PIMGAVir and Vir-MinION: Two Viral Metagenomic Pipelines for Complete Baseline Analysis of 2nd and 3rd Generation Data
Abstract
:1. Introduction
2. Objectives of PIMGAVir and Vir-MinION Pipelines
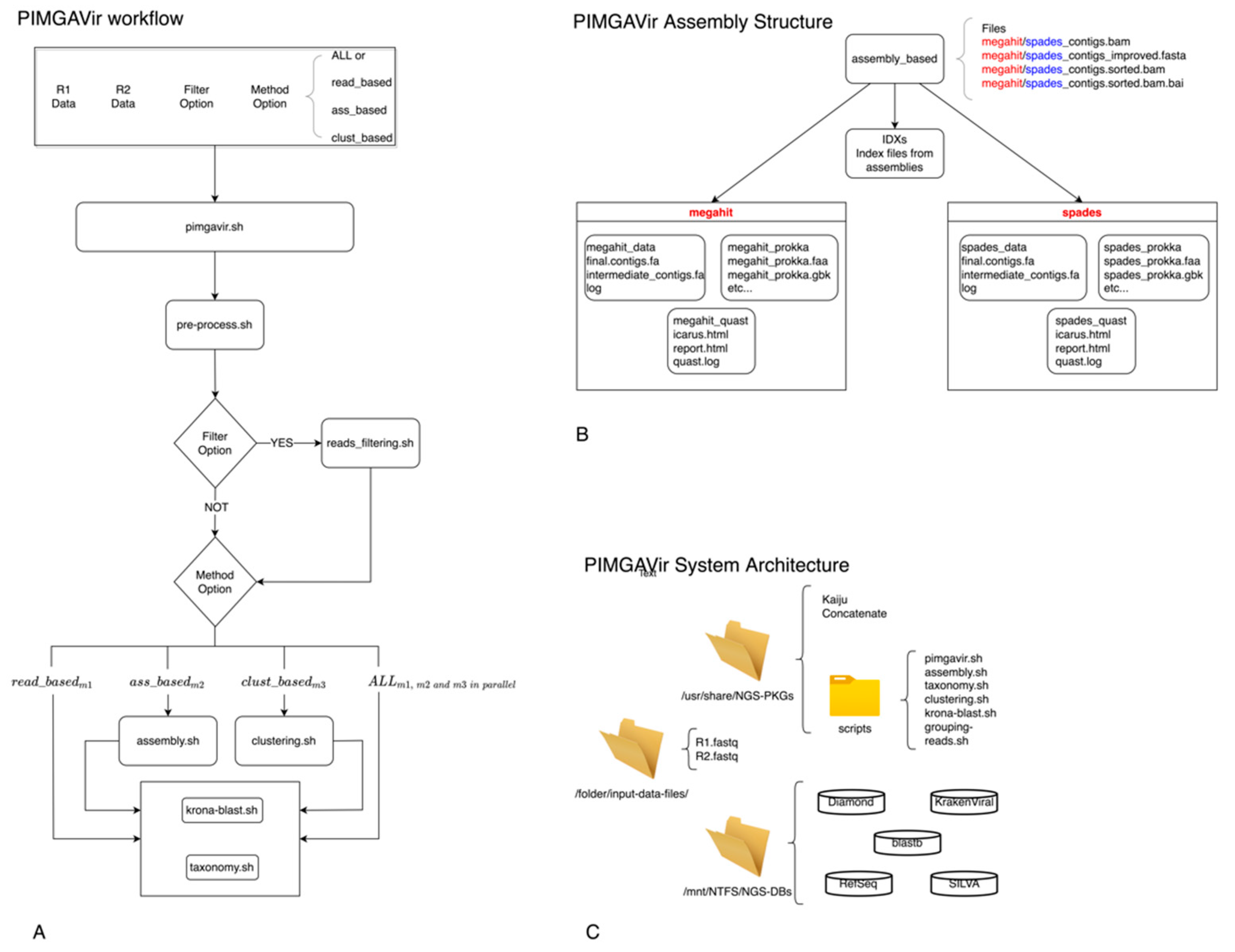
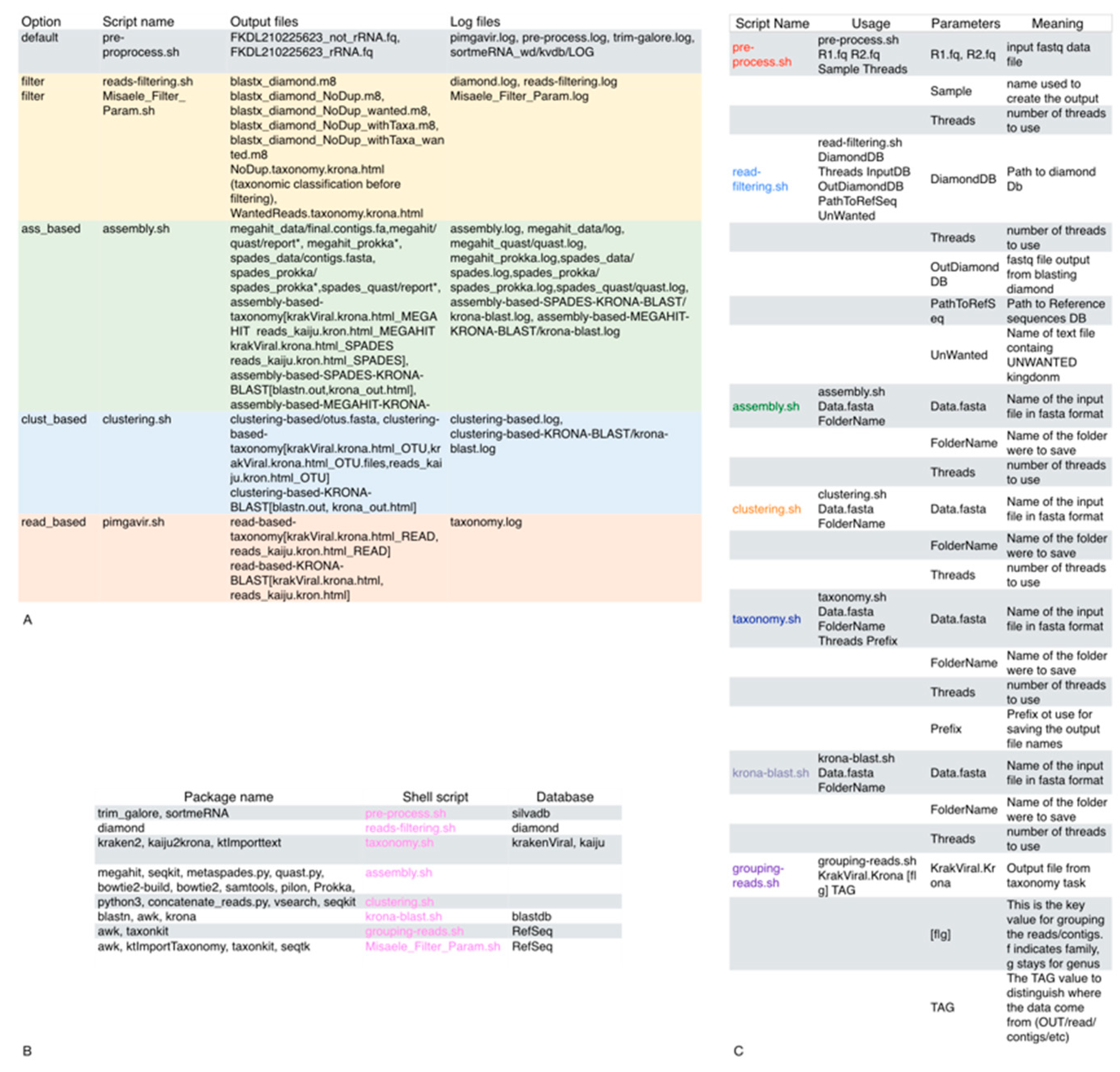
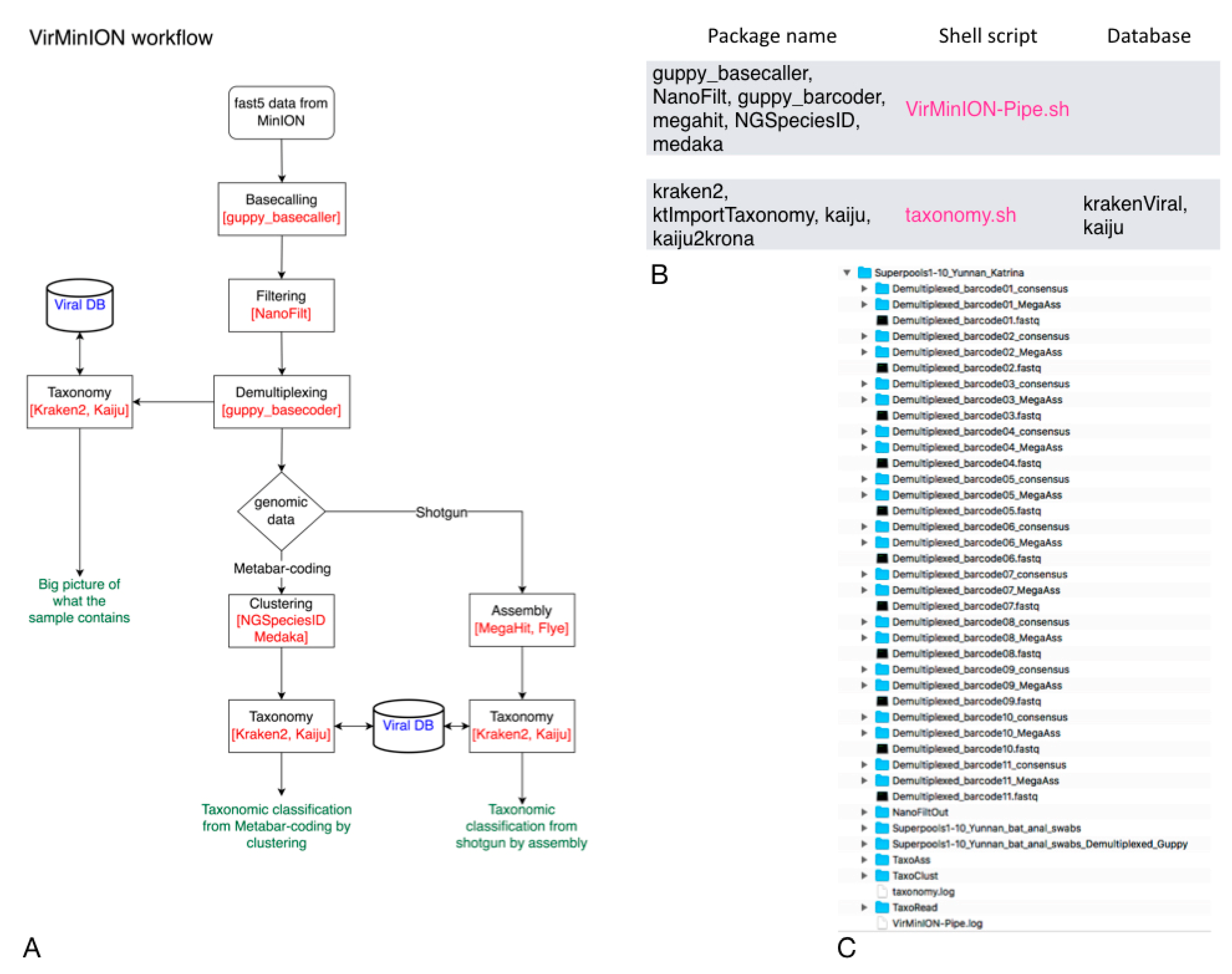
2.1. Description of the Two Pipelines
2.2. Test Pipelines
3. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gwinn, M.; MacCannell, D.R.; Khabbaz, R.F. Integrating Advanced Molecular Technologies into Public Health. J. Clin. Microbiol. 2017, 55, 703–714. [Google Scholar] [CrossRef] [Green Version]
- Lecuit, M.; Eloit, M. The diagnosis of infectious diseases by whole genome next generation sequencing: A new era is opening. Front. Cell Infect. Microbiol. 2014, 4, 25. [Google Scholar] [CrossRef] [Green Version]
- Schlaberg, R.; Chiu, C.Y.; Miller, S.; Procop, G.W.; Weinstock, G.; Professional Practice Committee and Committee on Laboratory Practices of the American Society for Microbiology; Microbiology Resource Committee of the College of American Pathologists. Validation of Metagenomic Next-Generation Sequencing Tests for Universal Pathogen Detection. Arch. Pathol. Lab. Med. 2017, 141, 776–786. [Google Scholar] [CrossRef] [Green Version]
