557 |
| Gender (n = 547) | |||
| Male | 146 (46%) | 115 (50%) | 261 (48%) |
| Female | 172 (54%) | 114 (50%) | 286 (52%) |
| Age (n= 546) | |||
| Mean (Standard deviation (SD)) | 26.8 (22.1) | 16.2 (15.3) | 22.4 (20.2) |
| Median (Interquartile range (IQR)) | 20.6 (39.5) | 10.2 (24.4) | 16.4 (33.6) |
| Range | 6 months–85 years | 2 months–77 years | 2 months–85 years |
| Age Groups (n = 545) | |||
| 0–5 years | 69 (22%) | 87 (39%) | 156 (29%) |
| 5–17 years | 81 (25%) | 45 (20%) | 126 (23%) |
| 18 years and above | 169 (53%) | 94 (41%) | 263 (48%) |
| Education Level in adults (n = 245) | |||
| None | 15 (10%) | 0 (0%) | 15 (6%) |
| Primary school | 77 (50%) | 27 (30%) | 104 (42%) |
| Secondary school | 29 (19%) | 36 (40%) | 65 (27%) |
| Tertiary level | 34 (22%) | 27 (30%) | 61 (25%) |
| History of travel (n = 552) | |||
| Any travel outside the study area | 54 (17%) | 88 (38%) | 142 (26%) |
| Travel to Nyanza or western Kenya | 9 (3%) | 56 (24%) | 65 (12%) |
| Travel to coastal Kenya | 37 (11%) | 2 (1%) | 39 (7%) |
| Contact with animals (n = 557) | |||
| Contact with goats | 134 (41%) | 25 (11%) | 159 (29%) |
| Contact with cattle | 96 (29%) | 26 (11%) | 122 (22%) |
| Contact with chicken | 190 (58%) | 43 (19%) | 233 (42%) |
| Contact with cats | 61 (19%) | 87 (38%) | 148 (27%) |
| Contact with rodents | 217 (67%) | 127 (55%) | 344 (62%) |
| Contact with bats | 94 (29%) | 6 (3%) | 100 (18%) |
| Signs and symptoms (n = 557) | |||
| Body Temperature in °C (Mean/SD) | 38.8 (0.56) | 38.4 (0.67) | 38.6 (0.63) |
| Median duration of fever (range) | 2 (1–7) days | 2 (1–14) | 2 (1–14) days |
| Joint pain | 139 (43%) | 70 (30%) | 209 (38%) |
| Myalgia | 123 (38%) | 65 (28%) | 188 (34%) |
| Headache | 53 (16%) | 108 (47%) | 161 (29%) |
| Cough | 44 (13%) | 95 (41%) | 139 (25%) |
| Vomiting | 53 (16%) | 68 (29%) | 121 (22%) |
| Diarrhoea | 29 (9%) | 47 (20%) | 76 (14%) |
| Rash | 18 (6%) | 16 (7%) | 34 (6%) |
| Sample Characteristics | n | CHIKV and ONNV Positive | CHIKV Positive | ONNV Positive | SINV Positive | Positive for Any Alphavirus | Negative |
|---|---|---|---|---|---|---|---|
| CHIKV IgG positive (IFA) | 31 | 12 (39%) | 0 (0%) | 13 (42%) | 0 (0%) | 25 (81%) | 6 (19%) |
| CHIKV IgM positive (ELISA) | 23 | 0 (0%) | 1 (4%) | 0 (0%) | 0 (0%) | 1 (4%) | 22 (96%) |
| Both CHIKV IgG and IgM positive | 4 | 2 (50%) | 0 (0%) | 1 (25%) | 0 (0%) | 3 (75%) | 1 (25%) |
| Negative | 11 | 0 (0%) | 0 (0%) | 1 (9%) | 0 (0%) | 1 (9%) | 10 (91%) |
| TOTAL | 69 | 14 (20%) | 1 (1%) | 15 (22%) | 0 (0%) | 30 (43%) | 39 (57%) |
| Characteristic | Alphavirus IgG Antibody Positive (PRNT) | Odds Ratio (95% CI) | p Value (Fisher’s Exact Test) |
|---|---|---|---|
| Age Group | |||
| Adults | 24/253 (9.5%) | 9.5 (2.83–31.96) | <0.001 |
| Children (<18 years) | 3/275 (1.1%) | ||
| Gender | |||
| Female | 15/274 (5.5%) | 1.1 (0.49–2.29) | 1.000 |
| Male | 13/253 (5.1%) | ||
| Facility Location | |||