- Afshinnekoo, E.; Chou, C.; Alexander, N.; Ahsanuddin, S.; Schuetz, A.N.; Mason, C.E. Precision Metagenomics: Rapid Metagenomic Analyses for Infectious Disease Diagnostics and Public Health Surveillance. J. Biomol. Tech. 2017, 28, 40–45. [Google Scholar] [CrossRef] [Green Version]
- Miller, S.; Chiu, C. The Role of Metagenomics and Next-Generation Sequencing in Infectious Disease Diagnosis. Clin. Chem. 2021, 68, 115–124. [Google Scholar] [CrossRef]
- Goldberg, B.; Sichtig, H.; Geyer, C.; Ledeboer, N.; Weinstock, G.M. Making the Leap from Research Laboratory to Clinic: Challenges and Opportunities for Next-Generation Sequencing in Infectious Disease Diagnostics. mBio 2015, 6, e01888-15. [Google Scholar] [CrossRef] [Green Version]
- d’Humieres, C.; Salmona, M.; Delliere, S.; Leo, S.; Rodriguez, C.; Angebault, C.; Alanio, A.; Fourati, S.; Lazarevic, V.; Woerther, P.L.; et al. The Potential Role of Clinical Metagenomics in Infectious Diseases: Therapeutic Perspectives. Drugs 2021, 81, 1453–1466. [Google Scholar] [CrossRef]
- Duan, H.; Li, X.; Mei, A.; Li, P.; Liu, Y.; Li, X.; Li, W.; Wang, C.; Xie, S. The diagnostic value of metagenomic next rectanglegeneration sequencing in infectious diseases. BMC Infect. Dis. 2021, 21, 62. [Google Scholar] [CrossRef]
- Lefterova, M.I.; Suarez, C.J.; Banaei, N.; Pinsky, B.A. Next-Generation Sequencing for Infectious Disease Diagnosis and Management: A Report of the Association for Molecular Pathology. J. Mol. Diagn. 2015, 17, 623–634. [Google Scholar] [CrossRef] [Green Version]
- Gwinn, M.; MacCannell, D.; Armstrong, G.L. Next-Generation Sequencing of Infectious Pathogens. JAMA 2019, 321, 893–894. [Google Scholar] [CrossRef] [Green Version]
- Bergner, L.M.; Orton, R.J.; da Silva Filipe, A.; Shaw, A.E.; Becker, D.J.; Tello, C.; Biek, R.; Streicker, D.G. Using noninvasive metagenomics to characterize viral communities from wildlife. Mol. Ecol. Resour. 2019, 19, 128–143. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.Y.; Miller, S.A. Clinical metagenomics. Nat. Rev. Genet. 2019, 20, 341–355. [Google Scholar] [CrossRef] [PubMed]
- Greninger, A.L.; Naccache, S.N.; Federman, S.; Yu, G.; Mbala, P.; Bres, V.; Stryke, D.; Bouquet, J.; Somasekar, S.; Linnen, J.M.; et al. Rapid metagenomic identification of viral pathogens in clinical samples by real-time nanopore sequencing analysis. Genome Med. 