| Kibera | 21/228 (9.2%) | 4.3 (1.83–10.48) | <0.001 |
| Taita-Taveta | 7/309 (2.3%) | ||
| History of Travel | |||
| Yes | 9/137 (6.6%) | 1.5 (0.64–3.36) | 0.369 |
| No | 18/395 (4.6%) | ||
| Travel to Western Kenya | |||
| Yes | 7/62 (11.3%) | 2.8 (1.12–6.77) | 0.032 |
| No | 21/475 (4.4%) | ||
| Travel to Coastal Kenya | |||
| Yes | 1/38 (2.6%) | 0.5 (0.06–3.58) | 0.712 |
| No | 27/499 (5.4%) | ||
| Joint Pain | |||
| Yes | 17/201 (8.5%) | 2.7 (1.25–5.95) | 0.015 |
| No | 11/336 (3.3%) | ||
| Myalgia | |||
| Yes | 12/180 (6.7%) | 1.5 (0.70–3.29) | 0.307 |
| No | 16/357 (4.5%) | ||
| Headache | |||
| Yes | 16/157 (10.2%) | 3.5 (1.61–7.54) | 0.002 |
| No | 12/380 (3.2%) | ||
| Cough | |||
| Yes | 5/136 (3.7%) | 0.6 (0.23–1.68) | 0.503 |
| No | 25/401 (6.2%) | ||
| Contact with goats | |||
| Yes | 4/148 (2.7%) | 0.4 (0.14–1.24) | 0.130 |
| No | 24/389 (6.2%) | ||
| Contact with cattle | |||
| Yes | 4/117 (3.4%) | 0.6 (0.20–1.72) | 0.480 |
| No | 24/420 (5.7%) | ||
| Contact with rodents | |||
| Yes | 15/331 (4.5%) | 0.7 (0.33–1.51) | 0.426 |
| No | 13/206 (6.3%) | ||
| Contact with bats | |||
| Yes | 3/91 (3.3%) | 0.6 (0.17–1.94) | 0.450 |
| No | 25/446 (5.6%) | ||
| Any animal contact | |||
| Yes | 19/419 (4.5%) | 0.6 (0.25–1.31) | 0.238 |
| No | 9/118 (7.6%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Masika, M.M.; Korhonen, E.M.; Smura, T.; Uusitalo, R.; Ogola, J.; Mwaengo, D.; Jääskeläinen, A.J.; Alburkat, H.; Gwon, Y.-D.; Evander, M.; et al. Serological Evidence of Exposure to Onyong-Nyong and Chikungunya Viruses in Febrile Patients of Rural Taita-Taveta County and Urban Kibera Informal Settlement in Nairobi, Kenya. Viruses 2022, 14, 1286. https://doi.org/10.3390/v14061286
Masika MM, Korhonen EM, Smura T, Uusitalo R, Ogola J, Mwaengo D, Jääskeläinen AJ, Alburkat H, Gwon Y-D, Evander M, et al. Serological Evidence of Exposure to Onyong-Nyong and Chikungunya Viruses in Febrile Patients of Rural Taita-Taveta County and Urban Kibera Informal Settlement in Nairobi, Kenya. Viruses. 2022; 14(6):1286. https://doi.org/10.3390/v14061286
Chicago/Turabian StyleMasika, Moses Muia, Essi M. Korhonen, Teemu Smura, Ruut Uusitalo, Joseph Ogola, Dufton Mwaengo, Anne J. Jääskeläinen, Hussein Alburkat, Yong-Dae Gwon, Magnus Evander, and et al. 2022. "Serological Evidence of Exposure to Onyong-Nyong and Chikungunya Viruses in Febrile Patients of Rural Taita-Taveta County and Urban Kibera Informal Settlement in Nairobi, Kenya" Viruses 14, no. 6: 1286. https://doi.org/10.3390/v14061286
APA StyleMasika, M. M., Korhonen, E. M., Smura, T., Uusitalo, R., Ogola, J., Mwaengo, D., Jääskeläinen, A. J., Alburkat, H., Gwon, Y.-D., Evander, M., Anzala, O., Vapalahti, O., & Huhtamo, E. (2022). Serological Evidence of Exposure to Onyong-Nyong and Chikungunya Viruses in Febrile Patients of Rural Taita-Taveta County and Urban Kibera Informal Settlement in Nairobi, Kenya. Viruses, 14(6), 1286. https://doi.org/10.3390/v14061286