2015, 7, 99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petersen, L.M.; Martin, I.W.; Moschetti, W.E.; Kershaw, C.M.; Tsongalis, G.J. Third-Generation Sequencing in the Clinical Laboratory: Exploring the Advantages and Challenges of Nanopore Sequencing. J. Clin. Microbiol. 2019, 58, e01315-19. [Google Scholar] [CrossRef] [PubMed]
- Gautam, A.; Tiwari, A.K.; Malik, Y.S. Bioinformatics Applications in Advancing Animal Virus Research. In Recent Advances in Animal Virology; Springer: Singapore, 2019; pp. 447–471. [Google Scholar]
- Holzer, M.; Marz, M. Software Dedicated to Virus Sequence Analysis “Bioinformatics Goes Viral”. Adv. Virus Res. 2017, 99, 233–257. [Google Scholar]
- Ciuffreda, L.; Rodriguez-Perez, H.; Flores, C. Nanopore sequencing and its application to the study of microbial communities. Comput. Struct. Biotechnol. J. 2021, 19, 1497–1511. [Google Scholar] [CrossRef]
- Nooij, S.; Schmitz, D.; Vennema, H.; Kroneman, A.; Koopmans, M.P.G. Overview of Virus Metagenomic Classification Methods and Their Biological Applications. Front. Microbiol. 2018, 9, 749. [Google Scholar] [CrossRef]
- FastQC, v. 0.11.9. 2015. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 29 April 2022).
- Bushnell, B.; Rood, J.; Singer, E. BBMerge—Accurate paired shotgun read merging via overlap. PLoS ONE 2017, 12, e0185056. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Meyer, F.; Paarmann, D.; D’Souza, M.; Olson, R.; Glass, E.M.; Kubal, M.; Paczian, T.; Rodriguez, A.; Stevens, R.; Wilke, A.; et al. The metagenomics RAST server—A public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinform. 2008, 9, 386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Liu, C.M.; Luo, R.; Sadakane, K.; Lam, T.W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langmead, B.; Wilks, C.; Antonescu, V.; Charles, R. Scaling read aligners to hundreds of threads on general-purpose processors. Bioinformatics 2019, 35, 421–432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, D.D.; Li, F.; Kirton, E.; Thomas, A.; Egan, R.; An, H.; Wang, Z. MetaBAT 2: An adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 2019, 7, e7359. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.W.; Simmons, B.A.; Singer, S.W. MaxBin 2.0: An automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 2016, 32, 605–607. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [Green Version]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Chen, I.A.; Chu, K.; Palaniappan, K.; Ratner, A.; Huang, J.; Huntemann, M.; Hajek, P.; Ritter, S.; Varghese, N.; Seshadri, R.; et al. The IMG/M data management and analysis system v.6.0: New tools and advanced capabilities. Nucleic Acids Res. 2021, 49, D751–D763. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahe, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Budkina, A.Y.; Korneenko, E.V.; Kotov, I.A.; Kiselev, D.A.; Artyushin, I.V.; Speranskaya, A.S.; Khafizov, K.; Akimkin, V.G. Utilizing the VirIdAl Pipeline to Search for Viruses in the Metagenomic Data of Bat Samples. Viruses 2021, 13, 2006. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Holt, K.E. Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome Biol. 2019, 20, 129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wick, R.R.; Judd, L.M.; Holt, K.E. Deepbinner: Demultiplexing barcoded Oxford Nanopore reads with deep convolutional neural networks. PLoS Comput. Biol. 2018, 14, e1006583. [Google Scholar] [CrossRef] [Green Version]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Completing bacterial genome assemblies with multiplex MinION sequencing. Microb. Genom. 2017, 3, e000132. [Google Scholar] [CrossRef]
- Kim, D.; Song, L.; Breitwieser, F.P.; Salzberg, S.L. Centrifuge: Rapid and sensitive classification of metagenomic sequences. Genome Res. 2016, 26, 1721–1729. [Google Scholar] [CrossRef] [Green Version]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef] [Green Version]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Loman, N.J.; Quick, J.; Simpson, J.T. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat. Methods 2015, 12, 733–735. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Medaka, NanoporeTech: Sequence Correction Provided by ONT Research. 2022. Available online: https://github.com/nanoporetech/medaka (accessed on 2 June 2022).
- Sahlin, K.; Lim, M.C.W.; Prost, S. NGSpeciesID: DNA barcode and amplicon consensus generation from long-read sequencing data. Ecol. Evol. 2021, 11, 1392–1398. [Google Scholar] [CrossRef] [PubMed]
- Murigneux, V.; Roberts, L.W.; Forde, B.M.; Phan, M.D.; Nhu, N.T.K.; Irwin, A.D.; Harris, P.N.A.; Paterson, D.L.; Schembri, M.A.; Whiley, D.M.; et al. MicroPIPE: Validating an end-to-end workflow for high-quality complete bacterial genome construction. BMC Genom. 2021, 22, 474. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Perez, H.; Ciuffreda, L.; Flores, C. NanoCLUST: A species-level analysis of 16S rRNA nanopore sequencing data. Bioinformatics 2021, 37, 1600–1601. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [Green Version]
- A Wrapper around Cutadapt and FastQC to Consistently Apply Adapter and Quality Trimming to FastQ files, with EXTRA FUNCTIONAlity for RRBS Data. Available online: https://github.com/FelixKrueger/TrimGalore (accessed on 26 April 2022).
- Kopylova, E.; Noe, L.; Touzet, H. SortMeRNA: Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 2012, 28, 3211–3217. [Google Scholar] [CrossRef] [PubMed]
- Ondov, B.D.; Bergman, N.H.; Phillippy, A.M. Interactive metagenomic visualization in a Web browser. BMC Bioinform. 2011, 12, 385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, W.; Ren, H. TaxonKit: A practical and efficient NCBI taxonomy toolkit. J. Genet. Genom. 2021, 48, 844–850. [Google Scholar] [CrossRef]
- Toolkit for Processing Sequences in FASTA/Q Formats. Available online: https://github.com/lh3/seqtk (accessed on 2 June 2022).
- Mikheenko, A.; Saveliev, V.; Gurevich, A. MetaQUAST: Evaluation of metagenome assemblies. Bioinformatics 2016, 32, 1088–1090. [Google Scholar] [CrossRef] [Green Version]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Wood, D.E.; Salzberg, S.L. Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014, 15, R46. [Google Scholar] [CrossRef] [Green Version]
- Menzel, P.; Ng, K.L.; Krogh, A. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat. Commun. 2016, 7, 11257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, W.; Le, S.; Li, Y.; Hu, F. SeqKit: A Cross-Platform and Ultrafast Toolkit for FASTA/Q File Manipulation. PLoS ONE 2016, 11, e0163962. [Google Scholar] [CrossRef] [PubMed]
- De Coster, W.; D’Hert, S.; Schultz, D.T.; Cruts, M.; Van Broeckhoven, C. NanoPack: Visualizing and processing long-read sequencing data. Bioinformatics 2018, 34, 2666–2669. [Google Scholar] [CrossRef] [PubMed]
- Fritz, A.; Hofmann, P.; Majda, S.; Dahms, E.; Droge, J.; Fiedler, J.; Lesker, T.R.; Belmann, P.; DeMaere, M.Z.; Darling, A.E.; et al. CAMISIM: Simulating metagenomes and microbial communities. Microbiome 2019, 7, 17. [Google Scholar] [CrossRef] [Green Version]
- Ni, P.; Huang, N.; Zhang, Z.; Wang, D.P.; Liang, F.; Miao, Y.; Xiao, C.L.; Luo, F.; Wang, J. DeepSignal: Detecting DNA methylation state from Nanopore sequencing reads using deep-learning. Bioinformatics 2019, 35, 4586–4595. [Google Scholar] [CrossRef]
- Kieft, K.; Zhou, Z.; Anantharaman, K. VIBRANT: Automated recovery, annotation and curation of microbial viruses, and evaluation of viral community function from genomic sequences. Microbiome 2020, 8, 90. [Google Scholar] [CrossRef]
- Roux, S.; Enault, F.; Hurwitz, B.L.; Sullivan, M.B. VirSorter: Mining viral signal from microbial genomic data. PeerJ 2015, 3, e985. [Google Scholar] [CrossRef]
| Package Name | Version | Pipeline | Task |
|---|---|---|---|
| TrimGalore [48] | 0.6.5-1 | PIMGAVir | preprocessing |
| SortMeRNA [49] | 4.3.4 | PIMGAVir | filtering |
| diamond [22] | 2.0.11.149 | PIMGAVir | filtering |
| KronaTools [50] | 2.8.1 | PIMGAVir | filtering |
| Taxonkit [51] | 0.10.1 | PIMGAVir | filtering |
| seqtk [52] | 1.3 | PIMGAVir | filtering |
| megahit [24] | v1.2.9 | PIMGAVir/Vir-MinION | assembly |
| flye [40] | v2.9 | Vir-MinION | assembly |
| quast [53] | v5.0.2 | PIMGAVir | assembly |
| spades [25] | 3.13.1 | PIMGAVir | assembly |
| bowtie2 [26] | 2.4.4 | PIMGAVir | assembly |
| samtools [54] | 1.10-3 | PIMGAVir | assembly |
| pilon [42] | 1.23 | PIMGAVir | assembly |
| Prokka [30] | 1.14.6 | PIMGAVir | assembly |
| kraken2 [55] | 2.1.2 | PIMGAVir/Vir-MinION | taxonomy |
| kaiju [56] | 1.8.2 | PIMGAVir/Vir-MinION | taxonomy |
| blastn [21] | 2.9.0+ | PIMGAVir/Vir-MinION | taxonomy |
| seqkit [57] | 2.0.0 | PIMGAVir | clustering |
| vsearch [32] | v2.18.0 | PIMGAVir | clustering |
| guppy_basecaller [35] | 5.0.13 | Vir-MinION | basecalling |
| NanoFilt [58] | 2.3.0 | Vir-MinION | filtering |
| guppy_barcoder [35] | 5.0.13 | Vir-MinION | demultiplexing |
| NGSpeciesID [44] | 0.1.2.1 | Vir-MinION | clustering |
| medaka [43] | 0.11.5 | Vir-MinION | clustering |
| Database Name | Version | Build Up Date |
|---|---|---|
| diamond/refseq_protein_nonredund | Refseq protein non redundant genomes Database format version = 3 | 6–14 May 2022 |
| krakendb/SILVA_138.1_SSURef_NR99_tax_silva | Silva DB v. 138.1 | 17 May 2021 |
| krakenviral/database.kraken | Kraken Viral DB v2.0.8 | 17 May 2021 |
| NCBI-RefSeq/viralseq_2021-12-14_14-45-53 | Refseq viruses’ representative genomes. BLASTDB Version: 5 | 14 December 2021 |
| SILVA/ssr138, slr138 | Ribosomal DB for SSR138 and SLR138 | 27 August 2020 |
| Reference Genome | Fragment Mean Size | Total Number of Reads | Number of Reads per Genome | Coverage with the Reference (%) | Average Depth per Genome | ||
|---|---|---|---|---|---|---|---|
| Simulated data type | 2G | Ippy virus segment S | 270 | 666,618 | 800 | 0.12 | 34.31 |
| Ippy virus segment L | 666 | 0.01 | 1.35 | ||||
| Hepatitis A virus | 733 | 0.11 | 15.12 | ||||
| Helicobacter hepaticus | 655,218 | 98.29 | 55.44 | ||||
| 3G | Ippy virus segment S | 500 | 13,499 | 5382 | 0.20 | 3348.52 | |
| Ippy virus segment L | 423 | 0.03 | 198.974 | ||||
| Hepatitis A virus | 5485 | 0.18 | 2571.08 | ||||
| Helicobacter hepaticus | 2205 | 97.30 | 5.07815 | ||||
| Task | Starting Number of Reads | Ending Number of Reads | Removed (Number) | Removed (Percentage) |
|---|---|---|---|---|
| Trimming | 666,618 | 642,614 | 24254 | 3.64 |
| Ribosomal removing | 642,614 | 640,614 | 1750 | 0.26 |
| Filtering unwanted | 640,614 | 170,960 | 469,654 | 70.45 |
| MEGAHIT | 170,960 | 884 | 170,086 | 99.48 |
| SPADES | 170,960 | 1354 | 169,616 | 99.20 |
| clustering | 170,960 | 44,783 | 126,187 | 73.80 |
| Approach | DB | Hepatitis A Virus—taxid: 12092 | Ippy Virus (Segment S + L)—taxid: 55096 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | Total of Reads/Cluster/Contigs Analyzed by the DB | Average Size (bp per Read) | Total of Reads/Cluster/Contigs Classified by the DB | % of Reads/Contigs Collected | Coverage of Genome (%) | Accuracy % of Align | Total of Reads/Contigs Classified by the DB | % of Reads/Contigs Collected | S (Ippy-01) | L (Ippy-02) | |||
| % of Coverage | Accuracy %of Align | % of Coverage | Accuracy %of Align | ||||||||||
| read-based | kraken | 170,960 | 139.9 | 648 | 0.38 | 91 | 100 | 790 | 0.46 | 98 | 91.65 | 69 | 7.97 |
| Kaiju | 170,960 | 139.9 | 644 | 0.37 | 91 | 38.69 | 780 | 0.45 | 98 | 44.42 | 72 | 3.92 | |
| BLASTN | 170,690 | 139.9 | 648 | 0.37 | 100 | 100 | 793 | 0.46 | 98 | 91.42 | 72 | 8.07 | |
| clustering-based | kraken | 44,783 | 145.7 | 247 | 0.55 | 100 | 100 | 237 | 0.55 | 97 | 76.25 | 70 | 22.50 |
| kaiju | 44,783 | 145.7 | 244 | 0.54 | 90 | 100 | 233 | 0.52 | 97 | 75.97 | 72 | 22.75 | |
| BLASTN | 44,783 | 145.7 | 247 | 0.55 | 90 | 100 | 241 | 0.54 | 97 | 76.25 | 72 | 22.50 | |
| assembly-based-megahit | kraken | 884 | 728.6 | 4 | 0.45 | 90 | 100 | 1 | 0.11 | 96 | 100 | 0 | 0 |
| kaiju | 884 | 728.6 | 4 | 0.45 | 90 | 100 | 1 | 0.11 | 96 | 100 | 0 | 0 | |
| BLASTN | 884 | 728.6 | 4 | 0.45 | 90 | 100 | 1 | 0.11 | 96 | 100 | 0 | 0 | |
| assembly-based-spades | kraken | 1354 | 617 | 1 | 0.07 | 91 | 100 | 7 | 0.52 | 98 | 14.29 | 23 | 85.71 |
| kaiju | 1354 | 617 | 1 | 0.07 | 91 | 100 | 7 | 0.52 | 98 | 14.29 | 23 | 85.71 | |
| BLASTN | 1354 | 617 | 1 | 0.07 | 91 | 100 | 7 | 0.52 | 98 | 14.29 | 23 | 85.71 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mastriani, E.; Bienes, K.M.; Wong, G.; Berthet, N. PIMGAVir and Vir-MinION: Two Viral Metagenomic Pipelines for Complete Baseline Analysis of 2nd and 3rd Generation Data. Viruses 2022, 14, 1260. https://doi.org/10.3390/v14061260
Mastriani E, Bienes KM, Wong G, Berthet N. PIMGAVir and Vir-MinION: Two Viral Metagenomic Pipelines for Complete Baseline Analysis of 2nd and 3rd Generation Data. Viruses. 2022; 14(6):1260. https://doi.org/10.3390/v14061260
Chicago/Turabian StyleMastriani, Emilio, Kathrina Mae Bienes, Gary Wong, and Nicolas Berthet. 2022. "PIMGAVir and Vir-MinION: Two Viral Metagenomic Pipelines for Complete Baseline Analysis of 2nd and 3rd Generation Data" Viruses 14, no. 6: 1260. https://doi.org/10.3390/v14061260
APA StyleMastriani, E., Bienes, K. M., Wong, G., & Berthet, N. (2022). PIMGAVir and Vir-MinION: Two Viral Metagenomic Pipelines for Complete Baseline Analysis of 2nd and 3rd Generation Data. Viruses, 14(6), 1260. https://doi.org/10.3390/v14061260






